Abstract
Analysis of stable carbon and nitrogen isotope values (δ13C and δ15N) of animal tissues can provide important information about diet, physiology, and movements. Interpretation of δ13C and δ15N values, however, is influenced by factors such as sample lipid content, tissue-specific isotope discrimination, and tissue turnover rates, which are typically species- and tissue-specific. In this study, we generated lipid normalization models for δ13C and investigated the effects of chemical lipid extractions on δ13C and δ15N in Pacific walrus (Odobenus rosmarus divergens) muscle, liver, and skin. We also evaluated tissue-specific isotope discrimination in walrus muscle, liver, skin, and bone collagen. Mean δ13Clipid-free of skin and bone collagen were similar, as were mean δ15N of muscle and liver. All other tissues differed significantly for both isotopes. Differences in δ13Clipid-free and δ15N among tissues agreed with published estimates of marine mammal tissue-specific isotope discrimination factors, with the exception of skin. The results of this work will allow researchers to gain a clearer understanding of walrus diet and the structure of Arctic food webs, while also making it possible to directly compare the results of contemporary walrus isotope research with those of historic and paleoecological studies.
Similar content being viewed by others
Introduction
Stable isotope analysis is widely-used as a tool to study animal movements, diet, and food web structure1,2,3,4. Carbon and nitrogen stable isotope values (δ13C and δ15N) are particularly useful when evaluating animal diet and trophic position3. Consumer δ13C is driven mainly by the type of primary production at the base of the food web, as uptake of CO2 for use in photosynthesis involves discrimination of carbon isotopes5. Trophic discrimination of 13C is typically small (~1‰ per trophic level), but accounts for some variation in stable carbon isotope values1,3. In contrast, variability in δ15N is strongly influenced by trophic discrimination, which results in an increase of ~3.0‰ per trophic level2,3,6. By interpreting δ13C and δ15N values from animal tissues in the context of these sources of isotopic discrimination, information can be inferred about diet, trophic position, and the flow of energy through biological systems.
Additional processes that result in discrimination of 13C and 15N can complicate reconstructions of animal diet and trophic position. For example, discrimination against the heavier 13C during lipid synthesis results in differences of ~6–8‰ between lipids and proteins7,8. Lipid content therefore affects tissue δ13C and may lead to observed variability in δ13C among animals resulting from differences in lipid storage, rather than diet. Because lipids are not always homogenously distributed within a tissue9, lipid content can be an important source of isotopic variability even among multiple samples of a single tissue from one individual10. Furthermore, δ13C varies among fatty acids11,12, thus the δ13C of lipids depends on their fatty acid composition and uneven distribution of different types of lipids within a tissue may lead to heterogeneity in δ13C.
Chemical lipid extraction is one way to account for differences in sample lipid content. In this process, lipids are typically removed using polar organic solvents (often a mixture of chloroform and methanol13,14) and stable isotope values of the lipid-free sample are measured. This process allows for direct instrumental measurement of the lipid-free sample (δ13Clipid-free), but requires more time and effort. Additionally, in some (but not all15,16) cases, chemical lipid extraction can alter sample δ15N by removing amino acids17. Thus, to ensure comparable measurements of both δ13C and δ15N across tissues, it is recommended that two aliquots of each sample be analyzed: one non-lipid extracted (bulk) and the other with lipids removed17. The δ15N value of the non-lipid extracted aliquot (δ15Nbulk) and the δ13Clipid-free can then be interpreted together; however, this approach increases the time and effort required for sample preparation and doubles the analytical cost.
Lipid normalization is an alternative to chemical lipid extraction that allows δ13Clipid-free to be approximated mathematically. This approach, sometimes called arithmetic or mathematical lipid correction, involves modeling the expected δ13Clipid-free based on the lipid content of the bulk sample. These models have been widely used for multiple tissues from a variety of taxa and can be quite accurate in their estimation of δ13Clipid-free (Supplementary Table 1). Typically, these models rely on the relationship between the carbon:nitrogen ratio of the non-lipid extracted sample (C:Nbulk; a proxy for the lipid content of the sample8,18), and the difference between δ13C values of the lipid-free and non-lipid extracted aliquots of each sample (δ13Clipid-free − δ13Cbulk = Δδ13C). This relationship tends to be species- and tissue-specific, thus, one limitation of lipid normalization models is that they must be parameterized for each species and tissue by performing chemical lipid extractions and comparing δ13Cbulk to δ13Clipid-free. Consequently, there are many species and tissues for which lipid normalization models are not available in the literature.
Tissue-specific isotope discrimination is another source of variability in stable isotope values that can impact diet reconstructions and the interpretation of δ13C and δ15N values. Stable isotope values in the tissues of an individual animal may vary substantially due to metabolic discrimination and differences in tissue composition2,19. Additionally, the degree of δ13C and δ15N discrimination between an animal’s diet and its tissues may be impacted by diet quality (i.e., protein and lipid content20). Finally, metabolic routing of different diet components (i.e., the pathways taken by dietary macromolecules such as proteins, fats, and carbohydrates during tissue synthesis and metabolism) impacts diet-tissue isotope discrimination and may mean that some tissues primarily reflect individual diet components (e.g., dietary protein), while others may be representative of whole diet21. Diet-tissue discrimination has been studied in a variety of animals and varies widely among species22. Thus, as with lipid normalization models, species- and tissue-specific discrimination factors are not currently available for many species.
Understanding diet-tissue discrimination, as well as the relationships between δ13C and δ15N of different tissues of individual animals, is critical for adequate interpretation of stable isotope studies. Furthermore, quantifying the degree of tissue-specific isotope discrimination allows researchers to more directly compare the results of studies analyzing δ13C and δ15N in different tissues. This is particularly valuable for comparing modern studies, which typically sample soft tissues, to studies of historic and paleoecology, for which bone collagen is the primary tissue available for analysis23. It is also important because analyses of various soft tissues provide different advantages. For many vertebrates, skin can be obtained using a biopsy, which is typically non-lethal and relatively non-invasive24,25. Muscle is often analyzed because this tissue has relatively slow metabolic turnover, can provide diet information over a period of weeks or months, and is the most commonly used vertebrate tissue in the published stable isotope literature26,27. Tissues with more rapid metabolic turnover rates can provide shorter-term information about diet, and are particularly informative when analyzed in tandem with tissues that turn over more slowly. Blood has a rapid turnover rate26, but obtaining it can be logistically difficult for some animals (e.g., free-ranging and subsistence harvested marine mammals). Liver also exhibits high metabolic turnover26, and can provide short-term diet information. Collection of liver samples can be logistically challenging, but may be feasible in instances where proper sampling and storage of blood are not.
Arctic marine mammals are ideal candidates for stable isotopic research. Biological systems in the Arctic have been historically understudied, and are currently undergoing rapid changes associated with warming of the regional climate28,29. Stable isotopes may provide a useful tool for understanding these changes. Examining δ13C and δ15N in marine mammal tissues can not only provide insight into the diet and movements of the animals themselves, but can also give important information about the food webs in which these animals feed8. Arctic marine mammals, including Pacific walruses (Odobenus rosmarus divergens), are important subsistence resources for Alaska Native communities, and monitoring the responses of these animals to climate change is critical to food security in the region. The overarching goal of this study was to address some of the factors that might confound the interpretation of stable isotope values in the tissues of Pacific walruses, with the goal of improving the ability of future research to draw conclusions about these animals and the food webs in which they feed. To accomplish this, δ13C and δ15N in walrus muscle, liver, skin, and bone collagen were analyzed to (1) assess the importance of lipid removal when comparing δ13C of various walrus tissues, (2) parameterize lipid normalization models for estimating Δδ13C based on C:Nbulk, and (3) estimate tissue-specific discrimination factors for Pacific walruses.
