Abstract
Nitrogen (N) and carbon(C) metabolisms in plants were investigated to assess different responses of Bt and non-Bt rice to different N treatments. T2A-1 (Bt rice variety) inserted with Cry2A* protein to resist Lepidoptera and its parental line MH63 was adopted in this study. The total N accumulation presented no statistical difference. But nitrogen contents in different parts of rice plant were significantly different between the two lines, especially on leaf and spike part. This study revealed that the nitrogen in leaf of T2A-1 was far more than that of MH63; however, the nitrogen in spike of T2A-1 was less than that of MH63. In addition, MH63 assimilated more carbon than T2A-1. However, the distribution proportion of carbon in leaf, stem and spike of T2A-1 and MH63 were both 1:1:1. What’s more, our study of the difference in metabolism pathway based on proteomics analysis provided more insights on the responses of two lines of Bt and non-Bt rice to different N treatments. And amino acid metabolism, energy metabolism, and carbohydrate metabolism presented significant difference between two lines. In addition, the number of differentially expressed proteins with N deficiency treatment was almost twice as many as that with normal N treatment. It could be inferred that the insertion of Cry2A* in T2A-1 may bring about effects on carbon and nitrogen allocation and related metabolisms, especially under N deficiency environment.
Similar content being viewed by others
Introduction
Since rice yield suffers great loss due to lepidopteran pests1, scientists worked on possible control measures including developing transgenic Bt rice in the last few decades2,3,4,5,6,7. Cultivation area of transgenic Bt crop was up to 98.5 million hectares worldwide in 20168. In recent years, planting Bt crops has been widely recognized as a pest prevention measure. Although many Bt genes have been found, only a few of them were selected for developing transgenic crops. Cry2A protein was reported to have different binding regions in brush border membrane vesicles and to present no cross-resistance9,10,11,12. Cry2A gene could be applied to develop marker-free Bt transgenic rice as a resistance source of gene pyramid. As a result, the evolution of insects’ resistance to toxicity of Bt rice would be postponed. Thus, Cry2A was considered as an important protein for bio-breeding scientists. However, several previous studies demonstrated the differences in the agronomy traits and physiological metabolism in some rice lines inserted by exogenous insect-resistant genes13,14,15,16,17,18,19. These differences included reduced plant heights and root lengths14, fewer grains per panicle17, decreased setting rates5,14,17,18,20, and so on, which in turn led to reductions in grain yield. The obvious advantage of Bt rice in pest resistance does not balance out its unintended effects resulting in lower yield21. These unintended effects brought by transgenic crop were supposed to have three sources: the disruption of endogenous genetic background22, the somaclonal variation happened during tissue culture processes23, and “bio-burden” brought by additional transgenic protein synthesis process24. “Bio-burden” was described as burden on material and energy, which could influence N and C metabolisms in plant.
The N amount demanded by plants is highest among all mineral nutrients. And it was one of the most important elements influencing crop yield directly. Generally, with the increase of N application, more Bt protein in plant tissues could be detected20,25,26. According to Bruns and Abel25, there was no significant difference in the total N uptake between Bt maize and their non-Bt counterparts. However, some other Bt hybrid maize strains were demonstrated to have lower N accumulation in grains but higher N accumulation in straws compared with their non-Bt control lines27. Moreover, Pioneer 38W36Bt, a Bt hybrid maize, accumulated more N in its kernels and entire maize plant than its conventional control Pioneer 38W36. But the two maize lines showed similar N and leaf chlorophyll contents at the late growth stages28. Besides, Bt cotton was reported to have higher N content and N metabolism enzymes than its non-Bt counterpart29. While, relatively less reports have been made about N metabolism in Bt rice. Thus, studies on responses of Bt rice lines to different N treatments are of great necessity.
Gurr and Rushton (2005) proposed that the difference between transgenic and non-transgenic lines might have been brought about by extra consumption of material and energy to maintain exogenous metabolism in plants24. Based on their hypothesis, C and N metabolism in transgenic plants were likely to change compared with their non-transgenic counterparts. C and N contents and ratio of C and N were assumed to be the indicators representing C and N metabolism in plants30. Therefore, whether the distribution and allocation of C and N in transgenic Cry2A* rice are different from those of its parent line could provide essential evidence for Bt rice breeding. This study was aimed to (1) explore the distribution and allocation of C and N in transgenic Cry2A* rice (T2A-1) and its conventional (non-transgenic) parent line MH63, a widely used indica cytoplasm male sterile (CMS) restorer rice line in Asia5; (2) attempt to discover the differences of proteins expression related to C and N metabolism under different N treatments combining physiological indicators of C and N metabolism with proteomics analysis.
