Abstract
We investigated the role of the resistance-nodulation-cell division superfamily (RND) efflux system on intrinsic multidrug resistance in Serratia marcescens. We identified eight putative RND efflux system genes in the S. marcescens Db10 genome that included the previously characterized systems, sdeXY, sdeAB, and sdeCDE. Six out of the eight genes conferred multidrug resistance on KAM32, a drug hypersensitive strain of Escherichia coli. Five out of the eight genes conferred resistance to benzalkonium, suggesting the importance of RND efflux systems in biocide resistance in S. marcescens. The energy-dependent efflux activities of five of the pumps were examined using a rhodamine 6 G efflux assay. When expressed in the tolC-deficient strain of E. coli, KAM43, none of the genes conferred resistance on E. coli. When hasF, encoding the S. marcescens TolC ortholog, was expressed in KAM43, all of the genes conferred resistance on E. coli, suggesting that HasF is a major outer membrane protein that is used by all RND efflux systems in this organism. We constructed a sdeXY deletion mutant from a derivative strain of the clinically isolated multidrug-resistant S. marcescens strain and found that the sdeXY deletion mutant was sensitive to a broad spectrum of antimicrobial agents.
Similar content being viewed by others
Introduction
Serratia marcescens, a Gram-negative bacilli, is widely distributed in the environment. Although not initially regarded as a pathogen, S. marcescens is associated with occasional hospital-related outbreaks. The treatment of S. marcescens infections with antimicrobial agents is becoming more challenging because clinically isolated strains that exhibit elevated resistance against β-lactams, aminoglycosides, and fluoroquinolones have been reported1,2.
Resistance-nodulation-cell division superfamily (RND) efflux systems play a major role in multidrug resistance in Gram-negative bacteria3,4,5,6,7,8,9. RND-type efflux systems consist of three components: the inner membrane protein (IMP), periplasmic membrane fusion protein (MFP), and outer membrane protein (OMP). The electrochemical potential of H+ across the cell membrane appears to be the driving force for drug efflux associated with RND efflux systems. Three RND efflux systems in S. marcescens, SdeXY10, SdeAB11, and SdeCDE11, have been characterized to date. SdeXY was the first multidrug efflux system to be characterized from S. marcescens, and showed broad substrate specificity when expressed and characterized in Escherichia coli10. The gene expression of sdeXY was up-regulated in a tigecycline-resistant clinically isolated strain of S. marcescens12. The gene inactivation of sdeXY from the environmentally isolated S. marcescens-type strain, NCTC10211, increased susceptibilities to tigecycline, tetracycline, ciprofloxacin, and cefpirome. SdeAB also showed broad substrate specificity when expressed in E. coli11. Gene knockout analyses revealed that SdeAB conferred intrinsic multidrug resistance to fluoroquinolones, chloramphenicol, novobiocin, sodium dodecyl sulphate (SDS), and ethidium bromide in S. marcescens13. In a separate study, a S. marcescens cetylpyridinium chloride mutant showed the up-regulated expression of sdeAB and also became resistance to fluoroquinolones, tetracycline, chloramphenicol, and benzalkonium chloride14. In contrast to SdeXY and SdeAB, SdeCDE did not exhibit broad substrate specificity and only conferred novobiocin resistance to S. marcescens15.
In the present study, we aimed to identify uncharacterized S. marcescens RND efflux systems that have the potential to render S. marcescens with multidrug resistance. To achieve this, we examined putative S. marcescens RND efflux system genes from S. marcescens Db10 and characterized their substrate specificities in drug-hypersensitive E. coli strain KAM3216. We identified an additional three uncharacterized RND efflux systems with broad substrate specificities. A gene deletion analysis revealed that SdeXY conferred intrinsic multidrug resistance to S. marcescens.
Results
Cloning of putative RND-type efflux pumps from S. marcescens
When we initiated this study, the S. marcescens Db11 genomic sequence database (http://www.sanger.ac.uk/resources/downloads/bacteria/serratia-marcescens.html) was the only publicly available resource for the genomic sequence of this bacterium. Using the S. marcescens Db11 genomic sequence, we searched for RND-type efflux systems in the S. marcescens Db11 genome and identified eight RND-type efflux systems (Fig. 1). These included three characterized S. marcescens RND efflux systems, SdeXY (SMA0370-0369)10, SdeAB (SMA1197-1196)11, and SdeCDE (SMA2945-2946-2947)11, and five putative RND-type efflux systems. We designated these putative RND efflux systems as shown in Fig. 1. The putative outer membrane protein (OMP) gene, omsA, was located adjacent to sdePQ. The other RND efflux systems did not contain the adjacent OMP gene. All of the RND efflux system genes contained the periplasmic membrane fusion protein (MFP) gene, except for sdeS. SdeCDE contained the two inner membrane protein (IMP) genes, sdeD and sdeE.
