Abstract
Myopia, commonly referred to as nearsightedness, is one of the most common causes of visual disability throughout the world. It affects more people worldwide than any other chronic visual impairment condition. Although the prevalence varies among various ethnic groups, the incidence of myopia is increasing in all populations across globe. Thus, it is considered a pressing public health problem. Both genetics and environment play a role in development of myopia. To elucidate the epigenetic mechanism(s) underlying the pathophysiology of high-myopia, we conducted methylation profiling in 18 cases and 18 matched controls (aged 4–12 years), using Illumina MethylationEPIC BeadChips array. The degree of myopia was variable among subjects, ranging from −6 to −15D. We identified 1541 hypermethylated CpGs, representing 1745 genes (2.0-fold or higher) (false discovery rate (FDR) p ≤ 0.05), multiple CpGs were p < 5 × 10−8 with a receiver operating characteristic area under the curve (ROC-AUC) ≥ 0.75 in high-myopia subjects compared to controls. Among these, 48 CpGs had excellent correlation (AUC ≥ 0.90). Herein, we present the first genome-wide DNA methylation analysis in a unique high-myopia cohort, showing extensive and discrete methylation changes relative to controls. The genes we identified hold significant potential as targets for novel therapeutic intervention either alone, or in combination.
Similar content being viewed by others
Introduction
Myopia, or nearsightedness, the most prevalent form of refractive error, is caused by excessive axial elongation of the eye as a major mechanism in children1. According to recent reports2, myopia is becoming an epidemic in the developed countries of East and South-East Asia, where the prevalence reaches 80–90% in children aged 17–18 attending secondary school. Concomitantly, the European Eye Epidemiology Consortium (E3) showed that the prevalence of myopia is also dramatically growing in the Western countries3.
A recent study by Holden et al. estimated the huge growth in the world population of myopia and high myopia, defined in his study as loss of 6.00 diopters (D) or more, as an increase from 1406 million and 163 million in 2000 to 4758 million and 938 million in 2050, for the two forms respectively4. These alarming data imply that by 2050 half of the world’s population may be affected by myopia, representing a significant social and economic burden to the global healthcare systems5.
The condition usually first appears between 8 to 12 years and as the child ages, vision can change rapidly, requiring a corrective prescription every few years. Myopia usually stabilizes by the age of approximately 20 years as the eyeball reaches its full size6. If myopia appears before the start of schooling (very early onset myopia), it is generally more severe and more likely to be genetic in origin. In addition, myopia developing at school age is less likely to progress to high myopia until the age of 11–13, and high myopia which appears after those ages is likely to be driven by environmental factors.
Around 30 distinct genetic risk loci have been identified for both high and mild myopia; however, the pathophysiology of variation(s) causing disease is obscure7,8. There is compelling evidence that both environmental and genetic factors are involved in the etiology of myopia9,10,11,12. Although environmental factors along with genetic predisposition are associated with the increasing prevalence of myopia amongst children, the mechanism through which they act is moderately understood. An epigenetic event such as DNA methylation could be one of the mechanisms through which these environmental factors influence the development of myopia. Previous studies have identified epigenetic analysis as a tool to reveal the causative mechanisms of ocular diseases including myopia13,14, but until now no genome-wide epigenetic association studies related to high myopia subjects have been reported. To elucidate potential epigenetic mechanism(s) underlying the pathophysiology of high myopia, we conducted a genome-wide methylation analysis on 18 high myopia subjects and an equal number of controls.
In this study, we investigated DNA methylation on a genome-wide scale using the Infinium MethylationEPICBeadChip-array technology in a unique cohort of children with non-syndromic high myopia, and identified associated biological pathways implicated in the development of high myopia. This study creates the preliminary awareness required to understand the influence of various factors that can contribute to the development of myopia.
Results
Differentially methylated CpG sites identification
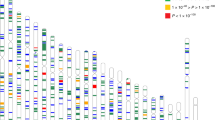
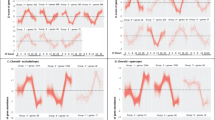
We identified 1541 CpG sites in 1745 unique genes that were differentially methylated (2.0-fold or higher) in high myopia subjects compared to controls without high myopia. All genes were hypermethylated and no CpG site was observed with significant hypomethylation. The detailed list of the most significant differentially methylated CpG sites based on FDR-corrected p-values, fold change and AUC for high myopia detection is shown in Supplementary Table S1. A total of 48 cytosine loci had excellent accuracy (AUC ≥ 0.90) for the detection of high myopia. A positive ‘% Methylation Change’ value indicates an average increase in methylation in high myopia subjects compared to control samples. The p-value indicates the significance of the differential methylation levels. Among the 1541 unique targets, the top 10 hypermethylated targets based on fold change were cg26526312 (LINGO1), cg27541540 (PTPN11), cg00609363 (ZNRD1), cg24877391 (PEX13; PUS10), cg21790796 (KIF20A; BRD8), cg14282407 (TRAPPC1; CNTROB), cg18191664 (ERLIN2), cg08048517 (KIAA0528), cg10646633 (ZNF224) and cg14694176 (TCEA1). The results for the four AUC ROC outcome measures are shown in Fig. 1. The study also identified 81 open reading frame (ORF) genes and 41 LOC genes associated with high myopia (Supplementary Tables S2 and S3). Both ORFs and LOC genes were highly significant between 2 to 7-fold difference and AUC value of >0.75 (FDR p-value < 0.01).
(A–D) Receiver operating characteristic curve analysis of methylation profiles. ROC analysis for four excellent CpG sites [(A) cg12711743: LRRC8C; (B) cg05367846: MICAL3; (C) cg00765710: PGBD2; and (D) cg16232979: TPM4]. At each locus, the FDR p-value for methylation difference between myopia subjects and controls was significantly different. ROC: Receiver operating characteristic; AUC: Area Under Curve; 95% CI: 95% Confidence Interval.
Cluster analysis of differentially methylated targets
Hierarchical clustering was performed using β-values for commonly methylated genes that showed differential methylation between the high myopia group and controls. Twenty target genes significantly hypermethylated in myopic subjects comparing to the control group were used in the clustering analysis. These 20 targets were displayed in two distinct clusters, indicating that methylation was associated with altered expression of these genes in high myopia individuals (Fig. 2). PCA results showed complete separation between high myopia samples and control sample sets in the PCA distribution 3D plot (PCA 3D, Fig. 3). Cluster data correlates with PCA.
Heatmap and hierarchical clustering of myopia cases based on DNA methylation. Heat map showing hypermethylation variation in myopia cases compared with controls. The Heat map in hierarchical clustering analysis represented DNA methylation levels from completely methylated (red) to unmethylated (green).
Functional enrichment analysis of differentially methylated genes is shown in Fig. 4. Forty two percent of those genes are under the transcription binding activity category, followed by 38% genes with functional catalytic activity. The smallest category represents genes with translation regulator activity, comprising 1% of genes identified. Ingenuity Pathway Analysis identified 10 important canonical signaling pathways associated with high myopia gene enrichment at probability values ≤ 0.01. The identified canonical signaling pathways include: Wnt/β-catenin signaling, insulin receptor signaling, protein kinase A signaling, actin cytoskeleton signaling, ILK signaling, signaling by Rho family GTPases, IGF-1 signaling, the opioid signaling pathway, axonal guidance signaling, and G-protein coupled receptor signaling (Fig. 5).