Methods
Skin, muscle, liver, and bone samples were collected from 30 adult walruses (20 males, 10 females) taken as part of the Alaska Native subsistence harvests on St. Lawrence Island, Alaska, from 2014–2016. Additional muscle (n = 95), liver (n = 5), and skin (n = 5) samples from the 2012–2016 subsistence harvests were analyzed to generate mathematical lipid-corrections and test the effects of chemical lipid extraction on δ13C and δ15N. Samples were collected in the field (typically in May in June) by Alaska Native subsistence hunters using guidelines developed by researchers at the Alaska Department of Fish and Game (ADF&G). At the time of harvest, skin, blubber and muscle were collected in a single, full-thickness sample, and a single, large section of the liver was taken. These tissues were frozen and shipped to ADF&G, in Fairbanks, Alaska, before being transferred to the University of Alaska Fairbanks, where they were stored at −80 °C. Bone samples were also collected by subsistence hunters at the time of harvest. Samples consisted mostly of cranium and mandible fragments; however, the skeletal element from which a sample was taken was not always identifiable. Samples that appeared to come from distal limb bones (i.e., carpals, tarsals, phalanges) were not used for this study, as the stable isotope values of these bones may not be representative of the skeleton as a whole30. Bones were cleaned of adherent tissue with a wire brush, dried, and stored at room temperature. All tissues used in this study were obtained under a letter of authorization to L. Horstmann from the United States Fish and Wildlife Service.
In preparation for stable isotope analysis, ~10 g of each soft tissue was subsampled, freeze-dried for 48 hours, and homogenized using a Wig-L-Bug® grinding mill. Hair was typically relatively sparse on the skin samples, and surface hair was removed with a razor blade prior to homogenization. Any remaining pieces of hair were typically resistant to homogenization, thus remained intact and could be avoided during subsampling. Homogenized samples were divided into two equal parts. A 0.2–0.4 mg subsample was taken from one half and submitted for stable isotope analysis. The other half was lipid-extracted using methods modified from Folch et al.14 and Bligh and Dyer13. Briefly, samples were immersed in 2:1 chloroform:methanol and mixed thoroughly using a vortex mixer. Samples were then allowed to sit for 15 minutes before they were centrifuged and the chloroform:methanol was removed by pipet. This process was repeated until the supernatant remained clear, at which point samples were allowed to air dry overnight. A 0.2–0.4 mg subsample of the lipid-extracted sample was then submitted for stable isotope analysis.
Collagen was extracted from bone samples using the methods described by Misarti et al. (2009)31, as modified from Matheus (1995)32. A ~0.4 g subsample of solid, cortical bone was removed from each specimen using handheld cutting tools, cleaned in a sonic bath, and allowed to air dry for 1–2 days. Lipids were removed by soaking the bone in 2:1 chloroform:methanol for 8 hours and discarding the solvent. The mineral component of the bone was then removed using a mixture of 6N hydrochloric acid and ultrapure water. After demineralization, the sample was gelatinized at 65 °C, filtered through a 0.45 μm filter to remove any insoluble particles and non-collagen organic compounds, and freeze-dried. A 0.2–0.4 mg subsample of the resulting purified collagen was submitted for stable isotope analysis.
Stable carbon and nitrogen isotope values of tissue samples were analyzed in the Alaska Stable Isotope Facility at the University of Alaska Fairbanks, using a Costech ECS 4010 elemental analyzer and ThermoScientific Conflo IV, interfaced with a ThermoScientific DeltaV isotope ratio mass spectrometer. Stable isotopic compositions were calibrated relative to Vienna Pee Dee Belemnite (VPDB) and atmospheric nitrogen gas (air) using USGS40 and USGS41 as calibration standards. Results were reported in parts per thousand (‰) using δ notation. A commercially available peptone standard (No. P-7750 bovine based protein, Sigma Chemical Company, lot #76f-0300; δ13C: −15.8‰, δ15N: 7.0‰, %C: 44.3, %N: 15.3) was analyzed as a check standard after every 10 samples to estimate uncertainty. Precision (±1 standard deviation; SD) of these analyses was ±0.1‰ for both δ13C and δ15N, based on repeated measurements of this standard across all analytical runs (n = 81). Measurements were accurate (±1 SD) to within ±0.03‰ for δ13C and ±0.05‰ for δ15N, based on differences between observed and known values of the peptone standard. Sample composition (%C and %N) was measured during stable isotope analysis, and was used to calculate the C:N ratio of the sample using the formula:
Paired t-tests were used to determine whether changes in δ13C and δ15N associated with lipid-extraction (Δδ13C) were significant for walrus muscle (n = 125), liver (n = 35), and skin samples (n = 35). To generate arithmetic lipid-corrections, Δδ13C was regressed against the carbon:nitrogen ratio of the non-lipid extracted sample (C:Nbulk). Lipid-correction models from the literature were fitted to the relationship, using the nlstools package in R33 to estimate parameters. These non-linear least squares models assume that residual error is normally distributed, and this assumption was met for all three tissue types.
The first model fitted was Equation 1a from Logan et al. (2008)34, which was derived from McConnaughey and McRoy’s (1979)8 Equation 1:
In this model, the x-intercept (−b/a) represents the estimated C:Nlipid-free, whereas the maximum Δδ13C is represented by a. The second model tested was Equation 2 from Logan et al. (2008)34, as derived from Fry (2002)35:
The model-estimated C:Nlipid-free in this equation is represented by ƒ, while p represents protein-lipid δ13C discrimination. The third model tested was Equation 3 from Logan et al. (2008)34:
In this model, the estimated C:NLipid-free is represented by \({e}^{\frac{-{\beta }_{0}}{{\beta }_{1}}}\). Finally, a linear model was fitted to the data for comparison with the above equations.
Predictive error was assessed using leave-one-out cross validation36. For each model, this technique was used to calculate mean squared error (MSE), mean absolute error (MAE), and the proportion of the samples for which the arithmetically estimated Δδ13C was within 0.5‰ of the measured Δδ13C. This 0.5‰ value represents roughly twice the measurement error commonly reported in ecological studies measuring δ13C, thus differences below this threshold are often not considered biologically important37,38. These metrics were compared to assess model fit, with the best fitting model defined as that with the lowest MSE and MAE, and the greatest proportion of samples for which Δδ13C was estimated to within 0.5‰ of measured Δδ13C.
Comparisons among walrus tissues were made using δ13Clipid-free. Because chemical lipid-extraction may impact stable nitrogen isotope values34, δ15N values from non-lipid extracted samples were used for these analyses. Linear mixed effects models were used to test whether δ13Clipid-free and δ15N differed among muscle, liver, skin, and bone collagen of 30 individual walruses. These models assume a Gaussian error distribution, and this assumption was met for all tissues. For each model, tissue was entered as a fixed effect. Individual was included as a by-subject random intercept to account for relationships among the isotope values of tissues from individual walruses. Models were fitted using the R package lme439, and p-values were obtained by likelihood ratio tests of the full model with the effect of tissue against the null model, which did not include tissue as an effect. Significance was assessed using an alpha of 0.05, and residuals were visually inspected for deviations from normality and homoscedasticity. Tukey’s Honest Significant Difference post hoc tests with a Holm-Bonferroni correction40 were conducted in the multcomp R package41 to examine differences among tissues. Tissue samples from female walruses were only available in 2016, and only 10 of the 30 animals sampled for this study were female. Additionally, there is no evidence to support the assumption that differences in stable isotope values between males and females observed in this study would remain consistent seasonally, inter-annually, and across broader time scales, thus sex of sampled walruses was not included in the models presented in here. However, visual examination of the data revealed apparent differences between the tissue stable isotope values of male and female walruses included in this study, thus additional linear mixed effects models including sex were run. The results of these models are presented and interpreted in the online supplementary material (Supplementary Discussion 1). All statistical analyses were conducted using R version 3.4.142 with RStudio version 1.0.15343.