We performed a dendrogram analysis of entire sequences of IMPs from S. marcescens, E. coli, Vibrio parahaemolyticus, Vibrio cholerae, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa, and revealed that the IMPs from these organisms were divided into five groups (Fig. 2). In contrast to E. coli that does not have IMP in Group 2 or 3, each group contained at least one S. marcescens IMP, indicating that S. marcescens has a wide variety of RND efflux systems.
Unrooted phylogenetic tree of the inner membrane protein of RND efflux pumps. The phylogenetic tree was obtained using CLUSTALW (https://clustalw.ddbj.nig.ac.jp). E. coli: AcrB, AcrD, AcrF, MdtB, MdtC, MdtF, CusA22,33,45,46. V. parahaemolyticus: VmeB, VmeD, VmeF, VmeI, VmeK, VmeM, VmeO, VmeQ, VmeS, VmeW, VmeV, VmeZ3,4,5. V. cholerae: VexB, VexD, VexF, VexH, VexK, VexM29,31. S. marcescens: SdeB, SdeD, SdeE, SdeY, SdeH, SdeJ, SdeO, SdeQ, SdeS, SM39_1914, SM39_195810,11,12,13,14,15. A baumannii: AdeB, AdeE, AdeG, AdeJ17,18,20,26. P aeruginosa: MexD, MexF, MexI, MexK, MexN, MexQ, MexW, MexY, MuxB, MuxC, TriC21,24,27,28,30,32,34,35,36. K pneumoniae: AcrB(K.P.), KexD, OqxB, KexC(KPN_RS15040), KexF(KPN_RS19875), KexH(KPN_RS21805), KexK(KPN_RS11560), KexM(KPN_RS25535), KexS(KPN_RS04245), KexU(KPN_RS03035), KexW(KPN_RS13595), KexX(KPN_RS13600)19,23,47.
We cloned all S. marcescens RND efflux system genes from the S. marcescens Db10 strain, the parental strain of Db11, and expressed them in the drug-hypersensitive E. coli strain, KAM32, for further characterization.
Substrate specificities of S. marcescens RND efflux systems
To assess the substrate specificity of each RND efflux system, we measured the MICs of various antimicrobial agents using strains expressing each RND efflux system gene(s) in E. coli KAM32 (Table 1).
Many of the RND efflux systems in Group 1 play a major role in intrinsic multidrug resistance due to their broad substrate specificities3,10,17,18,19,20,21,22,23,24,25. Group 1 also contains the RND efflux system genes that are not in an operon with an MFP gene (e.g. acrD from E. coli). This type of RND efflux system generally exhibits narrow substrate specificities and SdeS only conferred resistance to erythromycin, novobiocin, SDS, and deoxycholate. Consistent with our previous findings10, the KAM32 strain expressing sdeXY conferred resistance to a broad spectrum of antimicrobial agents (Table 1). When sdePQ was expressed in the KAM32 strain with the adjacent OMP gene, omsA, multidrug resistance against several antimicrobial agents was conferred.
The RND efflux systems categorized into Group 2 have relatively broad substrate specificities and generally confer acquired resistance4,11,13,14,26,27,28. SdeB was categorized into this group and conferred multidrug resistance to E. coli KAM32; however, the substrate specificity of SdeAB was not as broad as those of SdePQ-OmsA and SdeXY (Table 1). The amino acid sequence of SdeB was similar to the P. aeruginosa RND efflux pump, MexF. Similar to MexEF27, fluoroquinolone and chloramphenicol were good substrates for SdeAB. Consistent with previous findings11, the KAM32 strain expressing sdeAB was resistant to the quaternary ammonium compound benzalkonium chloride.