Discussion
Epigenetics plays a critical pathogenic role in the development of complex eye diseases including myopia13. Methylation profiling of DNA using array technology has been successfully used to explore CpG sites associated with various complex diseases15,16. Myopia is associated with number of other eye conditions such as cataracts17, glaucoma18, retinal detachment19, keratoconus20, macular degeneration, retinal holes, choroidal neovascularization21, diabetic retinopathy22 and retinitis pigmentosa23. Many studies have been performed to detect genes associated with myopia24,25. But studies on epigenetic factors, especially methylation studies, have not been performed to our knowledge. The current study identifies the global DNA methylation hotspots in the genome of high myopia patients under the age of 12 years.
We have identified differential methylation in genes those were previously suggested to be associated with myopia such as, PAX6, ZNRF3, PSEN1, SOCS1, GRB2, ADCY3, RGS5, SRF and AP1B1. Multiple variations of PAX6 were previously shown to be associated with myopia26,27. ZNRF3, which was identified as one of the loci in the Consortium for Refractive Error and Myopia (CREAM) study, was also identified in the present study as a hypermethylated gene28. Mutations were previously observed in PSEN1 in a patient with posterior cortical atrophy and myopia29 and we identified methylation on this gene in the present study. The differential expression of SOCS1 has been observed in corneal cells of myopic patients30 and SOCS1 deficient mice develop severe eye diseases31. In RPE cells, SOCS1 higher expression inhibits IFNγ-mediated responses which leads to uveitis32. GRB2 was found to be upregulated in chicks during imposed myopic defocus33 and also during retinal stress and neovascularization34. ADCY3 and RGS5 were previously noted to be upregulated in the retinal cells and scleral cells of myopic patients respectively, but without statistical significance30. SRF was shown to be upregulated in treated eyes versus control eyes of myopic guinea pigs35. A SNP (rs715494) within AP1B1 gene, which is located in the MYP6 region, was previously identified among myopic patients36. These results indicate the reproducibility of our study in identifying markers for myopia.
Oxidative stress
Oxidative stress is involved in myopia and other ocular diseases37. Besides inducing oxidative damage to various parts of eyes in myopia, high oxidative stress in various retinal structures leads to neovascularization in the choroid. Further, high myopia is associated with oxidative stress in various parts of eye in the development of glaucoma, cataract and retinal detachment. In the present study, we have identified several genes that are associated with regulation of oxidative stress including PEX1338, NNT39, OXT40, SOD341, PRDX142, PRDX543, CYB5B, CYB5R144 and COQ345. Interleukin-1alpha downregulates SOD3 and the SOD3 reduction leads to insufficient oxidative defense during keratoconus condition46.
We have also identified NADH-ubiquinone oxidoreductase genes that are involved in oxidative phosphorylation process such as NDUFA5, NDUFB2, NDUFB3, NDUFB7, NDUFB10, NDUFS8, and NDUFV2-AS147. Similarly, we have identified COX family member genes including COX6B1, COX18, and COX16; these take part in the physiological response to oxidative stress48,49. ACOX1 also plays a role in oxidative damage50; MCAT is a mitochondrial-specific catalase enzyme51 and PDIA4 expression increases with oxygen glucose deprivation52. NADK2 is one of the enzymes that catalyzes phosphorylation of NAD(+) to yield NADP(+)53. Interestingly, TRAF3IP2 and JUN genes were found to be hypermethylated in our study cases. TRAF3IP2 is one of the oxidative stress-responsive cytoplasmic adapters that regulates c-JUN54.
Axonal guidance
Axon guidance is vital for nerve growth in brain and sensory organs such as retina of eye. Retinal ganglion cells (RGC) project their axons to the visual cortex of the brain. There are number of proteins that act as axon guidance cues, such as netrins, semaphorins, slits, ephrins, L1CAM (L1), laminin, tenascin, chondroitin sulphate and Wnt proteins55,56,57,58. In high myopic individuals, thinning of the macular ganglion cell complex has been observed59. Lower macular thickness results in loss of RGCs and retinal nerve fibers60, resulting in disrupted signals to the visual cortex targets. Our study identified methylation differences in the semaphorins, such as SEMA3F, SEMA4B, SEMA4C and SEMA5A that were significantly associated with myopia. Among these, SEMA5A has been found to be specifically expressed during retinal axon outgrowth at the optic disc and along the optic nerve. Sema5A inhibits axon growth and retinal neurite outgrowth by retinal ganglion cells and Plexin family receptors respectively61,62. Even in the presence of growth-promoting axon guidance signaling molecules like laminin, L1 and netrins, the higher expression of SEMA5A could inhibit retinal growth cones63. Our data supports this mechanism for myopia, as we identified methylation at the gene body region that may support higher expression levels64 of SEMA5A and inhibition of retinal growth cones. PAX6 was also identified to be hypermethylated by 3-fold at the 5’UTR in association with myopia in our study. Genetic variation at the regulatory region and other regions on PAX6 expression is important for the development of axonal connections and PAX6 has been reported to play a role in development of myopia26,27. The same study noted diminished expression of SEMA5A65. Neurotrophic Tyrosine Kinase, Receptor, Type 2 (NTRK2), also called as Tyrosine Kinase Receptor B (TRKB) is a receptor for Brain-Derived Neurotrophic Factor (BDNF), which plays a crucial role in the activity-dependent refinement of synaptic connectivity of retinal ganglion cells. The BDNF/TRKB complex is vital for development of photoreceptors, and for synaptic communication between photoreceptors and second order retinal neurons66. The current study observed methylation of NTRK2 at the promoter region. Several other hypermethylated genes identified were ADAMTS1, ARPC5, BAIAP2, and TUBG1, for which further functional analysis is warranted. Among them, ADAMTS1 was previously found to be downregulated in corneas of keratoconus patients compared to control subjects67.
Growth factor signaling and cell differentiation
Studies on animal models of myopia have suggested a role for several different growth factors and their signaling in the development of myopia. TGFβ increases in the sclera of form-deprived myopic animals, while levels of bFGF were found to be decreased68. The influence for the secretion of VEGF, HGF and IGF growth factors is derived from their role in oxidative stress in different parts of eye, specifically the retina37.
Most growth factors exert their effects on cells through G-protein coupled receptors (GPCRs)69. Our study identified certain signaling molecules to be associated with myopia. We have found hypermethylation of Regulator of G-protein signaling 2 (RGS2), which was earlier reported to be upregulated in the sclera of form deprivation myopia (FDM). Zou and collaborators supported therapeutic strategies to control dysregulated RGS2 as a treatment for myopia70. ADORA2A was hypermethylated at the transcription start site in our study; this gene encodes the adenosine A2a receptor protein, which is a GPCR family member. ADORA2A is expressed in ocular tissues and modulates collagen synthesis and extracellular matrix production during eye growth. Exonic variants of this gene were previously identified in association with high myopia71. AdoRs showed to play role in growth regulation of eye and the variation in expression pattern of AdoR observed during form deprivation myopia confirms the pharmaceutical intervention may help in reducing myopia progression72.