Results
Chemical lipid extraction changed the C:N ratios, δ13C, and/or δ15N of walrus tissues (Table 1). Paired t-tests indicated that chemical lipid extraction significantly increased the δ13C values of walrus liver (mean change ± 1 SD = 0.9 ± 0.3‰, t34 = 17.42, p < 0.001) and skin (mean change ± 1 SD = 1.9 ± 1.8‰, t34 = 6.18, p < 0.001), but walrus muscle δ13C was unchanged (mean change ± 1 SD = 0.0 ± 0.5‰, t124 = 1.07, p = 0.287). Lipid extraction significantly increased the δ15N of muscle (mean change ± 1 SD = 0.1 ± 0.3‰, t124 = 3.05, p = 0.002) and liver (mean change ± 1 SD = 0.2 ± 0.2‰, t34 = 6.50, p < 0.001), whereas changes to skin δ15N (Δδ15N) were not statistically significant (mean change ± 1 SD = 0.1 ± 0.3‰, t34 = 1.46, p = 0.155). Regardless of whether mean δ15N was impacted by lipid extraction, the difference between δ15Nbulk and δ15Nlipid-free (Δδ15N = δ15Nbulk − δ15Nlipid-free) of a sample from an individual animal was frequently well beyond the range of analytical precision (muscle Δδ15N range = −0.7–1.0‰; liver Δδ15N range = −0.1–0.5‰; skin Δδ15N range = −0.6–0.8‰).
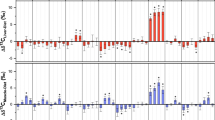
Lipid-normalization models were parameterized for walrus muscle, liver, and skin, but not for bone collagen, which is a purified protein. Of the three tissues examined in this portion of the study, only muscle and skin showed a relationship between Δδ13C and C:Nbulk (Table 2, Fig. 1). Liver Δδ13C was not related to C:Nbulk, thus a mathematical lipid correction could not be generated. For muscle, evaluation of model fit indicated that Equations 1–3 performed almost identically in their predictions of Δδ13C (Table 2, Fig. 1). Because lipid normalization models are primarily useful for predicting Δδ13C of samples with high C:Nbulk values, the fit of the three models was reevaluated by performing leave-one-out cross validation using only samples with C:Nbulk > 3.75 (n = 10). When model fit was considered for only these 10 samples, Equation 2 best predicted Δδ13C (MSE = 0.167, MAE = 0.307, P0.5 = 0.80), followed by Equations 3 (MSE = 0.192, MAE = 0.318, P0.5 = 0.70) and 1 (MSE = 0.198, MAE = 0.338, P0.5 = 0.70). The parameters p and ƒ were estimated for Equation 2 using walrus muscle δ13C values from this study, resulting in the following equation:
Non-lipid extracted carbon:nitrogen ratios (C:Nbulk) of walrus muscle (top, yellow triangles, n = 125), liver (middle, green circles, n = 35), and skin (bottom, orange diamonds, n = 35) plotted against the change in δ13C due to chemical lipid extraction (Δδ13C). Equations reflect the best fitting lipid-normalization models for muscle and skin, and solid black lines are predicted values for these models. Gray shaded areas represent 95% confidence envelopes for the predicted lines. Vertical dashed lines represent the carbon:nitrogen ratio of the lipid-free tissue (C:Nlipid-free) as predicted by the best-fitting lipid normalization model. It is important to note that the axes of each plot are scaled differently.
Which was simplified to:
The mean and median differences between the observed Δδ13C and the values predicted by this model were both 0.2‰. The maximum difference between the observed and predicted Δδ13C for a given individual was 1.2‰.
For walrus skin, the relationship between Δδ13C and C:Nbulk was best explained by Equation 2, which predicted Δδ13C to within 0.5‰ for >85% of samples (Table 3, Fig. 1). Walrus skin δ13C values from this study were used to estimate the parameters p and ƒ, which generated the following equation:
Which was simplified to:
For this model, the mean and median differences between the observed and predicted Δδ13C values were 0.3‰ and 0.2‰, respectively. The maximum difference between the observed and predicted Δδ13C for a given individual was 1.1‰.
Linear mixed effects models indicated that δ13Clipid-free and δ15N differed significantly among walrus tissues (δ13Clipid-free: χ23 = 282.87, p < 0.001; δ15N: χ23 = 193.86, p < 0.001; Tables 1 and 4, Fig. 2). Liver had the lowest δ13Clipid-free values (Tables 1, 4, and 5, Fig. 2). The output from the linear mixed effects models indicated that muscle δ13Clipid-free was, on average, 0.5 (95% CI: 0.3–0.7)‰ higher than liver. Skin and bone collagen were enriched in 13C by an additional 2.0 (95% CI: 1.8–2.2)‰ compared with muscle. Post hoc tests showed that the δ13Clipid-free of all tissues differed significantly, except for skin and bone collagen (Table 5). Bone collagen had the lowest δ15N values (Tables 1, 4, and 5, Fig. 2). Liver and muscle were enriched in 15N by 0.4 (95% CI: 0.2–0.6)‰ in relation to bone collagen. Skin δ15N values were an average of 2.1 (95% CI: 1.9–2.3)‰ higher than those of liver and muscle. For δ15N, all tissues were significantly different, except for muscle and liver (Table 5).
Scatterplot of δ13Clipid-free and δ15N values for muscle (yellow triangles), liver (green circles), skin (orange diamonds), and bone collagen (blue squares) from 30 individual walruses. Each walrus is represented by a complete set of tissues (muscle, liver, skin, and bone collagen). Mean values are represented by larger, darker symbols. Error bars represent ±1 SD from the mean.
Discussion
Accounting for sample lipid content (either by mathematical lipid correction or lipid extraction) and understanding tissue-specific discrimination are important for properly interpreting the results of stable isotope analyses of animal tissues. This is particularly true when δ13C and δ15N values are used to make quantitative diet reconstructions44,45, as well as when making decisions about which tissue to analyze or comparing stable isotope values among different tissues46. The results of this study provide quantitative estimates of the effects of chemical lipid extraction on δ13C and δ15N in walrus muscle, liver, and skin, lipid normalization models for walrus muscle and skin, and tissue-specific stable isotope discrimination factors for walrus muscle, liver, skin, and bone collagen.
Effects of Lipids on δ13C
Many studies have demonstrated the negative relationship between δ13C and lipid content in animal tissues1,17,34,44. In this study, chemical lipid extractions significantly increased the δ13C values of walrus liver and skin, indicating that lipids affected δ13C in these tissues. Muscle δ13C remained unchanged, likely because marine mammal muscle is typically comprised of lean protein with little lipid content47,48,49. Lipids are stored primarily in the blubber layer in these animals, rather than interspersed among the muscles. Depending on the degree of precision desired, future studies measuring δ13C in walrus muscle may choose to forego lipid extraction and/or lipid normalization altogether. Because an arithmetic lipid correction could not be generated for walrus liver, lipids should be chemically extracted from this tissue if liver δ13C is to be used for future studies.