The RND efflux systems in Group 3 have relatively broad substrate specificities and also confer acquired resistance4,29,30,31,32. Among S. marcescens RND IMPs, only SdeH was categorized into this group. The expression of sdeGH in E. coli KAM32 conferred multidrug resistance. The substrate specificity of SdeGH was broader than that of SdeAB, but was not similar to that of SdeXY or SdePQ-OmsA. Although SdeH showed amino acid sequence similarities to MexI and MexW, the substrate specificity of SdeGH was not similar to that of MexHI32 or MexVW30.
sdeCDE contained two IMP genes within its operon and SdeD and SdeE were both categorized into Group 4. As previously described15, novobiocin was the only substrate for SdeCDE (Table 1). To establish whether both of these IMP genes are required for this system, we constructed plasmids that expressed sdeCD or sdeCE and found that neither of these plasmids conferred novobiocin resistance in KAM32 (Table 1). This result indicated that SdeD and SdeE are both required for novobiocin resistance. This phenotype is similar to other RND efflux systems categorized into Group 4, such as MdtABCD from E. coli33 and MuxABC from P. aeruginosa34.
SdeJ and SdeO were categorized into Group 5. Although Group 5 contains many of the Vibrio RND efflux systems that show relatively wide substrate specificities4,5,29,31, the expression of sdeIJ conferred resistance to only benzalkonium and rhodamine 6 G, while sdeNO expression conferred resistance solely to SDS. TriC and MexK of P. aeruginosa were reported to contribute to the resistance of triclosan35,36. However, introduction of sdeIJ and sdeNO didn’t render triclosan resistance to host E. coli.
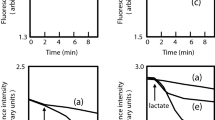
We measured the efflux of rhodamine 6 G to evaluate the activity of each RND efflux system because rhodamine 6 G is a good substrate for most of the S. marcescens RND efflux systems (SdeXY, SdeAB, SdeGH, SdeIJ, and SdePQ-OmsA) (Fig. 3). All of these five efflux systems showed higher rhodamine 6 G efflux activities when lactate was provided as the energy source, indicating that rhodamine 6 G efflux systems are energy-dependent.
Rhodamine 6 G efflux assay. Energy-starved cells of E. coli KAM32 strains that express S. marcescens RND efflux systems were prepared as described in the Materials and Methods. Energy-starved cells were resuspended in PBS containing 5 mM MgSO4 and 1 µM rhodamine 6 G. At the time point indicated by the arrow, 20 mM potassium lactate (K-Lactate) was added to energize cells. Intracellular rhodamine 6 G levels were monitored continuously by measuring the fluorescence of rhodamine 6 G at excitation and emission wavelengths of 529 and 553 nm, respectively. IJ; E. coli KAM32/pURS6 (sdeIJ), PQ; E. coli KAM32/pURS8 (sdePQ-omsA), XY; E. coli KAM32/pURS2 (sdeXY), GH; E. coli KAM32/pURS5 (sdeGH), AB; E. coli KAM32/pURS3 (sdeAB), control; E. coli KAM32/pUC19.
Requirement of OMP
Except for sdePQ-omsA, the other eight S. marcescens RND efflux systems did not contain the adjacent OMP gene. Thus, these RND efflux systems must rely on E. coli OMP(s) when expressed in E. coli. TolC is a major OMP in E. coli and all of the E. coli RND efflux systems require TolC for their activities37. To clarify whether S. marcescens RND efflux systems utilize TolC when expressed in E. coli, we introduced S. marcescens RND efflux system genes into E.coli KAM43, a tolC-deficient strain, and tested antimicrobial susceptibilities by measuring MICs. Except for sdePQ-omsA, none of the S. marcescens RND efflux systems showed increases in MICs when expressed in KAM43, indicating that S. marcescens RND efflux systems require TolC for their activities when expressed in E. coli (Table 2).
We then investigated whether SdePQ utilizes TolC as an OMP. We subcloned sdePQ and expressed it in E. coli KAM 32 and KAM43. When expressed in E. coli KAM32, sdePQ increased MICs for several antimicrobial agents, similar to E. coli KAM32 expressing sdePQ-omsA, except for SDS (Table 1). However, when expressed in KAM43, sdePQ didn’t increase MIC for any agents. These results indicated that SdePQ utilizes TolC when OmsA is absent; however, this interaction may be weaker than that with OmsA (Table 2).