In addition, we noted methylation alteration in the promoters of GPCR-associated Ras-regulating proteins such as GNAO173, PDPK1, RASGRP1, RASGRP2, RASGEF1B, and RASL10B74,75. Among them, RASGRP1 showed upregulation in human corneal epithelial cells and conjunctival epithelial cells upon IL-4 stimulation76,77. The Rho family of GTPases regulate growth and cellular transformation processes78. We found over 3-fold hypermethylation of promoters of two Rho family genes, RHOD and RHOF, to be significantly associated with myopia.
Wnt signaling
The Wnt family of proteins lead one of the most versatile regulatory pathways in developmental biology, cell cycle and tissue homeostasis79. Substantial literature is available to support the association of Wnt signaling in the pathophysiology of progression of cancer, chronic progressive disorders, cell fate specification and differentiation related disorders80. Comprehensive study has now revealed that Wnt signaling is initiated through receptors (the Frizzled family) to downstream β-catenin81. Between these two ends many proteins interact in a complex pathway of signal transduction.
Wnt signaling is a ubiquitous pathway that is influenced by the action of various growth factors such as TGFβ, bFGF, IGF, VEGF, HGF, etc., and its components are critical for the development of ocular tissues at various stages, including the formation of eye, retina, lens, ciliary body, iris and vascular development82,83. Activation of Wnt signaling leads to the progression of myopia in mice. Wnt2b and Wnt3 along with β-catenin were shown to be highly expressed in scleral fibroblast cells in FDM84,85. In our study, we observed hypermethylation of Catenin Beta Interacting Protein 1 (CTNNBIP1: p = 0.006) and Catenin Alpha-Like 1 (CTNNAL1: p = 2.6 × 10−38) with 2.3 and 3.19-fold changes respectively. We identified 22 genes which were hypermethylated (PPP3CA, RAC3, TCF7L1, CSNK2B, ROCK1, CTNNBIP1, PSEN1, BTRC, PPP3CC, TP53, AXIN1, PPP2R1B, CSNK1A1, TCF7L2, RBX1, FBXW11, TBL1XR1, NLK, PLCB4, PRKACB, JUN and SKP1) and pathway analysis showed significant effects of these genes on pathway outcome, via both pathway and physical interactions (Supplementary Fig. 1A,B). Xion et al. had reported dysregulated mir-203 and mir-350 that have downstream implications on the Nlk, Sema5a and Acer2 genes, vital in the regulatory network86. Our study also observed hypermethylation in NLK and SEMA5A, with significant methylation change (p = 3.3 × 10−38 and p = 2.7 × 10−9, respectively). Both genes had greater than 2-fold changes in methylation as compared to control subjects. NLK is known to interact with CTNNB1, TP53, TCF7L1, and TCF7L2, which were hypermethylated as compared to controls.
We have identified differentially methylated genes under the influence of Wnt/β-catenin, including JUN, MARK2, TGFBR3, NR5A2, SP1, NLK, CSNK1A1 and ACVR2A in myopic subjects. JUN is an oncogene shown to be upregulated in treated eyes versus control eyes of myopic guinea pigs35. Mitochondrial functioning is important for the refractive error in myopia87. Map/Microtubule Affinity-Regulating Kinase 2 (MARK2) has a strong role in maintaining the movement of mitochondria in retina/ganglion cells and helps in proper neuronal function88. TGFBR3 is associated with Primary Open Angle Glaucoma (POAG)89 and high myopia is one of the risk factors for POAG18. The methylation identified in our study in the promoter and first exon of TGFFBR3 may influence the expression of this gene, causing the myopic phenotype that may lead to POAG. The current study also shows hypermethylation on NR5A2, found to be linked to the SP1 gene which is hypermethylated at the transcription start site. SP1 is developmentally regulated and important for the corneal development90. The corneas in myopic patients tend to be thin compared to normal eyes91, implicating SP1 and other network genes in maintaining corneal structure. Sp1 was shown to participate in regulating type I collagen synthesis or degradation during myopic sclera remodeling, in association with TGF-β1 signaling, suggesting a role in the development of myopia92. Another key gene in the regulatory network of Wnt signaling is CSNK2B, which showed significant (p = 0.01) hypermethylation with a 2.45-fold change as compared to control subjects. Kloss et al. reported a nonsynonymous damaging variant in CSNK2B (c.473 A > G resulting in p. Tyr158Cys) among high myopia families, resulting in interference with the Wnt signaling pathway93. Hypermethylation of CSNK2B may result in low levels expression, affecting interaction with JUN, BTRC, PPP2R1B, and PPP3CC.
Thus, overall diminished expression of key genes is likely to result in underpowered Wnt signaling, and consequently repressed expression of genes responsive to possible pathophysiology of the retina in myopia.
Protein kinase A signaling activity
Protein kinase A signaling is an important process for retinal ganglion cell survival94,95. Hypermethylation in the 5’UTR of Dual-Specificity Phosphatase 16 (DUSP16) was significantly associated with myopia in our study. MAPKs regulate insulin sensitivity96, which is an activator of retinal transmitters resulting in eye growth97. Dual-specificity phosphatases inactivate MAPKs by dephosphorylation98. In addition, MAPK3 was also observed to be under a hypermethylation burden in our study. Several other genes seen to be hypermethylated are under protein kinase A signaling control, including PHKB, PHKG2, PTPN11 and CDC27. Among these, PTPN11 plays a role in lens and retinal development99,100 and found to be associated with myopia7, and two other hypermethylated genes in our study, FBN1 and OCA2, were also found to be associated with myopia in the same study7 and FBN1 is associated with refractive error101.
IGF-1 Signaling
IGF has been shown to be associated with myopia in a chick model. Insulin injections block hyperopia and induce axial myopia in chicks most likely because of its influence on the optics of the anterior segment of eye102. Forkhead Box O1A (FOXO1) is one of the major targets of insulin action103 and was hypermethylated at the transcription start site in the present study. This gene was previously identified in association with reduction of central corneal thickness104 and keratoconus105, a condition that exhibits increased axial length, which has significant relationship with axial myopia20. RPTOR and VAMP2 are two more genes identified in our study under insulin receptor signaling and were hypermethylated with a significant association with myopia.
The altered secretion of various growth factors leads to activation of intracellular signaling pathways such as MAPK/Erk, Wnt/β-catenin and PI3K/Akt97,106 and ultimately leads to altered organization of the cytoskeleton and secretion of extracellular matrix components. The actin cytoskeleton provides mechanical force for cell movement and division107,108. We have identified many genes in the canonical actin cytoskeleton pathway that are statistically significantly associated with high myopia. These are ACTB, MATK, MYH2, RB1, DIAPH1, FGF14, ARPC5L and WASF2I. Among them, MYH2 mutations are associated with ophthalmoplegia (paralysis or weakness of eye muscles)109. RB1 is associated with retinoblastoma110.
The LINGO1 gene is involved in actin dynamics111 and we found its promoter to be hypermethylated by 11-fold at the 5’UTR. ACTB codes for beta-actin, and in the presence of actin cue netrin-1, the translation of β-actin is initiated in the RGC growth cones112, suggesting the importance of this gene in maintenance of retinal structure.
Additional genes related to myopia and identified in this study are MATK that has shown differential expression in the retinal cells of myopic patients30. STAT3 signaling is critical for scleral remodeling and myopia development113, COL2A1 and BICC1 are candidate genes for myopia114,115.