Mathematical lipid correction requires that the change in δ13C associated with lipid removal (Δδ13C) be statistically related to the carbon:nitrogen ratio of the bulk sample. For walrus skin, Δδ13C was proportional to C:Nbulk, such that higher C:N ratios prior to lipid extraction were associated with larger changes to δ13C values as a result of extraction. Though chemical lipid extraction did not affect the mean δ13C of walrus muscle, samples with high C:Nbulk were associated with the largest changes in δ13C. C:Nbulk is indicative of the lipid content8,18, thus, as expected, samples with a greater lipid content exhibited a larger change in δ13C after the lipids were removed. The top-performing model for walrus muscle (Equation 5a) was parameterized from Equation 2 in Logan et al. (2008)34, using experimental data. In this equation, the C:Nlipid-free can be estimated by the term f, which resulted in a value of 3.45 (95% confidence interval [CI]: 3.41–3.48), indicating that a muscle sample with a C:Nbulk of ≤3.45 is comprised entirely of lean protein and need not be lipid extracted or corrected. Walrus muscle analyzed in this study appears to have contained very few lipids, with the model output indicating that more than 40% of samples had a carbon:nitrogen ratio less than or equal to 3.45. Some samples did contain a higher proportion of lipids, possibly as a result of the location from which the sample was taken (e.g., close to the blubber layer, where lipids may have leached into the muscle). Future studies may wish to sample muscle from locations far from the blubber to reduce the amount of lipids in the tissue. In instances where this is not possible, the equation presented here may be used to estimate Δδ13C of muscle with a greater-than-average lipid content (C:Nbulk > 3.45).
The C:N ratio of walrus skin (a proxy for lipid content8,18) ranged widely (C:Nbulk range = 2.7–10.5), and Δδ13C was proportional to C:Nbulk, making this tissue particularly well-suited for mathematical lipid correction. The top-performing lipid normalization model (Equation 6a) was adapted from the Logan et al. (2008)34 derivation of the Fry (2002)35 mass balance equation (Equation 2), which uses the C:Nbulk, C:Nlipid-free, and a term describing protein-lipid δ13C discrimination to estimate Δδ13C. The latter two parameters were estimated from the experimental data, providing valuable information about the characteristics of walrus skin. The C:Nlipid-free parameter from the fitted model had a value of 2.80 (95% CI: 2.74–2.87), indicating that a skin sample with a C:Nbulk of ~2.8 is already composed of lean protein and need not be lipid-extracted or corrected. The lipid-protein discrimination parameter was estimated to be 8.45, which was greater than published estimates for various fishes and invertebrates34, but similar to estimates for baleen whale skin and blubber38. The best-fitting model was effective at predicting Δδ13C for walrus skin samples based on a wide range of C:Nbulk values. These predicted values were within 0.5‰ for greater than 85% of samples. This level of precision is likely acceptable for many ecological studies; however, in situations where researchers seek to address questions requiring greater precision, chemical lipid extraction may be necessary.
Removal of lipids from walrus tissues sometimes resulted in unexpected changes in δ13C. For some samples, δ13Clipid-free was lower than δ13Cbulk, resulting in a negative value for Δδ13C. Chemical lipid extraction should not reduce δ13C, therefore these negative Δδ13C values likely resulted due to analytical error, within-tissue heterogeneity in δ13C, and/or incomplete homogenization of tissue samples before stable isotope analysis. Because only a small subsample (~0.2 mg) is analyzed for stable isotopes, it is possible or even likely that two subsamples taken from an incompletely homogenized tissue may differ in their composition (i.e., fatty acid and amino acid compositions), resulting in δ13C and δ15N differences. Most of the negative Δδ13C values were small (~−0.1‰); however, some muscle samples had δ13Clipid-free values that were much lower (~−0.6‰) than their δ13Cbulk values. Ensuring samples are completely homogenized before subsampling for isotope analysis may help reduce this type of error in future studies. Complete homogenization is particularly important because differences in stable isotope values may exist at small spatial scales within a tissue. Todd et al. (2010) found little variability in pinnipeds skin and muscle sampled at different sites on the body50; however, Wild et al. (2018) described differences in both δ13C and δ15N among cetacean skin layers51, indicating that this tissue represents a dietary time series, and stable isotope values may not be homogenous at different depths within a sample. Improper subsampling or incomplete homogenization prior to analysis could thus result in δ13C and δ15N differences driven by these spatial variations in isotope values within the skin. Further investigations into how δ13C and δ15N vary within individual tissues are warranted. In particular, it would be valuable to examine variations among different muscle groups, as well as differences between glycolytic and oxidative muscles.
Effects of Chemical Lipid Extraction on δ15N
A substantial drawback to chemical lipid extractions is that they may alter δ15N values, thus, a lipid extracted and non-lipid extracted aliquot of each sample must be run to ensure comparability across tissues and studies of both δ13C and δ15N values. Sotiropoulos et al. (2004)17 hypothesized that δ15N increases as the organic solvent removes amino acids contained within structural lipids; however, as Ryan et al. (2012)38 also noted for cetacean skin and blubber samples, changes in δ15N observed in our study were not unidirectional. The δ15N values of some walrus skin samples decreased by as much as 0.6‰, while others increased by as much as 0.8‰, indicating that the removal of 15N was not uniform across samples. The same pattern was observed for walrus muscle and liver, though the observed changes in δ15N were smaller. This may have resulted from the removal of amino acids from the sample during chemical lipid extraction17, but may also have been related to incomplete homogenization during sample processing and/or variation in sample lipid content. The effects of lipid extractions on δ15N in this study suggest that, for studies using chemical lipid extraction on walrus tissues, isotope values of samples must be analyzed both before and after extraction. This makes chemical lipid extraction at least twice as expensive as mathematical lipid correction. Though lipid extraction with chloroform:methanol did not significantly alter the mean δ15N values of walrus skin samples used in this study, the δ15N of the lipid-free samples was often different enough from the bulk sample (up to 0.8‰) to alter interpretation of the results. Such changes to δ15N would be particularly problematic for studies using mixing models to estimate animal diet52,53. In the future, researchers should assess the level of precision required to investigate their hypotheses, as well as the number of samples they are able to run, if laboratory costs are a limiting factor, before deciding whether to use chemical lipid extractions or the arithmetic lipid-correction for walrus tissues.