Results
Biomass accumulation and distribution in Bt (T2A-1) and non-Bt rice
Significant differences in dry matter of each part including stem, leaf and spike were observed between plants of MH63 and T2A-1(Table 1) under different nitrogen treatments. And dry matter of each part increased with the addition of nitrogen fertilizer under several treatments. Meanwhile, the total biomass of the two lines showed no significant difference under each N application treatment. For example, at the FS stage, the dry weight of the stem of T2A-1 was 21.2% lower than that of MH63 under N1 treatment, and the biomass of the spike of T2A-1 were 31.4% and 30.7% lower than that of MH63 under N3 and N4 treatments, respectively. What’s more, on day 15 after FS, similar results were observed in spike under N2, N3 and N4 treatments, while the opposite results were observed in leaf under the same treatments.
Carbon accumulation and distribution in Bt (T2A-1) and non-Bt rice
At two sampling stages, significant difference in carbon content of each part including stem, leaf and spike was found between the T2A-1 and MH63, and it was true with the total carbon content of the aboveground (Table 2). While, in flowering stage (FS), the mean carbon accumulation of the aboveground in T2A-1 was 610.6 mg under the four treatments N1–4, which was 221.6 mg lower than that of MH63. On day 15 after FS, this difference reached up to 447.9 mg. However, T2A-1 was similar to MH63 in the distribution of carbon in three parts (leaf, stem, spike) of the plant with the distribution ratio being 1:1:1.
Nitrogen accumulation and distribution in Bt (T2A-1) and non-Bt rice
There was no significant difference in nitrogen assimilation between Bt (T2A-1) and non-Bt rice. However, the distribution of nitrogen in leaf, stem and spike presented several differences at the two sampling times respectively on FS and day 15 after FS, especially on leaf and spike part (Table 3). To be more specific, mean value of nitrogen concentration in leaf of T2A-1 were 6.41% and 6.35% higher than that of MH63 respectively (Table 4). While the mean value in spike of T2A-1 were 4.74% and 8.38% lower than that of MH63. And the nitrogen content in each part of both lines T2A-1 and MH63 exhibited the same ranking, namely, leaf > stem > spike at FS. Whereas 15 days after FS, the ranking of the two lines was changed into stem > leaf > spike.
C/N ratio in different part of Bt (T2A-1) and non-Bt rice
Under the four nitrogen treatments, C/N ratio in different parts of T2A-1 and MH63 presented significant difference (Table 5). At both sampling time, i.e., FS and day 15 after FS, C/N ratio was the highest in stem, and the lowest in leaf. Furthermore, C/N ratio of T2A-1 was significantly lower under N4 treatment than under other N treatments. And the C/N ratio of MH63 didn’t show obvious difference under its four treatments. At FS, C/N ratio in stem of T2A-1 is higher under N1 treatment than under N2 and N3 treatments. While, on day 15 after FS, there was no difference on C/N ratio under all treatments N1–4 of T2A-1. And C/N ratio in leaf and spike was the lowest under N4 treatment, which was significantly lower than that under N1 treatment. The C/N ratio of whole aboveground part of T2A-1 was significantly lower than that of MH63, and so was the C/N ratio in leaf.
Differentially expressed proteins related to C and N metabolism in leaves of MH63 and T2A-1 by iTRAQ
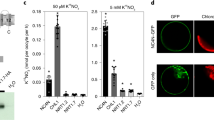
Flag leaves of MH63 and T2A-1 under RN(1 g N·plant−1) and N0 (0 g N·plant−1) treatments were collected for proteomics analysis in iTRAQ (isobaric tags for relative and absolute quantification) method. The analysis results indicated that of a total of 6040 proteins identified in the experiment, 206 proteins were up-regulated and 315 proteins were down-regulated under RN treatment (Fig. 1). While 320 up-regulated and 217 down-regulated proteins were identified in the leaves of two rice lines at N0 level. In addition, the number of those differentially expressed proteins related to C and N metabolism under the two N treatments were 55 and 61 (Tables 6 and 7), occupying 17.2% and 28.1% of the total difference at RN and N0 levels respectively, and the KEGG analysis of those differentially expressed proteins were shown in Fig. 2.