In summary, understanding various cellular and molecular pathways altered in patients with high myopia is essential in any attempts to block myopic growth, and hence can be useful in advancing treatment modalities for myopia. We have found different functional groups of genes that are significantly hypermethylated in the peripheral blood cells of high myopia patients. Whereas, we confirmed several previously reported candidate genes for myopia, many genes were newly identified with unknown mechanisms which further warrant functional studies. The replication of candidate gene association seen in this study adds to the weight of evidence that epigenetic methylation may be responsible for the development of myopic features seen in young children. In-depth bioinformatics network and pathway analysis identified significantly associated canonical pathways with eye development and function, allowing putative biological meaning to be assigned to the genes identified in the study. These genes were correlated to retinal ganglion cell development and maintenance, axial length difference, synaptic communication, corneal and scleral dysfunction. The pathway analysis also revealed possible interaction among the identified genes that may be involved in these mechanisms. One of the drawbacks of the study is that this analysis used blood DNA and not ocular tissue DNA. Nevertheless, this study provides confirmatory and novel data that should prompt further studies to address the functional role of the identified hypermethylated genes in the development of high myopia.
We proposed that methylation studies could help us to identify the genes associated with high myopia. Associated pathway analysis could provide insight into the mechanism of the development of myopia and might also help develop the preliminary awareness required to deter the influence of various factors contributing to the development of myopia. Identifying genes that contribute to the etiology of high myopia and elucidating the associated molecular mechanism(s) is the first step toward understanding the pathophysiology of this disease. This study details some of the etiology of myopia and eye growth patterns and thus has considerable public health application, as the high prevalence of myopia in the world is a formidable health care challenge. Furthermore, the identification of myopia-causing genes will be important for improving early myopia diagnosis and counseling and increasing our understanding of normal and abnormal development of the eye.
Methods
The present study was performed on the 18 Polish high myopia cases with refractive error ranging from −6.0 to −15.0 D in at least one eye with the axial length of the eye ranging from 26.22 mm to 27.85 mm (mean, 26.22 mm) and 18 controls without high myopia, with the axial length of the eye ranging from 22, 42 mm to 24.11 mm (mean, 22, 55 mm) with same ethnicity, age and gender. All cases were recruited at Department of Pediatric Ophthalmology Medical University of Bialystok and were aged between 4–12 years at the time of sample collection. All children had a comprehensive eye examination, including cycloplegic (cyclopentolate 1%) autorefraction to assess refractive error and ocular biometry to measure axial length of the eye. In all cases, the anterior segment of the eye, including cornea and lens, were normal. Clinical demographics are available for each high myopia subject of the study (Table 1). All subject identities were masked during experimental processing and analysis. This study was approved by the Institutional Review Boards at Poznan University of Medical Sciences in Poland and the informed consent was given by each subject’ parents, according to the Declaration of Helsinki. All methods were performed in accordance with the relevant guidelines and regulations.
Genome-wide methylation analysis
Genomic DNA from peripheral blood samples was extracted using Gentra Puregene Blood Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. DNA (500 ng) was bisulfite converted using the EZ DNA Methylation-Direct Kit (Zymo Research, Orange, CA) according to the manufacturer’s instruction.
Infinium MethylationEPIC BeadChip arrays (Illumina, Inc., California, USA) with 850,000 methylation sites were used for the analysis of genome-wide methylation according to manufacturer’s protocol. The methylation sites are distributed over regions including transcriptional start sites (TSS200, TSS1500), promoters, 5′UTR, exon boundaries, coding, and 3′UTR regions of autosomes and allosomes. Fluorescently-labeled BeadChips were imaged using iScan (Illumina, Inc.). Quality control, data preprocessing and background signal intensity correction was performed to yield a ratio of methylated and unmethylated signal intensities, followed by detailed bioinformatic and statistical analyses. 99% of the CpG loci were determined unequivocally and all the processing was done according to manufacturer’s instructions.
Bioinformatic and Statistical analysis
Gene-specific, genome-wide DNA methylation was quantified using the GenomeStudio methylation package (Illumina Software). DNA methylation ß-values were assigned to each CpG site. Differential methylation was identified by comparing these ß-values per individual nucleotide at each CpG site, between affected cases and normal controls. To avoid potential confounding factors, probes containing SNPs or within 10 bp of CpG sites listed on dbSNP entries were excluded from further analysis, as these interfere with determination of methylation at the corresponding sites116,117,118,119. Probes with SNPs over 10 bp from methylation sites or with an allelic frequency of ≤0.005 were further considered along with the remaining methylation sites for the analysis.
The fold change in CpG site variations were obtained by dividing the mean ß-values of the probes in each island by normal controls. Differentially methylated CpG sites were defined by the pre-set cutoff criteria of ≥2.0-fold increase and/or ≥2.0-fold decrease with False Discovery Rate (FDR) p < 0.05. Receiver Operating Characteristic (ROC) curves were generated for each CpG site, and the corresponding area under the curves (AUCs) were calculated to quantify diagnostic markers. To avoid potential experimental confounding, various statistical modeling was used. A heatmap was generated for the differentially-methylated genes, using ComplexHeatmap (v1.6.0)120 R package (v3.5.0). Ward’s minimum variance121 was used for the hierarchical clustering of samples. A multiple logistic regression analysis was done using more stringent criteria (FDR p ≤ 0.00001 and ≥2-fold change), to select candidate genes for high myopia biomarkers.
Principal Component Analysis (PCA)
Prior to analysis, we removed all CpG-probes with missing ß-values; the remaining ß-values of CpG targets were used for PCA. We analyzed PCA using the R function “prcomp” and used PC1, PC2 and PC3 for the PCA sharing plot. The 3D PCA distribution plot was created by using R package “ggplot2”. All CpG variables of myopic cases and controls were computed together to detect variations between myopia and controls.
Validation of methylation status by bisulfite pyrosequencing
Pyrosequencing was performed to verify the variable methylation status of important methylation variants, to determine that CHIP hybridizations were not artifacts but the true methylation differences. Forty-eight targets with AUC ≥ 0.9 were chosen for validation, comprising the 53 genes with the most significant methylation variation (p < 0.00001). 2 µg of genomic DNA was bisulfite-treated using the EZ methylation kit (Zymo Research) per the manufacturer’s instructions and sequencing was performed with appropriate oligos using the PyroMark Q24 System and advanced CpG reagents (Qiagen). An additional analysis replicated the top-25 differently methylated CpG sites in an independent second cohort of 24 high myopia cases and matched 24 controls and confirmed the top-ranking differentially-methylated CpG sites in whole blood DNA of our cohort samples.
Gene functional enrichment and pathway analysis
Genes found to be differentially methylated (FDR p-value < 0.00001) were subjected to functional enrichment analysis using the PANTHER Classification System122. Gene networks and pathways were analyzed using Ingenuity Pathway Analysis (IPA) software (Qiagen) to identify biological functions or interacting regulatory networks. All CpGs without mapping identifiers in IPA (GRCh37/hg19) were excluded from analysis. Only genes for which Entrez identifiers were available were further analyzed. Further, to understand the gene networks better, we performed gene enrichment analysis using WEB-based GEne SeT AnaLysis Toolkit and network analysis using GeneMANIA.