S. marcescens is known to possess the functional ortholog of TolC, HasF. Previous studies showed that SdeAB and SdeXY utilized HasF as their OMP component12,13. Since TolC may be utilized by all S. marcescens RND efflux systems, we hypothesized that the other S. marcescens RND efflux systems also utilize HasF as the OMP. To examine this, the hasF gene was cloned and expressed with S. marcescens RND efflux systems in the KAM43 strain. Consistent with previous findings, sdeXY and sdeAB both increased MICs when expressed with hasF in E. coli KAM43 (Table 2). When expressed with hasF, all of the S. marcescens RND efflux systems showed increased MICs, indicating that they also utilized HasF as the OMP. Compared Table 1 with Table 2, SdeAB-HasF and SdeNO-HasF showed higher MICs than SdeAB-TolC and SdeNO-TolC, whereas SdeIJ-HasF showed lower MICs than SdeIJ-TolC. These results indicated that compatibility between IMP and/or MFP and OMP is important.
Introduction of sdeS in KAM43 or KAM43/pSOS2 didn’t render the increase for any tested antimicrobial agents (Table 2), but in KAM32, increase of MICs for novobiocin, SDS, deoxycholate was observed (Table 1). Since no increase of MICs was observed in KAM33, an acrA disruptant of KAM32, SdeS would utilize AcrA as MFP in E. coli (Supplementary Table S1).
The sdeXY deletion mutant of S. marcescens is susceptible to a broad range of antimicrobial agents
Since the present results indicated that SdeXY has the broadest substrate specificity among the characterized S. marcescens efflux pumps10, we constructed a sdeXY mutant strain from S. marcescens. We attempted to construct the deletion strain from Db10, but were unsuccessful for an unknown reason. Therefore, we used another strain KS24, a derivative of the clinically isolated strain of S. marcescens SM3938. The sdeXY deletion strain of KS24 became more sensitive to a broad spectrum of antimicrobial agents than the parental strain (Table 3). The sdeXY mutant strain also showed the decreased energy-dependent efflux of ethidium (Fig. 4). These results indicated that SdeXY is a major RND efflux pump that confers intrinsic resistance to S. marcescens against multiple antimicrobial agents.
Ethidium efflux activity in S. marcescens cells. The cells of the S. marcescens KS24 strain (A) and its KS24∆sdeXY (B) were prepared as described in the Materials and Methods. Ethidium bromide was added to cell suspensions at a final concentration of 10 μM at the time point indicated by the first downward arrow. Intracellular ethidium levels were monitored continuously by measuring the fluorescence of ethidium at excitation and emission wavelengths of 500 and 580 nm, respectively. At the second downward arrow, CCCP was added to the suspensions at a final concentration of 100 µM. Assays were performed at 37 °C.
Discussion
Previous studies suggested that RND efflux systems play a major role in multidrug resistance in S. marcescens10,11,12,13,14. Since Gram-negative bacteria have been suggested to possess ‘multiple’ and ‘active’ RND efflux systems3,4,5,6,7,8,9, we hypothesized that S. marcescens has other ‘active’ RND efflux systems. To investigate this, we cloned all of the putative RND efflux systems from the S. marcescens Db10 strain, characterized them in E. coli, and further identified “active” S. marcescens RND efflux systems with broad substrate specificities.
A previous study suggested that SdeAB is the primary RND efflux system in S. marcescens, with the sdeB mutant becoming hypersensitive to multiple antimicrobial agents, similar to the hasF mutant13. However, the present study showed that SdeAB had narrower substrate specificities than SdeXY, SdePQ, and SdeGH. We also found that the sdeXY mutant became hypersensitive to a broad spectrum of antimicrobial agents. Furthermore, an independent study indicated that sdeAB was not expressed in the wild-type strain of S. marcescens and the expression of sdeAB was induced by the biocide, cetylpyridinium chloride14. Thus, SdeAB may play a primary role in multidrug resistance only in specific strains of S. marcescens and/or a strain that is exposed to a specific biocide.
We identified two previously uncharacterized S. marcescens RND efflux systems, SdePQ-OmsA and SdeGH, which exhibit broad substrate specificities. Our reverse transcription-PCR analyses on the S. marcescens Db10 and KS24 strains under normal growth conditions revealed that sdeQ and sdeH gene expression was not detected, while sdeY gene expression was observed in both strains (data not shown). When some mutations were occurred which caused the expression of SdePQ-OmsA and SdeGH systems, these pumps would contribute to the acquired resistance in S. marcescens.