Tissue-specific δ13C and δ15N Discrimination
Differences in δ13C and δ15N among animal tissues arise from multiple factors that impact stable isotope values, including tissue-specific isotope discrimination2,46,54, inter-tissue variability in turnover rates20,27,46, and differences in metabolic routing of dietary macromolecules, such as proteins, fats, and carbohydrates20,21,55. Disentangling the effects of these different processes is difficult, and many controlled feeding experiments have investigated the role that each of these processes plays in determining the stable isotope values of individual tissues within an organism27,46,54,56. For studies of free-ranging animals, these processes must be considered together and the likely importance of each weighed when interpreting stable isotope data. Though the specific factors driving inter-tissue variability in δ13C and δ15N may remain unknown, general patterns in the differences among tissues can be observed and compared with those reported in the literature. For example, the −0.5 (95% CI: −0.7–−0.3)‰ offset in δ13Clipid-free between walrus muscle and liver in the present study appears to be standard among marine mammals, for which reported values range from −0.8 to −0.1‰ (Supplementary Table 2 and references therein). Similarly, the +2.0 (95% CI: 1.8–2.2) ‰ difference between muscle and bone collagen δ13Clipid-free is typical of marine mammals (range: 1.3–2.8‰; Supplementary Table 2 and references therein). In contrast, walrus skin exhibits unusually high enrichment in 13C and 15N (+2.0 [95% CI: 1.8–2.2] ‰ and +2.1 [95% CI: 1.9–2.3]‰, respectively) as compared with muscle values (published ranges: δ13Clipid-free = −0.8–1.5‰; δ15N = −0.1–1.2‰; Supplementary Table 2 and references therein). Yurkowski et al. (2015)57 measured δ13C and δ15N in skin (n = 20) and liver (n = 19) of Atlantic walruses (Odobenus rosmarus rosmarus). Liver δ13Clipid-free averaged 0.4‰ lower than muscle δ13Clipid-free, closely matching the average difference of −0.5 (95% CI: −0.7–−0.3)‰ observed in this study. In contrast, Atlantic walrus liver δ15N was 0.4‰ greater than muscle δ15N, as compared to a difference of 0.0 (95% CI: −0.2–0.2)‰ in our study. The contrasting results for δ15N may have arisen due to a variety of factors, including sample size, timing of sample collection, and differences in the physiology, ecology, diet, and seasonal movements of Pacific and Atlantic walruses.
Examining the differences between paired samples from individual walruses on a tissue-by-tissue basis can provide insight into the factors driving inter-tissue variability in δ13Clipid-free and δ15N (Fig. 3). In our study, individual walruses with higher δ13Clipid-free and δ15N in one tissue generally had high δ13Clipid-free and δ15N in their other tissues. When lines are drawn between the paired tissue samples (Fig. 3), the lines are mostly parallel and generally of the same length. This suggests that the differences between these tissues are likely driven primarily by tissue-specific discrimination or metabolic routing, which would be expected to produce a consistent offset between the stable isotope values of individual tissues. Comparisons involving liver are the exception to this general pattern, with more variability in the differences between tissues from individual animals (Fig. 3); this is most apparent in the comparison between liver and muscle. Liver exhibits a small, but generally consistent offset from muscle towards more negative δ13Clipid-free; however, the differences between liver and muscle are highly variable in their magnitude and direction. The same is true, to a lesser extent, for the liver-skin and liver-bone collagen comparisons. This variability in the offset between paired tissue samples likely reflects differences in turnover rate among tissues. Comparisons between tissues with rapid turnover rates and those with longer turnover rates are more likely to result in isotopic variability among tissues due to the contrast between the short-term diet signal represented by the tissue with rapid turnover, and the integrated, longer-term signal reflected in the tissue with slow turnover. When the difference in turnover rates is large, and the relative importance of tissue-specific discrimination and metabolic routing are smaller, turnover rate will likely play a greater role in determining the magnitude and direction of differences between tissue samples from individual animals.
Comparisons of δ13Clipid-free and δ15N for muscle (yellow triangles), liver (green circles), skin (orange diamonds), and bone collagen (blue squares) from 30 individual walruses. Each walrus is represented by a complete set of tissues (muscle, liver, skin, and bone collagen). Paired data points from individual walruses are connected by dashed lines. It is important to note that the axes of each plot on identical scales.
Variation in turnover rate alone will not drive differences in the δ13C and δ15N of animal tissues. In the absence of other sources of variability in stable isotope values (i.e., tissue-specific discrimination and metabolic routing), animals consuming a monotonous diet would have consistent stable isotope values throughout their bodies, regardless of turnover rate. An animal’s diet must change for tissue turnover rates to drive differences in δ13C and δ15N among tissues. For walruses in this study, differences in δ13Clipid-free and δ15N of liver and muscle likely resulted from changes in diet (or prey isotope values) associated with the annual migration. Pacific walrus migration is sex-segregated58. The entire population winters together in the Bering Sea. In the summer, females and juveniles move northward into the Chukchi Sea, whereas most males move in the coastal waters of the Bering Sea, including Bristol Bay and the Russian coastline. Female walruses had liver δ15N values that were 1.4 (95% CI: 1.0–1.8) ‰ greater than those of males; whereas female δ15N was only 0.7 (95% CI: 0.3–1.1) to 0.8(95% CI: 0.4–1.2)‰ greater than that of males for all other tissues (Supplementary Discussion 1). Stomach content analysis indicates that the diets of male and female walruses are essentially identical when they are in the same location59, thus these sex-related differences are probably driven by changes in diet during the summers, when males and females are separated. These differences likely result from shifts in diet composition and geographic variability in baseline δ15N values, and provide further support for the hypothesis that the observed differences in isotope values between liver and other tissues were driven primarily by differences in turnover rate.
The relative turnover rates of animal tissues have been the subject of many studies60. Though direct estimates are not available for walruses, the turnover rates of muscle, liver, skin, and bone collagen have been established for many other species and typically follow the order liver <muscle/skin <bone collagen27,46. Turnover rate is fastest in liver, with a typical reported half-life of <10 days26,27,46,61, whereas bone collagen turns over more slowly and can represent an average of years of foraging26,27,62,63. The turnover rate of muscle is intermediate, with a half-life that is on the order of weeks to months26,27,61. When considering values from the published literature, it is critical to note that the rate of tissue turnover is directly related to the animal’s mass-specific metabolic rate64, and is proportional to body size60. Most of the studies cited above measured tissue turnover in small animals (e.g., rodents, small birds) that are readily studied in the lab. These animals have high mass-specific metabolic rates, and their tissues almost certainly turn over much faster than those of larger animals. For example, the stable carbon isotope half-lives of liver (37.3 days) and muscle (178.7 days) in alpacas (Lama pacos) are approximately six times those reported (6.4 days and 27.6 days, respectively) for gerbils46,64. Domestic cattle (Bos taurus) have similarly slow δ13C and δ15N turnover in their skeletal muscle (~134–157 days65). Tissue turnover rates in walruses are likely to be much closer to those of larger mammals than the values estimated for small animals, and may be even slower given their large body size.
In contrast to liver, muscle, and bone collagen, the metabolic turnover of skin is not well studied. Most estimates in the literature come from studies of marine mammals, for which skin biopsy is a commonly used sampling method66. Estimates for cetacean skin turnover range from roughly one to six months67,68,69,70. Research by Wild et al. (2018)51 showed that cetacean skin, which grows continuously at its innermost margin and is sloughed off upon reaching the surface, contains a time series of stable isotope values that may reflect diet over a longer period of time. They suggested that the inner layer of the skin alone might represent diet from an entire summer feeding period (~4 months). It is important to note that the structure and function of cetacean skin is different from that of pinnipeds58,71,72,73, and these estimates might not be appropriate for walruses.