The differentially depressed proteins were devoted to energy metabolism, carbohydrate and lipid metabolism, nucleotide metabolism, amino acid metabolism, glycan biosynthesis and metabolism, terpenoids and polyketides metabolism, and other secondary metabolites biosynthesis, all of which belonged to C and N metabolism. The concerning differentially expressed proteins were listed in Tables 6 and 7.
Discussion
This study found no significant difference in total biomass between T2A-1 and MH63 under different N treatments, indicating that the two lines had similar response to biomass assimilation. However, Bt rice line T2A-1 presented higher dry matter content in leaves and lower content in spike compared with its counterpart MH63. Previous studies reported other differences in agronomy traits. For example, lower yield caused by lower setting rates was also reported in several Bt rice lines introduced with a Cry2A*31, Cry1C*20, Cry1Ab17, Cry1Ac14, Xa214 or Bt/CpTI18 gene. And shorter plant heights and root lengths14, fewer grains per panicle17 and lower setting rates5,14,17,18,20 have also been reported in recent years. These differences in agronomy traits could not be ignored for optimizing cultivation of transgenic crops.
Besides, the results of accumulation and distribution of N examination were similar to those of biomass in the two lines, T2A-1 and MH63. N content in spike of T2A-1 was lower than that in spike of MH63, potentially resulting in the weight loss of spike of T2A-1. The experiment with maize indicated that lower one in grains but higher N accumulation in stovers were found in several Bt hybrid maize strains, compared with their non-Bt counterparts27. And more N in kernels was detected in Pioneer 38W36Bt, a Bt hybrid maize, compared with the control Pioneer 38W36 (non-Bt), while N concentration and leaf chlorophyll content at the silking and maturity stages in Bt hybrid maize were similar to that in control group28. Bt cotton was reported to have more active N metabolism in the vegetative stage than its non-Bt counterpart, resulting in a reduction in boll size29. It could be concluded that there existed differences in N metabolism between Bt crops and their non-Bt crops including rice, maize and cotton.
Additionally, the significant differences in the accumulation of C between the two lines under several N treatments were also observed. But C distribution ratios in plant parts of the two lines were the same, i.e., 1:1:1. Proteomics analysis results revealed that the greatest difference in the number of differentially expressed proteins related to carbohydrate metabolism was observed between RN and N0 treatment. As we know, C metabolism provides material and energy for all metabolism processes in plant. Thus, changes on C metabolism would have enormous influence on plant growth and development. Our proteomics analysis results obtained by iTRAQ method exhibited that differentially expressed proteins mainly functioned in energy metabolism, carbohydrate metabolism, and so on. Previous reports on Bt cotton revealed that differentially expressed proteins identified between transgenic Bt + CpTI cotton SGK321 and its counterpart SY321 accounted for approximate 10% of all proteins identified by using proteomics analysis. These differentially expressed proteins mainly involved in carbon fixation and photosynthesis, glyoxylate and dicarboxylate metabolism pathway, oxidative pentose phosphate pathway32. 2-D PAGE experiment by Ren et al. (2009) reported the detection of 102 differentially expressed proteins in 12 transgenic Arabidopsis, compared with their wild-type (ecotype Col-o)33. Their experiment also reported that most of these differentially expressed proteins were involved in energy transfer, oxidative respiration, and photosynthesis. In addition, Gong et al. used comparative proteomics approaches to determine proteome differences in seeds between 2 transgenic rice lines and their corresponding control groups, namely, D68 and MH8634. Mass spectrometry analysis exhibited that differentially expressed proteins participated in several cellular and metabolic processes, including protein synthesis and transportation, and defense response. And differentially expressed proteins were also detected in seeds of transgenic and non-transgenic soybean by comparative proteomics approach35. Therefore, it can be concluded that changes in C metabolism were also widely found in transgenic plants.