All of the S. marcescens RND efflux systems utilized TolC and HasF when expressed in E. coli. However, some RND pumps when expressed with TolC of E. coli or HasF of S. marcescens showed different substrate specificities. SdeNO expressed with HasF in KAM43 showed higher MICs than that expressed in KAM32 with TolC, whereas SdeIJ and SdeS showed lower MICs. These results suggest that compatibility between outer membrane proteins and other components in RND pumps is important for its efflux activity. As our group reported previously, only VmeAB in V. parahaemolyticus showed high MICs expressed with TolC of E. coli, while other RND pumps in V. parahaemolyticus had markedly higher MICs when expressed with VpoC, an orthologue of TolC4. This result may be important for understanding the interaction between outer membrane proteins and other components.
In summary, the present study revealed that S. marcescens has multiple RND efflux systems that have the potential to confer multidrug resistance. Among these systems, SdeXY plays a major role in intrinsic multidrug resistance in S. marcescens. SdePQ-OmsA and SdeGH showed broad substrate specificities similar to SdeXY; however, these systems appear to be inducible and do not play major roles in the intrinsic multidrug resistance of S. marcescens.
Materials and Methods
Bacterial strains and growth conditions
The bacterial strains and plasmids used in the present study are listed in Supplementary Table S2. Unless otherwise noted, bacterial cells were grown in Luria (L) medium (1% polypeptone, 0.5% yeast extract, 0.5% NaCl, pH 7) at 37 °C under aerobic conditions. Antibiotics were supplemented when required as follows: ampicillin, 100 µg/ml; chloramphenicol, 20 µg/ml.
Phylogenetic tree of IMPs
Entire sequences of inner membrane protein were obtained from several database. The phylogenetic tree was obtained using CLUSTALW (https://clustalw.ddbj.nig.ac.jp).
Cloning, sequencing, and gene manipulation
We identified putative S. marcescens RND efflux system genes using the S. marcescens Db11 genomic sequence database (http://www.sanger.ac.uk/resources/downloads/bacteria/serratia-marcescens.html). DNA fragments, which contained the open reading frames (ORFs) of S. marcescens RND efflux system genes or S. marcescens OMP, hasF (SMA3509), were amplified by PCR using the chromosomal DNA of S. marcescens Db10, the parent strain of Db1139, as a template. Primers used for cloning are listed in Supplementary Table S3. Each primer included a restriction enzyme recognition site (underlined). The PCR products obtained were digested with the indicated restriction enzymes, gel-purified, and then ligated into the same restriction enzyme sites of the vector pUC18, pUC19, or pSTV28 (for hasF), which were located downstream of the lac promoter of the vector plasmid. Since PCR products did not include the promoter region of the genes, gene expression was controlled by the lac promoter.
The cloning of sdeCDE (SMA2945-2946-2947) was performed in two steps. The 5′ half fragment was amplified with two primers (SMA2945-2947F fw EcoRI and SMA2945-2947F re BamHI), and the 3′ half was then amplified with two primers (SMA2945-2947B fw EcoRI and SMA2945-2947B re BamHI). After digestion with EcoRI and BamHI, each fragment was individually inserted into pUC18. The resultant plasmids were designated as pURS4F and pURS4B. After the digestion of pURS4B with MluI and BamHI, the fragment was inserted into pURS4F at the same sites. It was named pURS4. The plasmid pURS44 carrying sdeC and sdeD, but not sdeE was also constructed. The plasmid pURS4 carrying sdeC-sdeE was digested with NcoI and self-ligated. Similarly, the plasmid pURS45 carrying sdeC and sdeE, but not sdeD was constructed. The plasmid pURS4 was digested with Tth111I, blunted, and self-ligated.
To evaluate the function of OmsA, the gene of which is located downstream of sdePQ (SM1743-1742), two types of plasmids were constructed. The fragment including sdePQ-omsA was amplified with PCR using SMA1743-1741 fw XbaI and SMA1743-1741 re EcoRI. After digestion with XbaI and EcoRI, the fragment was inserted at the same site in pUC19, and the resultant plasmid was named pURS8. To construct pURS82 carrying incomplete omsA, pURS8 was digested with HpaI and self-ligated.