Tissue-specific stable isotope discrimination typically results from differences in the chemical composition of tissues. Proteins that differ in the relative abundance of their constituent amino acids may be subject to varying degrees of isotopic discrimination due to the various biochemical synthetic pathways for amino acids74. These different pathways result in some amino acids exhibiting greater isotopic discrimination than others. The difference between the δ13Clipid-free values of walrus skin and bone collagen compared with those of liver and muscle likely results from variation in the amino acid composition of these tissues. Walrus skin contains an unusually high proportion of collagen fibrils71, making it especially tough. Skin and bone collagen share similar chemical compositions and both contain a large amount of glycine75. This amino acid tends to be enriched in 13C by ~8.0‰ compared with other amino acids and, as a result, collagen typically has higher δ13C values than other tissues76. This is likely why, in the present study, walrus skin and bone collagen δ13Clipid-free values were similar to one another, and were greater than those of muscle and liver by around 2.0 (95% CI: 1.8–2.2)‰ (Table 5). For δ15N, some amino acids exhibit strong 15N-enrichment with each trophic transfer (‘trophic amino acids’), whereas others do not (‘source amino acids’)77. Walrus skin δ15N values were 2.5 (95% CI: 2.3–2.7)‰ greater than those of bone collagen, indicating important differences with regards to discrimination of nitrogen isotopes. In humans, skin collagen has greater proportions of the trophic amino acids alanine, asparagine, and glutamine than bone collagen75. This difference may be responsible for the high δ15N values of walrus skin in this study, though this could also be due in part to dietary changes and differences in tissue turnover rates.
Quantifying diet-tissue discrimination is important for reconstructing animal diet and understanding food web structure. The results of this study provide information about the relative degree of isotope discrimination among tissues, but their relationship to walrus diet remains unknown. Estimates from other marine mammals indicate an average diet-muscle δ13C discrimination factor (Δ13C) of +1.2‰ (Supplementary Table 2), with little variation among species (range: +1.0–1.3‰). For δ15N, marine mammals average an estimated discrimination factor (Δ15N) of +2.8‰ between diet and muscle; however the range of these estimates is much greater (range: +1.7–4.3‰). A rough estimate of diet-muscle discrimination can be made for walruses using published prey stable isotope values. Average reported δ13C and δ15N of bivalve species common in walrus diet (Serripes groenlandicus and Macoma calcarea) are ~−18.8‰ and ~9.2‰, respectively48,49,78,79. Using these values to estimate diet-muscle discrimination estimates for walruses in this study results in a value of +1.8‰ (Δ13C) and +3.3‰ (Δ15N). Though bivalves comprise a large portion of their diet, walruses consume a wide variety of prey items, including higher trophic level prey such as predatory snails, seabirds, and seals49,59,80. The estimate for Δ15Nmuscle-diet may thus be high, as inclusion of these higher trophic level prey items would increase the estimated average δ15N of walrus prey. These estimates are generated from a limited number of prey items, collected in different years than this study’s specimens, so these values are approximations and should be interpreted with caution. Further research is warranted to more precisely estimate diet-tissue discrimination for walruses.
Conclusions
Stable isotopic analysis of walrus tissues indicates considerable differences among the δ13Clipid-free and δ15N of muscle, liver, skin, and bone collagen. The differences in δ13Clipid-free and δ15N among tissues observed in this study generally agree with published values for mammals, with the exception of skin, which differed more substantially from muscle for both δ13Clipid-free and δ15N. Lipid normalization models were parameterized for walrus muscle and skin. Walrus liver Δδ13C was not related to C:Nbulk, so a mathematical lipid correction could not be generated. Both lipid normalization models performed well, and can be used in future studies as an alternative to chemical lipid extractions. Many walrus muscle samples analyzed in this study had a C:Nbulk that indicated they were composed primarily of lean protein (≤3.4) and did not require chemical lipid extraction or mathematical lipid extraction.
The results of the present research will allow for better comparison of δ13C and δ15N in walrus tissues, giving researchers more flexibility in stable isotope studies of subsistence harvested walruses. Our results will also make studies of δ13C and δ15N using non-lethal sample collection (i.e., remote skin biopsy) a more viable approach, by decreasing the costs of analysis and providing context for interpreting the results. Finally, the results of this study will be particularly useful for comparing contemporary studies, which typically analyze soft tissues, with historic and paleoecological studies, which generally measure δ13C and δ15N in bone collagen23. The δ13C values of walrus skin and bone collagen were indistinguishable, indicating that these tissues likely both primarily represent dietary protein and are suitably comparable. Skin δ15N was 2.5 (95% CI: 2.3–2.7)‰ greater than that of bone collagen on average, suggesting differences in nitrogen isotope discrimination between these tissues or a change in diet. The results of this study will also help refine stable isotope mixing models and provide better estimates of walrus diet. Future research measuring the degree of trophic discrimination between the δ13C and δ15N values of walrus prey and tissues will be important for further improving these reconstructions, as well as for a better understanding of Arctic food web structure and energy flow.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Change history
17 January 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
DeNiro, M. J. & Epstein, S. Influence of diet on the distribution of carbon isotopes in animals. Geochim. Cosmochim. Acta 42, 495–506 (1978).
DeNiro, M. J. & Epstein, S. Influence of diet on the distribution of nitrogen isotopes in animals. Geochim. Cosmochim. Acta 45, 341–351 (1981).
Peterson, B. J. & Fry, B. Stable isotopes in ecosystem studies. Annu. Rev. Ecol. Syst. 18, 293–320 (1987).
Hobson, K. A. Tracing origins and migration of wildlife using stable isotopes: A review. Oecologia 120, 314–326 (1999).
Fry, B. Stable isotope ecology. 521 (Springer, 2006).
Minagawa, M. & Wada, E. Stepwise enrichment of 15N along food chains: Further evidence and the relation between δ15N and animal age. Geochim. Cosmochim. Acta 48, 1135–1140 (1984).
DeNiro, M. J. & Epstein, S. Mechanism of carbon isotope fractionation associated with lipid synthesis. Science (80-.). 197, 261–263 (1977).
McConnaughey, T. & McRoy, C. P. Food-web structure and the fractionation of carbon isotopes in the Bering Sea. Mar. Biol. 53, 257–262 (1979).
Blumer, T. N., Craig, H. B., Pierce, E. A., Smart, W. W. G. Jr. & Wise, M. B. Nature and variability of marbling deposits in longissimus dorsi muscle of beef carcasses. J. Anim. Sci. 21, 935–942 (1962).
Kurle, C. M. & Worthy, G. A. J. Stable nitrogen and carbon isotope ratios in multiple tissues of the northern fur seal Callorhinus ursinus: implications for dietary and migratory reconstructions. Mar. Ecol. Prog. Ser. 236, 289–300 (2002).
Wang, S. W., Budge, S. M., Gradinger, R. R., Iken, K. & Wooller, M. J. Fatty acid and stable isotope characteristics of sea ice and pelagic particulate organic matter in the Bering Sea: Tools for estimating sea ice algal contribution to Arctic food web production. Oecologia 174, 699–712 (2014).
Oxtoby, L. E., Mathis, J. T., Juranek, L. W. & Wooller, M. J. Estimating stable carbon isotope values of microphytobenthos in the Arctic for application to food web studies. Polar Biol. 39, 473–483 (2016).
Bligh, E. G. & Dyer, W. J. A rapid method of total lipid extraction and purification. Biochem. Cell Biol. 37, 911–917 (1959).
Folch, J., Lees, M. & Stanley, G. H. S. A simple method for the isolation and purification of total lipides from animal tissues. Journal of Biological Chemistry 1, 497–509 (1957).
Bodin, N., Le Loc’h, F. & Hily, C. Effect of lipid removal on carbon and nitrogen stable isotope ratios in crustacean tissues. J. Exp. Mar. Bio. Ecol. 341, 168–175 (2007).