It is well known that C metabolism and N metabolism are always intertwined in plant36. Up to 55% of net plant carbon was devoted to nitrogen assimilation and metabolism in some tissues37. Carbon and nitrogen metabolism are intimately linked. This study revealed different C/N ratios in leaf between T2A-1 and MH63. The C/N ratio in whole aboveground part of T2A-1 was significantly lower than that of MH63. C/N ratio has influence on glucose metabolism, assimilate transportation, senescence process of leaf, etc. Thus, as plant material and energy suppliers, C and N metabolism ought to be taken into consideration simultaneously in their function assessment.
Until now, there were three hypotheses of unintended effects brought by transgenic technology. But insertion position of transgenic rice line T2A-1 used in our study was noncoding region, and it was chosen to exhibit the best field resistance and excellent agronomy traits by several field experiment among 102 transgenic rice lines6. Thus, we are inclined to attribute these effects to bio-burden caused by additional Bt protein synthesis. As a soluble protein, Bt protein would consume extra N and energy in Bt crops. And C and N metabolism were assumed as basic processes regulating N allocation and energy distribution in plant. Thus, it has drawn a wide concern whether the Bt protein synthesis process will influence the fundamental C and N metabolism in Bt crops resulting in a possible “bio-burden” affecting plant growth and development18,24,38,39. Poor adaptabilities to nutrient deficiency and several abiotic stresses were reported in some Bt cotton lines in previous studies. For example, experiment conducted by Wei et al. revealed that root activity (less root organic acid exudation) in several Bt cotton lines under low N supply condition was lower than that in their non-Bt wild types40. And lower biomass was also observed in some Bt cotton lines under low K application condition than their non-Bt counterparts41. Besides, some Bt cotton lines were reported to have suffered more from pathogens stress42 and CeO2 nanoparticles stress43 than their non-Bt wild types. Another experiment by Li et al. (2015) indicated that Bt cottons adapted poorly to drought stress or elevated O3 stress42. And inhibited nitrogen metabolism by salinity, waterlogging and the combined stress was observed to result in the decline of Bt protein expression, causing bollworm control reduction44.
Based on the discussion above, it could be concluded that the phenotypic and yield changes of Bt crops under different conditions were frequently occurring phenomena. However, the mechanism of such changes has not been known well until now. Our study revealed that differences in C and N partitioning could be one of possible reasons accounting for changes in yield and phenotypic traits in T2A-1. Future study could make a further exploration of energy utilization related to C and N metabolism under the framework of the “bio-burden” hypothesis comparing more transgenic plants and their wild types.
Materials and Methods
Materials
Bt rice line T2A-1, expressing Cry2A* protein, was used for the assessment of C and N metabolism processes in the plant. And its conventional (non-Bt) parent line MH63, an excellent three-line rice restorer line, served as a control in this study. Both of the two rice lines were provided by National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University. During the transformation process, a series of Cry2A* rice lines were obtained from the modification of the Cry2A* gene with no amino acid sequence increased. Besides, the insertion position of Cry2A* in T2A-1 was a noncoding region, which theoretically had few effects on exogenous gene expression. And T2A-1 was found to exhibit the best field resistance and excellent agronomy traits, therefore it was selected for the experiment among 102 independent transformants obtained6.
Soils
Considering its low nitrogen content, loess soil was chosen for this nitrogen concentration gradient experiment at the Agricultural Experiment Station of Huazhong Agriculture University, Wuhan, China. The soil was first air-dried in open air environment at normal temperature, and then ground, finally mixed with sand before use. The soil contained 18.48 g kg−1 of organic matter, 0.11 g kg−1 of total N, and 1.19 g kg−1 of C with a pH being 6.56.
Experimental Design
The nitrogen gradient experiment using T2A-1 and MH63 was carried out from May to September 2016 in net house. Four nitrogen fertilizer treatments were performed in this study, namely, supplying 0.2 g (N1 treatment), 0.35 g (N2 treatment), 0.5 g (N3 treatment), and 1 g (N4 treatment) nitrogen, respectively, in the whole growing period of rice. Nitrogen fertilizer in form of urea was applied three times during the whole developing period with applying ratio being 50% as base fertilizer, 30% at mid-tillering stage, and 20% at young panicle differentiation stage, respectively. In addition, potassium and phosphorus fertilizers were applied in a common way. Seedlings were transplanted to PVC pipes (16 cm × 55 cm). One seedling was planted into each pipe, then three pipes were placed into one bucket as a group. And there were three buckets as three replicates for each nitrogen fertilizer treatment using T2A-1 and MH63. These treatments were arranged in a completely random arrangement. One whole plant would be sampled and separated into leaf, stem and spike part for C and N content measurement at each sampling time. And two sampling times were set during reproductive periods including the flowering stage (FS), and 15 days after the flowering, named as FS, and 15 d after FS, respectively.