Data Availability
All data is appended in the manuscript.
References
Fledelius, H. C. Ophthalmic changes from age of 10 to 18 years. A longitudinal study of sequels to low birth weight. IV. Ultrasound oculometry of vitreous and axial length. Acta Ophthalmol (Copenh) 60, 403–411 (1982).
Dolgin, E. The myopia boom. Nature 519, 276–278, https://doi.org/10.1038/519276a (2015).
Delcourt, C. et al. Ophthalmic epidemiology in Europe: the “European Eye Epidemiology” (E3) consortium. Eur J Epidemiol 31, 197–210, https://doi.org/10.1007/s10654-015-0098-2 (2016).
Fricke, T. R. et al. Global prevalence of visual impairment associated with myopic macular degeneration and temporal trends from 2000 through 2050: systematic review, meta-analysis and modelling. Br J Ophthalmol 102, 855–862, https://doi.org/10.1136/bjophthalmol-2017-311266 (2018).
Holden, B. A. et al. Global Prevalence of Myopia and High Myopia and Temporal Trends from 2000 through 2050. Ophthalmology 123, 1036–1042, https://doi.org/10.1016/j.ophtha.2016.01.006 (2016).
Vitale, S., Ellwein, L., Cotch, M. F., Ferris, F. L. 3rd & Sperduto, R. Prevalence of refractive error in the United States, 1999–2004. Arch Ophthalmol 126, 1111–1119, https://doi.org/10.1001/archopht.126.8.1111 (2008).
Flitcroft, D. I. et al. Novel Myopia Genes and Pathways Identified From Syndromic Forms of Myopia. Invest Ophthalmol Vis Sci 59, 338–348, https://doi.org/10.1167/iovs.17-22173 (2018).
Ratnamala, U. et al. Refinement of the X-linked nonsyndromic high-grade myopia locus MYP1 on Xq28 and exclusion of 13 known positional candidate genes by direct sequencing. Invest Ophthalmol Vis Sci 52, 6814–6819, https://doi.org/10.1167/iovs.10-6815 (2011).
Farbrother, J. E., Kirov, G., Owen, M. J. & Guggenheim, J. A. Family aggregation of high myopia: estimation of the sibling recurrence risk ratio. Invest Ophthalmol Vis Sci 45, 2873–2878, https://doi.org/10.1167/iovs.03-115545/9/2873 (2004).
Guggenheim, J. A., Kirov, G. & Hodson, S. A. The heritability of high myopia: a reanalysis of Goldschmidt’s data. J Med Genet 37, 227–231 (2000).
Hammond, C. J., Snieder, H., Gilbert, C. E. & Spector, T. D. Genes and environment in refractive error: the twin eye study. Invest Ophthalmol Vis Sci 42, 1232–1236 (2001).
Lyhne, N., Sjolie, A. K., Kyvik, K. O. & Green, A. The importance of genes and environment for ocular refraction and its determiners: a population based study among 20–45 year old twins. Br J Ophthalmol 85, 1470–1476 (2001).
He, S., Li, X., Chan, N. & Hinton, D. R. Review: Epigenetic mechanisms in ocular disease. Mol Vis 19, 665–674 (2013).
Ramessur, R., Williams, K. M. & Hammond, C. J. Risk factors for myopia in a discordant monozygotic twin study. Ophthalmic Physiol Opt 35, 643–651, https://doi.org/10.1111/opo.12246 (2015).
Docherty, L. E. et al. Genome-wide DNA methylation analysis of patients with imprinting disorders identifies differentially methylated regions associated with novel candidate imprinted genes. Journal of medical genetics 51, 229–238, https://doi.org/10.1136/jmedgenet-2013-102116 (2014).
Pan, H. et al. Measuring the methylome in clinical samples: improved processing of the Infinium Human Methylation450 BeadChip Array. Epigenetics: official journal of the DNA Methylation Society 7, 1173–1187, https://doi.org/10.4161/epi.22102 (2012).
Brown, N. A. & Hill, A. R. Cataract: the relation between myopia and cataract morphology. Br J Ophthalmol 71, 405–414 (1987).
Chen, S. J., Lu, P., Zhang, W. F. & Lu, J. H. High myopia as a risk factor in primary open angle glaucoma. Int J Ophthalmol 5, 750–753, https://doi.org/10.3980/j.issn.2222-3959.2012.06.18 (2012).
Moisseiev, E. & Yiu, G. Retinal detachment in severe myopia. The Lancet 389, 1133, https://doi.org/10.1016/S0140-6736(16)31407-6 (2017).
Touzeau, O., Scheer, S., Allouch, C., Borderie, V. & Laroche, L. The relationship between keratoconus and axial myopia. J Fr Ophtalmol 27, 765–771 (2004).
Saw, S. M. How blinding is pathological myopia? Br J Ophthalmol 90, 525–526, https://doi.org/10.1136/bjo.2005.087999 (2006).
Bazzazi, N., Akbarzadeh, S., Yavarikia, M., Poorolajal, J. & Fouladi, D. F. HIGH MYOPIA AND DIABETIC RETINOPATHY: A Contralateral Eye Study in Diabetic Patients With High Myopic Anisometropia. Retina 37, 1270–1276, https://doi.org/10.1097/IAE.0000000000001335 (2017).
Sheth, S., Rush, R. & Narayanan, R. Retinitis pigmentosa inversa with unilateral high myopia with fellow eye optic disc pitting. Eur J Ophthalmol 21, 509–512, https://doi.org/10.5301/EJO.2011.6264 (2011).
Young, T. L. Molecular genetics of human myopia: an update. Optom Vis Sci 86, E8–E22, https://doi.org/10.1097/OPX.0b013e3181940655 (2009).
Rydzanicz, M. et al. Identification of novel suggestive loci for high-grade myopia in Polish families. Mol Vis 17, 2028–2039 (2011).
Hammond, C. J., Andrew, T., Mak, Y. T. & Spector, T. D. A susceptibility locus for myopia in the normal population is linked to the PAX6 gene region on chromosome 11: a genomewide scan of dizygotic twins. Am J Hum Genet 75, 294–304, https://doi.org/10.1086/423148 (2004).
Tang, S. M. et al. PAX6 gene associated with high myopia: a meta-analysis. Optom Vis Sci 91, 419–429, https://doi.org/10.1097/OPX.0000000000000224 (2014).
Cheng, C. Y. et al. Nine loci for ocular axial length identified through genome-wide association studies, including shared loci with refractive error. Am J Hum Genet 93, 264–277, https://doi.org/10.1016/j.ajhg.2013.06.016 (2013).
Sitek, E. J. et al. A patient with posterior cortical atrophy possesses a novel mutation in the presenilin 1 gene. PLoS One 8, e61074, https://doi.org/10.1371/journal.pone.0061074 (2013).
Hawthorne, F. Convergence of Genetic Disease Association and Ocular Expression. DUKE University library, 1–198 (2012).
Yu, C. R. et al. SOCS1 regulates CCR7 expression and migration of CD4+ T cells into peripheral tissues. J Immunol 181, 1190–1198 (2008).
Bazewicz, M. et al. Effect of SOCS1 overexpression on RPE cell activation by proinflammatory cytokines. Neurosci Lett 630, 209–215, https://doi.org/10.1016/j.neulet.2016.07.054 (2016).