Ricca, M. A., Miles, A. K., Anthony, R. G., Deng, X. & Hung, S. S. O. Effect of lipid extraction on analyses of stable carbon and stable nitrogen isotopes in coastal organisms of the Aleutian archipelago. Can. J. Zool. 85, 40–48 (2007).
Sotiropoulos, M. A., Tonn, W. M. & Wassenar, L. I. Effects of lipid extraction on stable carbon and nitrogen isotope analyses of fish tissues: potential consequences for food web studies. Ecol. Freshw. Fish 13, 155–160 (2004).
Schmidt, K. et al. Trophic relationships among Southern Ocean copepods and krill: Some uses and limitations of a stable isotope approach. Limnol. Oceanogr. 48, 277–289 (2003).
Hobson, K. A., Alisauskas, R. A. Y. T. & Clark, R. G. Stable-nitrogen isotope enrichment in avian tissues due to fasting and nutritional stress: Implications for isotopic analyses of diet. Condor 95, 388–394 (1993).
Mohan, J. A. et al. Tissue-specific isotope turnover and discrimination factors are affected by diet quality and lipid content in an omnivorous consumer. J. Exp. Mar. Bio. Ecol. 479, 35–45 (2016).
Wolf, N., Newsome, S. D., Peters, J. & Fogel, M. L. Variability in the routing of dietary proteins and lipids to consumer tissues influences tissue-specific isotopic discrimination. Rapid Commun. Mass Spectrom. 29, 1448–1456 (2015).
Caut, S., Angulo, E. & Courchamp, F. Variation in discrimination factors (Δ15N and Δ13C): The effect of diet isotopic values and applications for diet reconstruction. J. Appl. Ecol. 46, 443–453 (2009).
Hedges, J. E. M., Stevens, R. E. & Koch, P. L. Isotopes in bones and teeth. In Isotopes in Palaeoenvironmental Research (ed. Leng, M. J.) 117–145, https://doi.org/10.1007/1-4020-2504-1_03 (Springer, 2006).
Noren, D. P. & Mocklin, J. A. Review of cetacean biopsy techniques: Factors contributing to successful sample collection and physiological and behavioral impacts. Mar. Mammal Sci. 28, 154–199 (2012).
Gemmell, N. J. & Majluf, P. Projectile biopsy sampling of fur seals. Mar. Mammal Sci. 13, 512–516 (1997).
Boecklen, W. J., Yarnes, C. T., Cook, B. A. & James, A. C. On the use of stable isotopes in trophic ecology. Annu. Rev. Ecol. Evol. Syst. 42, 411–440 (2011).
Hobson, K. A. & Clark, R. G. Assessing avian diets using Stable Isotopes .1 Turnover of C-13 in tissues. Condor 94, 189–197 (1992).
Kȩdra, M. et al. Status and trends in the structure of Arctic benthic food webs. Polar Res. 34, 1–23 (2015).
Grebmeier, J. M. et al. A major ecosystem shift in the northern Bering Sea. Science (80-.). 311, 1461–1464 (2006).
Clark, C. T., Horstmann, L. & Misarti, N. Quantifying variability in stable carbon and nitrogen isotope ratios within the skeletons of marine mammals of the suborder Caniformia. J. Archaeol. Sci. Reports 15, 393–400 (2017).
Misarti, N., Finney, B., Maschner, H. & Wooller, M. J. Changes in northeast Pacific marine ecosystems over the last 4500 years: evidence from stable isotope analysis of bone collagen from archeological middens. The Holocene 19, 1139–1151 (2009).
Matheus, P. E. Diet and co-ecology of Pleistocene short-faced bears and brown bears in eastern Beringia. Quaternary Research 44, 447–453 (1995).
Baty, F. et al. A Toolbox for Nonlinear Regression in R: The Package nlstools. J. Stat. Softw. 66, 1–21 (2015).
Logan, J. M. et al. Lipid corrections in carbon and nitrogen stable isotope analyses: Comparison of chemical extraction and modelling methods. J. Anim. Ecol. 77, 838–846 (2008).
Fry, B. Stable isotopic indicators of habitat use by Mississippi River fish. J. North Am. Benthol. Soc. 21, 676–685 (2002).
Friedman, J., Hastie, T. & Tibshirani, R. The elements of statistical learning. 1 (Springer series in statistics New York, 2001).
Ehrich, D. et al. Stable isotope analysis: Modelling lipid normalization for muscle and eggs from arctic mammals and birds. Methods Ecol. Evol. 2, 66–76 (2011).
Ryan, C. et al. Accounting for the effects of lipids in stable isotope (δ13C and δ15N values) analysis of skin and blubber of balaenopterid whales. Rapid Commun. Mass Spectrom. 26, 2745–2754 (2012).
Bates, D., Mächler, M., Bolker, B. M. & Walker, S. C. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015).
Holm, S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6, 65–70 (1979).
Hothorn, T., Bretz, F. & Westfall, P. Simultaneous inference in general parametric models. Biometrical J. 50, 346–363 (2008).
R Core Team. R: A language and environment for statistical computing (2014).
RStudio Team. RStudio: Integrated development environment for R (2015).
Kiljunen, M. et al. A revised model for lipid-normalizing δ13C values from aquatic organisms, with implications for isotope mixing models. J. Appl. Ecol. 43, 1213–1222 (2006).
Bond, A. L. & Diamond, A. W. Recent Bayesian stable-isotope mixing models are highly sensitive to variation in discrimination factors. Ecol. Appl. 21, 1017–1023 (2011).
Tieszen, L. L., Boutton, T. W., Tesdahl, K. G. & Slade, N. A. Fractionation and turnover of stable carbon isotopes in animal tissues: Implications for δ13C analysis of diet. Oecologia 57, 32–37 (1983).
Hoekstra, P. F. et al. Trophic ecology of bowhead whales (Balaena mysticetus) compared with that of other arctic marine biota as interpreted from carbon-, nitrogen-, and sulfur-isotope signatures. Can. J. Zool. 80, 223–231 (2002).
Dehn, L. A. et al. Feeding ecology of phocid seals and some walrus in the Alaskan and Canadian Arctic as determined by stomach contents and stable isotope analysis. Polar Biol. 30, 167–181 (2007).
Seymour, J., Horstmann-Dehn, L. & Wooller, M. J. Proportion of higher trophic-level prey in the diet of Pacific walruses (Odobenus rosmarus divergens). Polar Biol. 37, 941–952 (2014).
Todd, S. K., Holm, B., Rosen, D. A. S. & Tollit, D. J. Stable isotope signal homogeneity and differences between and within pinniped muscle and skin. Mar. Mammal Sci. 26, 176–185 (2010).
Wild, L. A., Chenoweth, E. M., Mueter, F. J. & Straley, J. M. Evidence for dietary time series in layers of cetacean skin using stable carbon and nitrogen isotope ratios. Rapid Commun. Mass Spectrom, https://doi.org/10.1002/rcm.8168 (2018).
Giménez, J. et al. Lipid effects on isotopic values in bottlenose dolphins (Tursiops truncatus) and their prey with implications for diet assessment. Mar. Biol. 164, 1–9 (2017).
Tarroux, A. et al. Sensitivity of stable isotope mixing models to variation in isotopic ratios: evaluating consequences of lipid extraction. Methods Ecol. Evol. no–no https://doi.org/10.1111/j.2041-210x.2010.00033.x (2010).
Arneson, L. S. & MacAvoy, S. E. Carbon, nitrogen, and sulfur diet-tissue discrimination in mouse tissues. Can. J. Zool. 83, 989–995 (2005).