Since S. marcescens showed higher β-lactam resistance, we were unable to utilize pURS2 carrying sdeXY. To complement sdeXY, we constructed another plasmid pSMXY. PCR was initially performed with two primers SMA0370-0369 fw EcoRI and SMA0370-0369 re XbaI using the Db10 genome as a template. PCR was then performed with two different primers, SMA0370-0369 fw EcoRI and SMA0370-0369 re BglII, to create the BglII site instead of the XbaI site. The PCR products obtained were digested with EcoRI and BglII, gel-purified, and then ligated into the EcoRI-BamHI sites of the vector pSTV28.
Minimum inhibitory concentrations (MICs)
The MICs of various antimicrobial agents were assessed in Muller–Hinton broth (Difco) using the standard two-fold dilution method as previously described40.
Construction of the sdeXY deletion strain
S. marcescens KS24, a plasmid pSMC2-cured derivative of S. marcescens KS341 was used to construct a sdeXY deletion strain. The sdeXY deletion strain of S. marcescens KS24 was constructed by homologous recombination using the lambda Red recombinase system42. Two-step PCR of the gentamicin cassette flanked by long (1000 nt) homologous extensions of the target gene were essentially performed as previously described43 using pBRFRTGM and the genomic DNA of S. marcescens KS24 as the template and the primers listed in Supplementary Table S3. The resulting PCR product was separated on an agarose gel and purified using GENECLEAN II KIT (MP Biomedicals Inc.). sdeXY mutant strains were generated by electroporation of the purified PCR product into S. marcescens KS24/pKD46 as described previously42. The deletion of sdeXY in the mutant strain was verified by PCR.
Measurements of rhodamine 6 G and ethidium efflux activities
The efflux of rhodamine 6 G and ethidium was evaluated as previously described9,40. In the rhodamine6G efflux assay, E. coli KAM32 strains were grown in L media until O.D650 = 0.7. E. coli cells were harvested by centrifugation, washed twice using Potassium Phosphate Buffer (PPB) containing 5 mM MgSO4, and resuspended in the same buffer that contained 1 µM rhodamine 6 G and 40 µM carbonylcyanide-m-chlorophenylhydrazone (CCCP). The cell suspension was incubated at 37 °C for one hour to de-energize cells, washed twice using the same buffer that did not contain CCCP, and then resuspended in the same buffer. The resultant cell suspension was incubated on ice for two hours and used in the efflux assay.
In the ethidium efflux assay, the S. marcescens KS24 and S. marcescens KS24ΔsdeXY strains were grown in L media until O.D650 = 0.7. S. marcescens cells were harvested by centrifugation, washed twice using modified Tanaka Buffer44, and resuspended in the same assay. The resultant cell suspension was incubated at 37 °C in the presence of 20 mM lactate-tetramethylammonium hydroxide (pH 7.0) for 5 min and used in the efflux assay.
References
Hejazi, A. & Falkiner, F. R. Serratia marcescens. J. Med. Microbiol. 46, 903–912 (1997).
Jones, R. N. Important and emerging beta-lactamase-mediated resistances in hospital-based pathogens: the Amp C enzymes. Diagn. Microbiol. Infect. Dis. 31, 461–466 (1998).
Matsuo, T. et al. VmeAB, an RND-type multidrug efflux transporter in Vibrio parahaemolyticus. Microbiology 153, 4129–4137 (2007).
Matsuo, T. et al. Characterization of all RND-type multidrug efflux transporters in Vibrio parahaemolyticus. Microbiologyopen 2, 725–742 (2013).
Matsuo, T., Ogawa, W., Tsuchiya, T. & Kuroda, T. Overexpression of vmeTUV encoding a multidrug efflux transporter of Vibrio parahaemolyticus causes bile acid resistance. Gene 541, 19–25 (2014).
Morita, Y. et al. Construction of a series of mutants lacking all of the four major mex operons for multidrug efflux pumps or possessing each one of the operons from Pseudomonas aeruginosa PAO1: MexCD-OprJ is an inducible pump. FEMS Microbiol. Lett. 202, 139–143 (2001).
Nikaido, H. & Takatsuka, Y. Mechanisms of RND multidrug efflux pumps. Biochimica et biophysica acta 1794, 769–781 (2009).
Nishino, K., Latifi, T. & Groisman, E. A. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59, 126–141 (2006).
Nishino, K. & Yamaguchi, A. Overexpression of the response regulator evgA of the two-component signal transduction system modulates multidrug resistance conferred by multidrug resistance transporters. J. Bacteriol. 183, 1455–1458 (2001).