In addition, to explore the different expression of proteins related to C and N metabolism between MH63 and T2A-1, the other experiment concluding two nitrogen treatments was conducted for proteomics detection at the same time in the same screen house. And 0 g (N0 treatment) and 1 g (RN treatment) nitrogen fertilizer application treatments during the whole rice growth period were chosen to enlarge the difference between the two lines. Flag leaves were collected on day 15 after the flowering for proteomics testing.
Total Carbon and Nitrogen Measurement
Leaf, stem and spike parts were separated from one sampling plant. All samples were oven-dried at 70 °C to constant weight before testing, and the final content of C and N was obtained through the formula that content percentage measured by elemental analyzer (Vario ISOTOPE, Elementar) multiplied dry weight of each sample. Nitrogen accumulation equated to the sum of N content of leaf, stem with sheath, and spike.
Procedure of iTRAQ (isobaric tags for relative and absolute quantifica)Protein Extraction
The samples were frozen in liquid nitrogen and ground with pestle and mortar. Five times volume of TCA/acetone (1:9) was added to the powder and mixed by vortex. The mixture was placed at −20 °C for 4 h, and centrifuged at 6000 g for 40 min at 4 °C. The supernatant was discarded. The pre-cooling acetone was added and washed for three times. The precipitate was air dried. Then, 30 times volume of SDT buffer was added to 20–30 mg powder. After mixing and 5 min boiling, the lysate was sonicated and then boiled for another 15 min. Followed by being centrifuged at 14000 g for 40 min, the supernatant was filtered with 0.22 µm filters. The filtrate was quantified with the BCA Protein Assay Kit (Bio-Rad, USA). The sample was stored at −80 °C.
Protein preparation, digestion and iTRAQ labelling
200 μg of proteins from each sample were incorporated into 30 μl SDT buffer (4% SDS, 100 mM DTT, 150 mM Tris-HCl pH 8.0). The detergent, DTT, and other low-molecular-weight components were removed by using UA buffer (8 M Urea, 150 mM Tris-HCl pH 8.0) through repeated ultrafiltration (Microcon units, 10 kD). Then 100 μl of iodoacetamide (100 mM IAA in UA buffer) was added to block reduced cysteine residues and the samples were incubated for 30 min in darkness. The filters were washed first with 100 μl UA buffer three times and then with 100 μl of Dissolution buffer (DS buffer) twice. Finally, the protein suspensions were digested with 4 μg of trypsin (Promega) in 40 μl of DS buffer overnight at 37 °C, and the obtained peptides were collected as a filtrate. The peptides from each sample were desalted on C18 Cartridges (Empore™ SPE Cartridges C18 (standard density), bed I.D. 7 mm, volume 3 ml, Sigma), concentrated by vacuum centrifugation and reconstituted in 40 µl of 0.1% (v/v) formic acid. The peptide content was estimated by UV light spectral density at 280 nm using an extinctions coefficient of 1.1 of 0.1% (g/l) solution that was calculated on the basis of the frequency of tryptophan and tyrosine in vertebrate proteins.
An eight-plex iTRAQ was set for the proteomics analysis, including N and N0 (Table 8). And 1 and 2 represented the replicates. Each replicate entailed 4 biological sampling replicates. 100 μg of peptide mixture from each sample was labelled by using iTRAQ reagent according to the manufacturer’s instructions (Applied Biosystems).