Newsome, S. D., Wolf, N., Peters, J. & Fogel, M. L. Amino acid δ13C analysis shows flexibility in the routing of dietary protein and lipids to the tissue of an omnivore. Integr. Comp. Biol. 54, 890–902 (2014).
Kurle, C. M., Koch, P. L., Tershy, B. R. & Croll, D. A. The effects of sex, tissue type, and dietary components on stable isotope discrimination factors (∆13C and ∆15N) in mammalian omnivores. Isotopes Environ. Health Stud. 50, 307–321 (2014).
Yurkowski, D. J., Hussey, N. E., Semeniuk, C., Ferguson, S. H. & Fisk, A. T. Effects of lipid extraction and the utility of lipid normalization models on δ13C and δ15N values in Arctic marine mammal tissues. Polar Biol. 38, 131–143 (2015).
Fay, F. H. Ecology and biology of the Pacific walrus, Odobenus rosmarus divergens Illiger. North Am. Fauna 1–279 (1982).
Sheffield, G. & Grebmeier, J. M. Pacific walrus (Odobenus rosmarus divergens): Differential prey digestion and diet. Mar. Mammal Sci. 25, 761–777 (2009).
Vander Zanden, M. J., Clayton, M. K., Moody, E. K., Solomon, C. T. & Weidel, B. C. Stable isotope turnover and half-life in animal tissues: A literature synthesis. PLoS One 10, 1–16 (2015).
Miller, J. F., Millar, J. S. & Longstaffe, F. J. Carbon- and nitrogen-isotope tissue–diet discrimination and turnover rates in deer mice, Peromyscus maniculatus. Can. J. Zool. 86, 685–691 (2008).
Riofrío-Lazo, M. & Aurioles-Gamboa, D. Timing of isotopic integration in marine mammal skull: Comparative study between calcified tissues. Rapid Commun. Mass Spectrom. 27, 1076–1082 (2013).
Manolagas. Birth and death of bone cells: Basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr. Rev. 21, 115–137 (2000).
Sponheimer, M. et al. Turnover of stable carbon isotopes in the muscle, liver, and breath CO2 of alpacas (Lama pacos). Rapid Commun. Mass Spectrom. 20, 1395–1399 (2006).
Bahar, B. et al. Turnover of carbon, nitrogen, and sulfur in bovine longissimus dorsi and psoas major muscles: Implications for isotopic authentication of meat. J. Anim. Sci. 87, 905–913 (2009).
Noren, S. R., Udevitz, M. S. & Jay, C. V. Bioenergetics model for estimating food requirements of female Pacific walruses Odobenus rosmarus divergens. Mar. Ecol. Prog. Ser. 460, 261–275 (2012).
Browning, N. E., Dold, C., I-fan, J. & Worthy, G. A. J. Isotope turnover rates and diet-tissue discrimination in skin of ex situ bottlenose dolphins (Tursiops truncatus). J. Exp. Biol. 217, 214–221 (2014).
Busquets-Vass, G. et al. Estimating blue whale skin isotopic incorporation rates and baleen growth rates: Implications for assessing diet and movement patterns in mysticetes. PLoS One 12, 1–25 (2017).
St. Hicks, B. D., Aubin, D. J., Geraci, J. R. & Brown, W. R. Epidermal growth in the bottlenose dolphin, Tursiops truncatus. J. Invest. Dermatol. 85, 60–63 (1985).
Giménez, J., Ramírez, F., Almunia, J., G. Forero, M. & de Stephanis, R. From the pool to the sea: Applicable isotope turnover rates and diet to skin discrimination factors for bottlenose dolphins (Tursiops truncatus). J. Exp. Mar. Bio. Ecol. 475, 54–61 (2016).
Sokolov, W. The skin structure in Pinnipedia of the U.S.S.R. fauna. J. Morphol. 107, 285–296 (1960).
Geraci, J. R., St Aubin, D. J. & Hicks, B. D. The epidermis of odontocetes: a view from within. In Research on Dolphins, Part 1 (eds Bryden, M. & Harrison, R.) 1, 3–21 (Clarendon Press, 1986).
Reeb, D., Best, P. B. & Kidson, S. H. Structure of the integument of southern right whales, Eubalaena australis. Anat. Rec. 290, 596–613 (2007).
Gannes, L. Z., O’Brien, D. M. & Martínez del Rio, C. Stable isotopes in animal ecology: Assumptions, caveats, and a call for more laboratory experiments. Ecology 78, 1271–1276 (1997).
Corr, L. T. et al. Probing dietary change of the Kwädąy Dän Ts’ìnchį individual, an ancient glacier body from British Columbia: II. Deconvoluting whole skin and bone collagen δ13C values via carbon isotope analysis of individual amino acids. J. Archaeol. Sci. 36, 12–18 (2009).
Hare, P. E. et al. The isotopic composition of carbon and nitrogen in individual amino acids isolated from modern and fossil proteins. J. Archaeol. Sci. 18, 277–292 (1991).
Popp, B. N. et al. Insight into the trophic ecology of yellowfin tuna, Thunnus albacares, from compound‐specific nitrogen isotope analysis of proteinaceous amino acids. In Stable Isotopes as Indicators of Ecological Change 1, 173–190 (Elsevier, 2007).
Iken, K., Bluhm, B. & Dunton, K. Benthic food-web structure under differing water mass properties in the southern Chukchi Sea. Deep. Res. Part II Top. Stud. Oceanogr. 57, 71–85 (2010).
Nguyen, H. M. Spatial and temporal variations in benthic bivalve stable isotopic composition in the Chukchi Sea, Arctic Ocean. (University of Texas at Austin, 2017).
Lowry, L. F. & Fay, F. H. Seal eating by walruses in the Bering and Chukchi Seas. Polar Biol. 3, 11–18 (1984).
Acknowledgements
Thanks to the Alaska Native subsistence hunters of St. Lawrence Island, Eskimo Walrus Commission, USFWS, North Slope Borough Department of Wildlife Management, Alaska Department of Fish and Game, University of Alaska Museum, National Museum of Natural History (Smithsonian Institution), and the Alaska Stable Isotope Facility. Many thanks to B. Konar, P. Lemons, and K. Severin for providing input on this research, and to P. Charapata and H. Starbuck for field and laboratory support. This work was funded by the National Science Foundation Arctic SEES Program, Grant No. 1263848, with supplementary funds from the Bureau of Ocean Energy Management. Additional funding was provided by the North Pacific Research Board, and the National Institutes of Health Biomedical Learning and Student Training Program at the University of Alaska Fairbanks. This publication is the result in part of research sponsored by the Cooperative Institute for Alaska Research with funds from the National Oceanic and Atmospheric Administration under cooperative agreement NA13OAR4320056 with the University of Alaska. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Numbers UL1GM118991, TL4GM118992, or RL5GM118990. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The University of Alaska is an AA/EO employer and educational institution and prohibits illegal discrimination against any individual.
Author information
Authors and Affiliations
Contributions
C.T.C., L.H. and N.M. conceived and designed the study. C.T.C. performed the experiments, analyzed the data, and wrote the manuscript; L.H. and N.M. provided editorial advice.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Clark, C.T., Horstmann, L. & Misarti, N. Lipid normalization and stable isotope discrimination in Pacific walrus tissues. Sci Rep 9, 5843 (2019). https://doi.org/10.1038/s41598-019-42095-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-42095-z
This article is cited by
-
Variability in nitrogen-derived trophic levels of Arctic marine biota
Polar Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.