Schippert, R., Schaeffel, F. & Feldkaemper, M. P. Microarray analysis of retinal gene expression in chicks during imposed myopic defocus. Mol Vis 14, 1589–1599 (2008).
Burdon, K. P. et al. Genome-wide association study for sight-threatening diabetic retinopathy reveals association with genetic variation near the GRB2 gene. Diabetologia 58, 2288–2297, https://doi.org/10.1007/s00125-015-3697-2 (2015).
Pucker, A. D. Mechanotransduction in the Ciliary Muscle, The Ohio State University (2016).
Zayats, T. The Family study of high myopia: Association studies. http://orca.cf.ac.uk/54943/1/U585332.pdf (2010).
Francisco, B. M., Salvador, M. & Amparo, N. Oxidative stress in myopia. Oxid Med Cell Longev 2015, 750637, https://doi.org/10.1155/2015/750637 (2015).
Muller, C. C. et al. PEX13 deficiency in mouse brain as a model of Zellweger syndrome: abnormal cerebellum formation, reactive gliosis and oxidative stress. Dis Model Mech 4, 104–119, https://doi.org/10.1242/dmm.004622 (2011).
Roucher-Boulez, F. et al. NNT mutations: a cause of primary adrenal insufficiency, oxidative stress and extra-adrenal defects. Eur J Endocrinol 175, 73–84, https://doi.org/10.1530/EJE-16-0056 (2016).
Deing, V. et al. Oxytocin modulates proliferation and stress responses of human skin cells: implications for atopic dermatitis. Exp Dermatol 22, 399–405, https://doi.org/10.1111/exd.12155 (2013).
Scandalios, J. G. Oxidative stress: molecular perception and transduction of signals triggering antioxidant gene defenses. Brazilian Journal of Medical and Biological Research 38, 995–1014 (2005).
Ding, C., Fan, X. & Wu, G. Peroxiredoxin 1 - an antioxidant enzyme in cancer. J Cell Mol Med 21, 193–202, https://doi.org/10.1111/jcmm.12955 (2017).
De Simoni, S., Linard, D., Hermans, E., Knoops, B. & Goemaere, J. Mitochondrial peroxiredoxin-5 as potential modulator of mitochondria-ER crosstalk in MPP+ -induced cell death. J Neurochem 125, 473–485, https://doi.org/10.1111/jnc.12117 (2013).
Ozer, U., Barbour, K. W., Clinton, S. A. & Berger, F. G. Oxidative Stress and Response to Thymidylate Synthase-Targeted Antimetabolites. Mol Pharmacol 88, 970–981, https://doi.org/10.1124/mol.115.099614 (2015).
Grant, C. M., MacIver, F. H. & Dawes, I. W. Mitochondrial function is required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. FEBS Lett 410, 219–222 (1997).
Olofsson, E. M., Marklund, S. L., Pedrosa-Domellof, F. & Behndig, A. Interleukin-1alpha downregulates extracellular-superoxide dismutase in human corneal keratoconus stromal cells. Mol Vis 13, 1285–1290 (2007).
Sharma, L. K., Lu, J. & Bai, Y. Mitochondrial respiratory complex I: structure, function and implication in human diseases. Curr Med Chem 16, 1266–1277 (2009).
Bourens, M., Fontanesi, F., Soto, I. C., Liu, J. & Barrientos, A. Redox and reactive oxygen species regulation of mitochondrial cytochrome C oxidase biogenesis. Antioxid Redox Signal 19, 1940–1952, https://doi.org/10.1089/ars.2012.4847 (2013).
Burdon, C., Mann, C., Cindrova-Davies, T., Ferguson-Smith, A. C. & Burton, G. J. Oxidative stress and the induction of cyclooxygenase enzymes and apoptosis in the murine placenta. Placenta 28, 724–733, https://doi.org/10.1016/j.placenta.2006.12.001 (2007).
Chen, X. F. et al. SIRT5 inhibits peroxisomal ACOX1 to prevent oxidative damage and is downregulated in liver cancer. EMBO Rep 19, https://doi.org/10.15252/embr.201745124 (2018).
Dai, D. F. et al. Mitochondrial oxidative stress mediates angiotensin II-induced cardiac hypertrophy and Galphaq overexpression-induced heart failure. Circ Res 108, 837–846, https://doi.org/10.1161/CIRCRESAHA.110.232306 (2011).
Herrmann, A. G. et al. Adaptive changes in the neuronal proteome: mitochondrial energy production, endoplasmic reticulum stress, and ribosomal dysfunction in the cellular response to metabolic stress. J Cereb Blood Flow Metab 33, 673–683, https://doi.org/10.1038/jcbfm.2012.204 (2013).
Ohashi, K., Kawai, S., Koshimizu, M. & Murata, K. NADPH regulates human NAD kinase, a NADP(+)-biosynthetic enzyme. Mol Cell Biochem 355, 57–64, https://doi.org/10.1007/s11010-011-0838-x (2011).
Erikson, J. M. et al. Targeting TRAF3IP2 by Genetic and Interventional Approaches Inhibits Ischemia/Reperfusion-induced Myocardial Injury and Adverse Remodeling. J Biol Chem 292, 2345–2358, https://doi.org/10.1074/jbc.M116.764522 (2017).
Erskine, L. & Herrera, E. The retinal ganglion cell axon’s journey: insights into molecular mechanisms of axon guidance. Dev Biol 308, 1–14, https://doi.org/10.1016/j.ydbio.2007.05.013 (2007).
Bray, E. R. et al. 3D Visualization of Individual Regenerating Retinal Ganglion Cell Axons Reveals Surprisingly Complex Growth Paths. eNeuro 4, https://doi.org/10.1523/ENEURO.0093-17.2017 (2017).
Birgbauer, E. Lysophospholipids in retinal axon guidance: roles and cell signaling. Neural Regen Res 10, 1067–1068, https://doi.org/10.4103/1673-5374.160091 (2015).
Oster, S. F. & Sretavan, D. W. Connecting the eye to the brain: the molecular basis of ganglion cell axon guidance. Br J Ophthalmol 87, 639–645 (2003).
Szuminski, M. & Bakunowicz-Lazarczyk, A. [Assessment of retinal ganglion cells thickness in high myopia]. Klin Oczna 114, 180–183 (2012).
Bhagat, P. R., Deshpande, K. V. & Natu, B. Utility of Ganglion Cell Complex Analysis in Early Diagnosis and Monitoring of Glaucoma using a Different Spectral Domain Optical Coherence Tomography. J Curr Glaucoma Pract 8, 101–106, https://doi.org/10.5005/jp-journals-10008-1171 (2014).
Goldberg, J. L. et al. An oligodendrocyte lineage-specific semaphorin, Sema5A, inhibits axon growth by retinal ganglion cells. J Neurosci 24, 4989–4999, https://doi.org/10.1523/JNEUROSCI.4390-03.2004 (2004).
Matsuoka, R. L. et al. Class 5 transmembrane semaphorins control selective Mammalian retinal lamination and function. Neuron 71, 460–473, https://doi.org/10.1016/j.neuron.2011.06.009 (2011).