Peptide Fractionation with Strong Cation Exchange (SCX) Chromatography
iTRAQ-labelled peptides were fractionated by SCX chromatography using the AKTA Purifier system (GE Healthcare). The dried peptide mixture was reconstituted and acidified with buffer A (10 mM KH2PO4 in 25% of ACN, pH 3.0) and loaded onto a PolySULFOETHYL 4.6 × 100 mm column (5 µm, 200 Å, PolyLC Inc, Maryland, U.S.A.). The peptides were eluted at a flow rate of 1 ml/min with a gradient of 0–8% buffer B (500 mM KCl, 10 mM KH2PO4 in 25% of ACN, pH 3.0) for 22 min, 8–52% buffer B from 22 min to 47 min, 52–100% buffer B from 47 min to 50 min, 100% buffer B from 50 min to 58 min, and buffer B was reset to 0% after 58 min. The elution was monitored by absorbance at 214 nm, and fractions were collected every 1 min. The collected fractions were desalted on C18 Cartridges (Empore™ SPE Cartridges C18 (standard density), bed I.D. 7 mm, volume 3 ml, Sigma) and concentrated by vacuum centrifugation.
LC-MS/MS Analysis by Q Exactive
LC-MS/MS analysis was performed on a Q Exactive mass spectrometer (Thermo Scientific) that was coupled to Easy nLC (Proxeon Biosystems, now Thermo Fisher Scientific) for 60 min. The mass spectrometer was operated in positive ion mode. MS data was acquired by using a data-dependent top10 method dynamically choosing the most abundant precursor ions from the survey scan (300–1800 m/z) for HCD fragmentation. Automatic gain control (AGC) target was set to 3e6, and maximum inject time to 10 ms. Dynamic exclusion duration was 40.0 s. Survey scans were acquired at a resolution of 70,000 at m/z 200 and the resolution for HCD spectra was set to 17,500 at m/z 200, and isolation width was 2 m/z. Normalized collision energy was 30 eV and the underfill ratio, which specifies the minimum percentage of the target value likely to be reached at maximum fill time, was defined as 0.1%. The instrument was run with peptide recognition mode enabled.
Data analysis
SAS 9.1 software (SAS Institute, Inc., Cary, NC, USA) was used for the analysis of variance (ANOVA) of data referring to C and N distribution in this experiment. And these data are expressed as the means ± standard deviation (SD, n = 3). Differences of the means were statistically significant at α = 0.05.
Besides, to conduct proteomics analysis, MS/MS spectra were searched using MASCOT engine (Matrix Science, London, UK; version 2.2) embedded into Proteome Discoverer 1.4. Parameters were set as shown in Table 9. Database of uniprot _Oryza_sativa_168274 _ 20170123.fasta (http://www.uniprot.org) was used for proteomics analysis. All data related to this study has been public available on iProX (www.iprox.com) with ID IPX0001090000 (http://www.iprox.org/page/PDV014.html?projectId=IPX0001090000).
References
Pandi, V., Sundara, Babu, P. C. & Kailasam, C. Prediction of damage and yield caused by rice leaffolder at different crop Periods in a susceptible rice cultivar (IR50). J. Appl. Entomol. 122, 595–599 (2009).
Fujimoto, H., Itoh, K., Yamamoto, M., Kyozuka, J. & Shimamoto, K. Insect resistant rice generated by introduction of a modified δ-endotoxin gene of Bacillus thuringiensis. Nat. Biotechnol. 11, 1151–1155 (1993).
Tu, J. M. et al. Expression and function of a hybrid Bt toxin gene in transgenic rice conferring resistance to insect pest. Plant Biotechnol. 15, 195–203 (1998).
Tu, J. et al. Field performance of transgenic elite commercial hybrid rice expressing Bacillus thuringiensis δ-endotoxin. Nat. Biotechnol. 18, 1101–1104 (2000a).
Tu, J., Datta, K., Khush, G. S., Zhang, Q. & Datta, S. K. Field performance of Xa21 transgenic indica rice (Oryza sativa L.), IR72. Theor. Appl. Genet. 101, 15–20 (2000b).
Chen, H. et al. Transgenic indica rice plants harboring a synthetic cry2A* gene of Bacillus thuringiensis exhibit enhanced resistance against lepidopteran rice pests. Theor. Appl. Genet. 111, 1330–1337 (2005).
Tang, W. et al. Development of insect-resistant transgenic indica rice with a synthetic cry1C* gene. Mol. Breeding 18, 1–10 (2006).
Clive, J. Global status of commercialized biotech/GM crops: ISAAA Brief 52 (2016).