Chen, J., Kuroda, T., Huda, M. N., Mizushima, T. & Tsuchiya, T. An RND-type multidrug efflux pump SdeXY from Serratia marcescens. J. Antimicrob. Chemother. 52, 176–179 (2003).
Kumar, A. & Worobec, E. A. Cloning, sequencing, and characterization of the SdeAB multidrug efflux pump of Serratia marcescens. Antimicrob. Agents Chemother. 49, 1495–1501 (2005).
Hornsey, M. et al. Tigecycline resistance in Serratia marcescens associated with up-regulation of the SdeXY-HasF efflux system also active against ciprofloxacin and cefpirome. J. Antimicrob. Chemother. 65, 479–482 (2010).
Begic, S. & Worobec, E. A. The role of the Serratia marcescens SdeAB multidrug efflux pump and TolC homologue in fluoroquinolone resistance studied via gene-knockout mutagenesis. Microbiology 154, 454–461 (2008).
Maseda, H., Hashida, Y., Konaka, R., Shirai, A. & Kourai, H. Mutational upregulation of a resistance-nodulation-cell division-type multidrug efflux pump, SdeAB, upon exposure to a biocide, cetylpyridinium chloride, and antibiotic resistance in Serratia marcescens. Antimicrob. Agents Chemother. 53, 5230–5235 (2009).
Begic, S. & Worobec, E. A. Characterization of the Serratia marcescens SdeCDE multidrug efflux pump studied via gene knockout mutagenesis. Can. J. Microbiol. 54, 411–416 (2008).
Chen, J. et al. VmrA, a member of a novel class of Na+-coupled multidrug efflux pumps from Vibrio parahaemolyticus. J. Bacteriol. 184, 572–576 (2002).
Chau, S. L., Chu, Y. W. & Houang, E. T. Novel resistance-nodulation-cell division efflux system AdeDE in Acinetobacter genomic DNA group 3. Antimicrob. Agents Chemother. 48, 4054–4055 (2004).
Damier-Piolle, L., Magnet, S., Bremont, S., Lambert, T. & Courvalin, P. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob. Agents Chemother. 52, 557–562 (2008).
Li, D. W. et al. Properties and expression of a multidrug efflux pump AcrAB-KocC from Klebsiella pneumoniae. Biol. Pharm. Bull. 31, 577–582 (2008).
Magnet, S., Courvalin, P. & Lambert, T. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45, 3375–3380 (2001).
Mine, T., Morita, Y., Kataoka, A., Mizushima, T. & Tsuchiya, T. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43, 415–417 (1999).
Nishino, K. & Yamaguchi, A. Analysis of a complete library of putative drug transporter genes in. Escherichia coli. J. Bacteriol. 183, 5803–5812 (2001).
Ogawa, W., Onishi, M., Ni, R., Tsuchiya, T. & Kuroda, T. Functional study of the novel multidrug efflux pump KexD from Klebsiella pneumoniae. Gene 498, 177–182 (2012).
Poole, K. et al. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21, 713–724 (1996).
Poole, K., Krebes, K., McNally, C. & Neshat, S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol 175, 7363–7372 (1993).
Coyne, S., Rosenfeld, N., Lambert, T., Courvalin, P. & Perichon, B. Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 54, 4389–4393 (2010).
Kohler, T. et al. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23, 345–354 (1997).
Mima, T., Sekiya, H., Mizushima, T., Kuroda, T. & Tsuchiya, T. Gene cloning and properties of the RND-type multidrug efflux pumps MexPQ-OpmE and MexMN-OprM from Pseudomonas aeruginosa. Microbiol. Immunol. 49, 999–1002 (2005).
Bina, J. E., Provenzano, D., Wang, C., Bina, X. R. & Mekalanos, J. J. Characterization of the Vibrio cholerae vexAB and vexCD efflux systems. Arch. Microbiol. 186, 171–181 (2006).
Li, Y. et al. A new member of the tripartite multidrug efflux pumps, MexVW-OprM, in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 52, 572–575 (2003).
Rahman, M. M. et al. Molecular cloning and characterization of all RND-type efflux transporters in Vibrio cholerae non-O1. Microbiol. Immunol. 51, 1061–1070 (2007).
Sekiya, H. et al. Functional cloning and characterization of a multidrug efflux pump, mexHI-opmD, from a Pseudomonas aeruginosa mutant. Antimicrob. Agents Chemother. 47, 2990–2992 (2003).