Oster, S. F., Bodeker, M. O., He, F. & Sretavan, D. W. Invariant Sema5A inhibition serves an ensheathing function during optic nerve development. Development 130, 775–784 (2003).
Yang, X. et al. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 26, 577–590, https://doi.org/10.1016/j.ccr.2014.07.028 (2014).
Jones, L., Lopez-Bendito, G., Gruss, P., Stoykova, A. & Molnar, Z. Pax6 is required for the normal development of the forebrain axonal connections. Development 129, 5041–5052 (2002).
Grishanin, R. N. et al. Retinal TrkB receptors regulate neural development in the inner, but not outer, retina. Mol Cell Neurosci 38, 431–443, https://doi.org/10.1016/j.mcn.2008.04.004 (2008).
Bykhovskaya, Y., Gromova, A., Makarenkova, H. P. & Rabinowitz, Y. S. Abnormal regulation of extracellular matrix and adhesion molecules in corneas of patients with keratoconus. Int J Keratoconus Ectatic Corneal Dis 5, 63–70, https://doi.org/10.5005/jp-journals-10025-1123 (2016).
Seko, Y., Shimokawa, H. & Tokoro, T. Expression of bFGF and TGF-beta 2 in experimental myopia in chicks. Invest Ophthalmol Vis Sci 36, 1183–1187 (1995).
Perry, K. J. et al. The G-protein-coupled receptor, GPR84, is important for eye development in Xenopus laevis. Dev Dyn 239, 3024–3037, https://doi.org/10.1002/dvdy.22446 (2010).
Zou, L. et al. Upregulation of regulator of G-protein signaling 2 in the sclera of a form deprivation myopic animal model. Mol Vis 20, 977–987 (2014).
Chen, X. et al. Assessment of exonic single nucleotide polymorphisms in the adenosine A2A receptor gene to high myopia susceptibility in Chinese subjects. Mol Vis 17, 486–491 (2011).
Cui, D. et al. Adenosine receptor protein changes in guinea pigs with form deprivation myopia. Acta Ophthalmol 88, 759–765, https://doi.org/10.1111/j.1755-3768.2009.01559.x (2010).
Huang, L. et al. G protein subunit G gamma 13 is coexpressed with G alpha o, G beta 3, and G beta 4 in retinal ON bipolar cells. J Comp Neurol 455, 1–10, https://doi.org/10.1002/cne.10396 (2003).
Raaijmakers, J. H. & Bos, J. L. Specificity in Ras and Rap signaling. J Biol Chem 284, 10995–10999, https://doi.org/10.1074/jbc.R800061200 (2009).
Zou, H., Hu, L., Li, J., Zhan, S. & Cao, K. Cloning and characterization of a novel small monomeric GTPase, RasL10B, with tumor suppressor potential. Biotechnol Lett 28, 1901–1908, https://doi.org/10.1007/s10529-006-9176-6 (2006).
Ueta, M., Sotozono, C. & Kinoshita, S. Expression of interleukin-4 receptor alpha in human corneal epithelial cells. Jpn J Ophthalmol 55, 405–410, https://doi.org/10.1007/s10384-011-0030-6 (2011).
Ueta, M., Mizushima, K., Yokoi, N., Naito, Y. & Kinoshita, S. Expression of the interleukin-4 receptor alpha in human conjunctival epithelial cells. Br J Ophthalmol 94, 1239–1243, https://doi.org/10.1136/bjo.2009.173419 (2010).
Karnoub, A. E. & Der, C. J. Rho Family GTPases and Cellular Transformation. In: Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience (2000–2013).
Bryja, V., Cervenka, I. & Cajanek, L. The connections of Wnt pathway components with cell cycle and centrosome: side effects or a hidden logic? Crit Rev Biochem Mol Biol 52, 614–637, https://doi.org/10.1080/10409238.2017.1350135 (2017).
Shi, J., Li, F., Luo, M., Wei, J. & Liu, X. Distinct Roles of Wnt/beta-Catenin Signaling in the Pathogenesis of Chronic Obstructive Pulmonary Disease and Idiopathic Pulmonary Fibrosis. Mediators Inflamm 2017, 3520581, https://doi.org/10.1155/2017/3520581 (2017).
MacDonald, B. T., Tamai, K. & He, X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 17, 9–26, https://doi.org/10.1016/j.devcel.2009.06.016 (2009).
Fuhrmann, S. Wnt signaling in eye organogenesis. Organogenesis 4, 60–67 (2008).
Drenser, K. A. Wnt signaling pathway in retinal vascularization. Eye Brain 8, 141–146, https://doi.org/10.2147/EB.S94452 (2016).
Ma, M. et al. Wnt signaling in form deprivation myopia of the mice retina. PLoS One 9, e91086, https://doi.org/10.1371/journal.pone.0091086 (2014).
Li, M. et al. Expression of Wnt/beta-Catenin Signaling Pathway and Its Regulatory Role in Type I Collagen with TGF-beta1 in Scleral Fibroblasts from an Experimentally Induced Myopia Guinea Pig Model. J Ophthalmol 2016, 5126560, https://doi.org/10.1155/2016/5126560 (2016).
Xiong, F. et al. Altered retinal microRNA expression profiles in early diabetic retinopathy: an in silico analysis. Curr Eye Res 39, 720–729, https://doi.org/10.3109/02713683.2013.872280 (2014).
Riddell, N. & Crewther, S. G. Novel evidence for complement system activation in chick myopia and hyperopia models: a meta-analysis of transcriptome datasets. Scientific Reports 7, 9719, https://doi.org/10.1038/s41598-017-10277-2 (2017).
Matenia, D., Hempp, C., Timm, T., Eikhof, A. & Mandelkow, E. M. Microtubule affinity-regulating kinase 2 (MARK2) turns on phosphatase and tensin homolog (PTEN)-induced kinase 1 (PINK1) at Thr-313, a mutation site in Parkinson disease: effects on mitochondrial transport. J Biol Chem 287, 8174–8186, https://doi.org/10.1074/jbc.M111.262287 (2012).
Li, Z. et al. A common variant near TGFBR3 is associated with primary open angle glaucoma. Hum Mol Genet 24, 3880–3892, https://doi.org/10.1093/hmg/ddv128 (2015).
Nakamura, H., Ueda, J., Sugar, J. & Yue, B. Y. Developmentally regulated expression of Sp1 in the mouse cornea. Invest Ophthalmol Vis Sci 46, 4092–4096, https://doi.org/10.1167/iovs.05-0324 (2005).
Chang, S. W., Tsai, I. L., Hu, F. R., Lin, L. L. & Shih, Y. F. The cornea in young myopic adults. Br J Ophthalmol 85, 916–920 (2001).
Jiang, B. et al. Expression and role of specificity protein 1 in the sclera remodeling of experimental myopia in guinea pigs. Int J Ophthalmol 10, 550–554, https://doi.org/10.18240/ijo.2017.04.08 (2017).
Kloss, B. A. et al. Exome Sequence Analysis of 14 Families With High Myopia. Invest Ophthalmol Vis Sci 58, 1982–1990, https://doi.org/10.1167/iovs.16-20883 (2017).
Corredor, R. G. et al. Soluble adenylyl cyclase activity is necessary for retinal ganglion cell survival and axon growth. J Neurosci 32, 7734–7744, https://doi.org/10.1523/JNEUROSCI.5288-11.2012 (2012).