Lee, M. K., Aguda, R., Cohen, M., Gould, F. & Dean, D. (1997): Determination of Binding of Bacillus thuringiensis (delta)-Endotoxin Receptors to Rice Stem Borer Midguts. Appl. Environ. Microb. 63, 1453–1459 (2016).
Karim, S. & Dean, D. Toxicity and Receptor Binding Properties of Bacillus thuringiensis δ-Endotoxins to the Midgut Brush Border Membrane Vesicles of the Rice Leaf Folders, Cnaphalocrocis medinalis and Marasmia patnalis. Curr. Microbiol. 41, 276–283 (2000).
Alcantara, E., Aguda, R., Curtiss, A., Dean, D. & Cohen, M. Bacillus thuringiensis δ-endotoxin binding to brush border membrane vesicles of rice stem borers. Arch. Insect Biochem. Physiol. 55, 169–177 (2004).
Gouffon, C., Van Vliet, A., Van Rie, J., Jansens, S. & Jurat-Fuentes, J. Binding sites for Bacillus thuringiensis Cry2Ae toxin on heliothine brush border membrane vesicles are not shared with Cry1A, Cry1F, or Vip3A toxin. Appl. Environ. Microb. 77, 3182–3188 (2011).
Lynh, P., Jones, J. & Blackhall, N. The phenotypic characterization of R2 generation transgenic rice plants under field and glasshouse conditions. Euhytica 85, 395–401 (2005).
Shu, Q. et al. Agronomic and morphological characterization of Agrobacterium-transformed Bt rice plants. Euphytica 127, 345–352 (2002).
Jiang, G. et al. Pyramiding of insect-and disease-resistance genes into an elite indica, cytoplasm sterile restorer line of rice, “Minghui63”. Plant Breeding 123, 112–116 (2004).
Chen, L., Snow, A., Wang, F. & Lu, B. Effects of insect-resistance genes on fecundity in rice (Oryza Sativa, Poaceae): a test for underlying cost. Am. J. Bot. 93, 94–101 (2006).
Kim, S. et al. Inheritance and field performance of transgenic Korean Bt rice lines resistant to rice yellow stem borer. Euphytica 164, 829–839 (2008).
Xia, H., Chen, L., Wang, F. & Lu, B. R. Yield benefit and underlying cost of insect-resistance transgenic rice: implication in breeding and deploying transgenic crops. Field Crop. Res. 118, 215–220 (2010).
Wang, F. et al. Yield differences between Bt transgenic rice lines and their non-Bt counterparts, and its possible mechanism. Field Crop. Res. 74, 255–262 (2011).
Wang, F. et al. Yield differences between Bt transgenic rice lines and their non-Bt counterparts, and its possible mechanism. Field Crop. Res. 126, 8–15 (2012).
Jiang, Y. et al. Non-target effects of Bt transgenes on grain yield and related traits of an elite restorer rice line in response to nitrogen and potassium applications. Field Crop. Res. 169, 39–48 (2014).
Feldmann, K. A., Marks, M. D., Christianson, M. L. & Quatrano, R. S. A dwarf mutant of Arabidopsis generated by T-DNA insertion mutagenesis. Science 243, 1351–1354 (1989).
Larkin, P. J. & Scowcroft, W. R. Somaclonal variation-a novel source of variability from cell cultures for plant improvement. Theor. Appl. Genet. 60, 197–214 (1981).
Gurr, S. & Rushton, P. Engineering plants with increased disease resistance: what are we going to express? Trends Biotechnol. 23, 275–282 (2005).
Bruns, H. & Abel, C. Nitrogen fertility effects on Bt-endotoxin and nitrogen concentrations of maize during early growth. Agron. J. 95, 207–211 (2003).
Dong, H. & Li, W. Variability of endotoxin expression in Bt transgenic cotton. J. Agron. Crop Sci. 193, 21–29 (2007).
Ma, B. & Subedi, K. Development, yield, grain moisture and nitrogen uptake of Bt corn hybrids and their conventional near-isolines. Field Crop. Res. 93, 199–211 (2005).
Subedi, K. & Ma, B. Dry matter and nitrogen partitioning patterns in Bt and non-Bt near-isoline maize hybrids. Crop Sci. 47, 1186–1192 (2007).
Chen, D., Ye, G., Yang, C., Chen, Y. & Wu, Y. Effect after introducing Bacillus thuringiensis gene on nitrogen metabolism in cotton. Field Crop. Res. 87, 235–244 (2004).