Baranova, N. & Nikaido, H. The baeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J. Bacteriol. 184, 4168–4176 (2002).
Mima, T. et al. Gene cloning and characteristics of the RND-type multidrug efflux pump MuxABC-OpmB possessing two RND components in Pseudomonas aeruginosa. Microbiology 155, 3509–3517 (2009).
Chuanchuen, R., Narasaki, C. T. & Schweizer, H. P. The MexJK efflux pump of Pseudomonas aeruginosa requires OprM for antibiotic efflux but not for efflux of triclosan. J Bacteriol 184, 5036–5044 (2002).
Mima, T., Joshi, S., Gomez-Escalada, M. & Schweizer, H. P. Identification and characterization of TriABC-OpmH, a triclosan efflux pump of Pseudomonas aeruginosa requiring two membrane fusion proteins. J Bacteriol 189, 7600–7609 (2007).
Nishino, K., Yamada, J., Hirakawa, H., Hirata, T. & Yamaguchi, A. Roles of TolC-dependent multidrug transporters of Escherichia coli in resistance to β-lactams. Antimicrob. Agents Chemother. 47, 3030–3033 (2003).
Nakamura, T. et al. IMP-1 type metalo-beta-lactamase producing Serratia marcescens strains isolated from blood culture between 1991 to 2000. Kansenshogaku zasshi. The Journal of the Japanese Association for Infectious Diseases 76, 246–253 (2002).
Flyg, C., Kenne, K. & Boman, H. G. Insect pathogenic properties of Serratia marcescens: phage-resistant mutants with a decreased resistance to Cecropia immunity and a decreased virulence to Drosophila. J. Gen. Microbiol. 120, 173–181 (1980).
Minato, Y., Shahcheraghi, F., Ogawa, W., Kuroda, T. & Tsuchiya, T. Functional gene cloning and characterization of the SsmE multidrug efflux pump from Serratia marcescens. Biol. Pharm. Bull. 31, 516–519 (2008).
Iguchi, A. et al. Genome Evolution and Plasticity of Serratia marcescens, an Important Multidrug-Resistant Nosocomial Pathogen. Genome Biol. Evol. 6, 2096–2110 (2014).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645 (2000).
Kuwayama, H. et al. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res 30, E2 (2002).
Tanaka, S., Lerner, S. A. & Lin, E. C. Replacement of a phosphoenolpyruvate-dependent phosphotransferase by a nicotinamide adenine dinucleotide-linked dehydrogenase for the utilization of mannitol. J Bacteriol 93, 642–648 (1967).
Bleuel, C. et al. TolC is involved in enterobactin efflux across the outer membrane of Escherichia coli. J Bacteriol 187, 6701–6707 (2005).
Franke, S., Grass, G., Rensing, C. & Nies, D. H. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J Bacteriol 185, 3804–3812 (2003).
Yuan, J. et al. Prevalence of the oqxAB gene complex in Klebsiella pneumoniae and Escherichia coli clinical isolates. The Journal of antimicrobial chemotherapy 67, 1655–1659 (2012).
Acknowledgements
This work was supported by JSPS KAKENHI Grant Number JP24590080.
Author information
Authors and Affiliations
Contributions
S.T., Y.M., W.O., T.T. and T.K. planned this project. Y.M. and T.K. wrote the main manuscript. S.T., Y.M., Y.K. and D.M. constructed all plasmids and transformants. S.M. and N.G. cured plasmids from S. marcescens. S.T. and Y.M. constructed the sdeXY deletion strain, and evaluated R6G efflux activity. S.T., Y.M., Y.K., K.H., S.K. and D.M. evaluated MICs. Y.M., W.O. and T.K. prepared Figures 1–2. Y.M., W.O., T.K. and Y.M. had critical discussions with T.K. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Toba, S., Minato, Y., Kondo, Y. et al. Comprehensive analysis of resistance-nodulation-cell division superfamily (RND) efflux pumps from Serratia marcescens, Db10. Sci Rep 9, 4854 (2019). https://doi.org/10.1038/s41598-019-41237-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-41237-7
This article is cited by
-
Quaternary ammonium disinfectants and antiseptics: tolerance, resistance and potential impact on antibiotic resistance
Antimicrobial Resistance & Infection Control (2023)
-
The role of RND-type efflux pumps in multidrug-resistant mutants of Klebsiella pneumoniae
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.