Gentleman, S., Hemmings, B. A., Russell, P. & Chader, G. J. Developmental expression of the RI subunit of cyclic AMP-dependent protein kinase in retina. Exp Eye Res 48, 717–731 (1989).
Zhang, W., Thompson, B. J., Hietakangas, V. & Cohen, S. M. MAPK/ERK signaling regulates insulin sensitivity to control glucose metabolism in Drosophila. PLoS Genet 7, e1002429, https://doi.org/10.1371/journal.pgen.1002429 (2011).
Penha, A. M., Burkhardt, E., Schaeffel, F. & Feldkaemper, M. P. Effects of intravitreal insulin and insulin signaling cascade inhibitors on emmetropization in the chick. Mol Vis 18, 2608–2622 (2012).
Dreixler, J. C. et al. Mitogen-activated protein kinase phosphatase-1 (MKP-1) in retinal ischemic preconditioning. Exp Eye Res 93, 340–349, https://doi.org/10.1016/j.exer.2010.10.011 (2011).
Cai, Z., Feng, G. S. & Zhang, X. Temporal requirement of the protein tyrosine phosphatase Shp2 in establishing the neuronal fate in early retinal development. J Neurosci 30, 4110–4119, https://doi.org/10.1523/JNEUROSCI.4364-09.2010 (2010).
Li, H. et al. Frs2alpha and Shp2 signal independently of Gab to mediate FGF signaling in lens development. J Cell Sci 127, 571–582, https://doi.org/10.1242/jcs.134478 (2014).
Fan, Q. et al. Meta-analysis of gene-environment-wide association scans accounting for education level identifies additional loci for refractive error. Nat Commun 7, 11008, https://doi.org/10.1038/ncomms11008 (2016).
Feldkaemper, M. P., Neacsu, I. & Schaeffel, F. Insulin acts as a powerful stimulator of axial myopia in chicks. Invest Ophthalmol Vis Sci 50, 13–23, https://doi.org/10.1167/iovs.08-1702 (2009).
O-Sullivan, I. et al. FoxO1 integrates direct and indirect effects of insulin on hepatic glucose production and glucose utilization. Nat Commun 6, 7079, https://doi.org/10.1038/ncomms8079 (2015).
Gao, X. et al. Genome-wide association study identifies WNT7B as a novel locus for central corneal thickness in Latinos. Hum Mol Genet 25, 5035–5045, https://doi.org/10.1093/hmg/ddw319 (2016).
Bykhovskaya, Y., Margines, B. & Rabinowitz, Y. S. Genetics in Keratoconus: where are we? Eye Vis (Lond) 3, 16, https://doi.org/10.1186/s40662-016-0047-5 (2016).
Lin, H. J. et al. Role of Chronic Inflammation in Myopia Progression: Clinical Evidence and Experimental Validation. EBioMedicine 10, 269–281, https://doi.org/10.1016/j.ebiom.2016.07.021 (2016).
Rao, P. V. & Maddala, R. The role of the lens actin cytoskeleton in fiber cell elongation and differentiation. Semin Cell Dev Biol 17, 698–711, https://doi.org/10.1016/j.semcdb.2006.10.011 (2006).
Schoenwaelder, S. M. & Burridge, K. Bidirectional signaling between the cytoskeleton and integrins. Curr Opin Cell Biol 11, 274–286 (1999).
Tajsharghi, H. et al. Recessive myosin myopathy with external ophthalmoplegia associated with MYH2 mutations. Eur J Hum Genet 22, 801–808, https://doi.org/10.1038/ejhg.2013.250 (2014).
Yun, J., Li, Y., Xu, C. T. & Pan, B. R. Epidemiology and Rb1 gene of retinoblastoma. Int J Ophthalmol 4, 103–109, https://doi.org/10.3980/j.issn.2222-3959.2011.01.24 (2011).
Iobbi, C., Korte, M. & Zagrebelsky, M. Nogo-66 Restricts Synaptic Strengthening via Lingo1 and the ROCK2-Cofilin Pathway to Control Actin Dynamics. Cereb Cortex 27, 2779–2792, https://doi.org/10.1093/cercor/bhw122 (2017).
Ströhl, F. et al. Single Molecule Translation Imaging Visualizes the Dynamics of Local β-Actin Synthesis in Retinal Axons. Scientific Reports 7, 709, https://doi.org/10.1038/s41598-017-00695-7 (2017).
Zhu, Z. C. et al. Insulin-like growth factor-1 induced activation and expression of signal transducers and activators of transcription-3 in scleral fibroblast of guinea pigs. Zhonghua Yan Ke Za Zhi 43, 1125–1129 (2007).
Mutti, D. O. et al. Candidate gene and locus analysis of myopia. Mol Vis 13, 1012–1019 (2007).
Hepei, L., Mingkun, X., Li, W. & Jin, W. Assessment of BicC family RNA binding protein 1 and Ras protein specific guanine nucleotide releasing factor 1 as candidate genes for high myopia: A case-control study. Indian J Ophthalmol 65, 926–930, https://doi.org/10.4103/ijo.IJO_625_16 (2017).
Liu, Y. et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol 31, 142–147, https://doi.org/10.1038/nbt.2487 (2013).
Zhang, C. et al. Two novel mutations of the NCSTN gene in Chinese familial acne inverse. J Eur Acad Dermatol Venereol 27, 1571–1574, https://doi.org/10.1111/j.1468-3083.2012.04627.x (2013).
Wilhelm-Benartzi, C. S. et al. Review of processing and analysis methods for DNA methylation array data. Br J Cancer 109, 1394–1402, https://doi.org/10.1038/bjc.2013.496 (2013).
Daca-Roszak, P. et al. Impact of SNPs on methylation readouts by Illumina Infinium HumanMethylation450 BeadChip Array: implications for comparative population studies. BMC Genomics 16, 1003, https://doi.org/10.1186/s12864-015-2202-0 (2015).
Gu, Z. ComplexHeatmap: Making Complex Heatmaps. R package version 1.6.0. https://github.com/jokergoo/ComplexHeatmap (2015).
Murtagh, F. & Legendre, P. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? Journal of Classification 31, 274–295, https://doi.org/10.1007/s00357-014-9161-z (2014).
Mi, H. et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45, D183–D189, https://doi.org/10.1093/nar/gkw1138 (2017).
Acknowledgements
We thank the patients for their cooperation in the study.
Author information
Authors and Affiliations
Contributions
U. Radhakrishna designed and conducted the study, analyzed the data, critically edited the manuscript. S.V. and U. Ratnmala, performed the experiments, data analysis and drafted the manuscript. J.S. and M.G. participated in the design and execution of the study, examined the clinical data, supervised the clinical data interpretation. S.S.C., K.R.J., N.K.M. and C.G. performed the statistical and bioinformatics data analysis. M.M. ascertained the patients, J.A.K. handled and prepared the DNA samples. All authors read and critically revised the manuscript and approved the final version.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Vishweswaraiah, S., Swierkowska, J., Ratnamala, U. et al. Epigenetically dysregulated genes and pathways implicated in the pathogenesis of non-syndromic high myopia. Sci Rep 9, 4145 (2019). https://doi.org/10.1038/s41598-019-40299-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-40299-x
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.