Martin, M. E. & Aber, J. High-spectral resolution remote sensing of forest canopy ligin, nitrogen and ecosystem process. Ecol. Appl. 7, 431–443 (1997).
Jiang, Y. et al. Yield changes of Bt-MH63 with cry1C* or cry2A* genes compared with MH63 (Oryza sativa) under different nitrogen levels. Field Crop. Res. 151, 101–106 (2013).
Wang, L. et al. Comparative proteomics of Bt-transgenic and non-transgenic cotton leaves. Proteome Sci. 13, 15 (2015).
Ren, Y. et al. A comparative proteomics approach to detect unintended effects in transgenic Arabidopsis. J. Genet. Genomics 36, 629–39 (2009).
Gong, C., Li, Q., Yu, H., Wang, Z. & Wang, T. Proteomics insight into the biological safety of transgenic modification of rice as compared with conventional genetic breeding and spontaneous genotypic variation. J. Proteome Res. 11, 3019–3029 (2012).
Brandao, A., Barbosa, H. & Arruda, M. Image analysis of two-dimensional gel electrophoresis for comparative proteomics of transgenic and non-transgenic soybean seeds. J. Proteomics 73, 1433–1440 (2010).
Kumar P. A., Polisetty R. & Abrol Y. P. Interaction between Carbon and Nitrogen Metabolism. In: Abrol Y. P., Mohanty P., Govindjee (eds) Photosynthesis: Photoreactions to Plant Productivity. Springer, Dordrecht (1993).
Huppe, H. & Turpin, D. Integration of carbon and nitrogen metabolism in plant and algal cells. Annu. Rev. Plant Boil. 45, 577–607 (1994).
Gahakwa, D. et al. Transgenic rice as a system to study the stability of transgene expression: multiple heterologous transgenes show similar behaviour in diverse genetic backgrounds. Theor. Appl. Genet. 101, 388–399 (2000).
Marrelli, M. T., Moreira, C. K., Kelly, D., Alphey, L. & Jacobs-Lorena, M. Mosquito transgenesis: what is the fitness cost? Trends Parasitol. 22, 197–202 (2006).
Wei, D. Y., Wei, M. S., Bao, H. L. & Zhang, M. Overexpression of a foreign Bt gene in cotton affects the low-molecular-weight components in root exudates. Pedosphere 17, 324–330 (2007).
Zhang, Z. et al. Differential responses of conventional and Bt-transgenic cotton to potassium deficiency. J. Plant Nutr. 30, 659–670 (2007).
Li, X., Ding, C., Wang, X. & Liu, B. Comparison of the physiological characteristics of transgenic insect-resistant cotton and conventional lines. Sci. Rep-UK. 5, 8739 (2015).
Ma, C. et al. Phytotoxic mechanism of nanoparticles: destruction of chloroplasts and vascular bundles and alteration of nutrient absorption. Sci. Rep-UK. 5, 116–118 (2015).
Luo, Z., Dong, H., Li, W., Ming, Z. & Zhu, Y. Individual and combined effects of salinity and waterlogging on Cry1Ac expression and insecticidal efficacy of Bt cotton. Crop Prot. 27, 1485–1490 (2008).
Acknowledgements
This work was supported by the State Key Special Program (No. 2017YFD0301400), the National Natural Science Foundation of China (No. 31701359), the Fundamental Research Funds for the Central Universities (No. 2662017JC007) and China Postdoctoral Science Foundation (No. 2017M612477). We express our gratitude to the National Key Laboratory of Crop Genetic Improvement laboratory for providing the Bt rice lines used in this study. Many thanks also to Mr. Jin for managing our pot experiment.
Author information
Authors and Affiliations
Contributions
L.L., Y.J., M.C. and C.C. designed the experiments. L.L., X.L. and K.W. performed the experiments, collected and analyzed the data. L.L. and Y.J. wrote the manuscript text.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ling, L., Li, X., Wang, K. et al. Carbon and nitrogen partitioning of transgenic rice T2A-1 (Cry2A*) with different nitrogen treatments. Sci Rep 9, 5351 (2019). https://doi.org/10.1038/s41598-019-41267-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-41267-1
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.