Abstract
Cetuximab is a standard-of-care treatment for RAS wild-type metastatic colorectal cancer (mCRC) but not for those harbor a KRAS mutation since MAPK pathway is constitutively activated. Nevertheless, cetuximab also exerts its effect by its immunomodulatory activity despite the presence of RAS mutation. The aim of this study was to determine the impact of polymorphism FcγRIIIa V158F and killer immunoglobulin-like receptor (KIR) genes on the outcome of mCRC patients with KRAS mutations treated with cetuximab. This multicenter Phase II clinical trial included 70 mCRC patients with KRAS mutated. We found KIR2DS4 gene was significantly associated with OS (HR 2.27; 95% CI, 1.08–4.77; P = 0.03). In non-functional receptor homozygotes the median OS was 2.6 months longer than in carriers of one copy of full receptor. Multivariate analysis confirmed KIR2DS4 as a favorable prognostic marker for OS (HR 6.71) in mCRC patients with KRAS mutation treated with cetuximab. These data support the potential therapeutic of cetuximab in KRAS mutated mCRC carrying non-functional receptor KIR2DS4 since these patients significantly prolong their OS even after heavily treatment. KIR2DS4 typing could be used as predictive marker for identifying RAS mutated patients that could benefit from combination approaches of anti-EGFR monoclonal antibodies and other immunotherapies to overcome the resistance mediated by mutation in RAS.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide and the second most common in Europe with 447,136 new cases in 20121. Specifically in Spain the incidence was 32,240 new cases. If trends continue by December 31, 2020, Spain will have approximately 37,229 new CRC cases2. Approximately 20% of patients present metastatic disease (mCRC) at diagnosis3.
Deregulation of the mitogen-activated protein kinase pathway is required for the development of CRC and it is activated by binding of growth factor ligands to the epidermal growth factor receptor (EGFR)4. Cetuximab is a monoclonal antibody that binds to the extracellular domain of EGFR, causing inhibition of the downstream signaling pathway, and is a widely used biological treatment for mCRC5. Mutations in KRAS result in constitutive activation of the pathway, since it is expected that patients with mutations in KRAS will not benefit from cetuximab treatment6. Despite this rationale, approximately 30% of mCRC patients with mutations in KRAS do benefit from cetuximab7. Understanding the mechanisms underlying the response of this subgroup to cetuximab could benefit these hard-to-treat patients.
Because cetuximab is an IgG1 monoclonal antibody it also exerts its effect by its immunomodulatory activity in part via antibody-dependent cell-mediated cytotoxicity (ADCC)8,9,10. Cetuximab stimulates ADCC activity when its constant region (Fc) binds to a Natural Killer (NK) cell receptor (CD16/FcγRIIIa) leading their own lytic activity on tumor cells9,11. Also important downstream immunological responses are possible when CD32A/ FcγRIIa receptors are present on dendritic cells and neutrophils leading to priming of tumor antigen-specific cellular immunity12. Previous reports suggested that specific polymorphic variants of FcγR are associated with the clinical outcome of cancer patients13,14. In particular, the FcγRIIa H131 allele has been shown to increase the ability of cetuximab to control disease in patients with mutated KRAS8. The role of FcγRIIIa in the ADCC induction by cetuximab in colorectal cancer patients is controversial. While in one study no statistically significant associations between the presence of FcγRIIIa and response, progression-free survival (PFS) or RAS status in mCRC patients were found15, in a subsequent study a tendency for a higher disease control rate (DCR) was shown in patients with the FcγRIIIa V-containing genotype8. Recently, a different study demonstrated that the FcγRIIIa V158V genotype was correlated with a higher ADCC16, although sample size was too small to confirm the impact of this association.
A clinical trial with KRAS wt mCRC patients treated with cetuximab + Irinotecan showed that ADCC response was not affected by BRAF or NRAS mutations17. Another phase III clinical trial with very similar patients demonstrated that ADCC response scores were higher in patients carrying FcγRIIa 131H allele vs. 131R/R18. Furthermore, FcγRIIIa 158V carriers were associated with higher cetuximab-mediated ADCC compared to 158F/F. Objective response was significantly higher in both patients carrying the FcγRIIa 131H allele and those carrying the FcγRIIIa 158V allele. However, survival analysis only revealed longer progression-free survival in patients carrying FcγRIIIa 158V allele18.
Besides FcγRIIIa, Killer immunoglobulin-like receptors (KIRs) are also essential in the immune response against tumor cells17,18. They are located on the surface of NK cells and regulate their killing function in the ADCC response19. To date, 14 functional KIR genes and two pseudogenes located in the leukocyte receptor complex on chromosome 19q13.4 have been identified. They present a very high degree of structural homology and can be classified as inhibitors or activators of the immune response depending on their cytoplasmic tails. Products of inhibitory KIR genes are characterized by long cytoplasmic tails (“L” KIR genes) and transmit inhibitory signals leading to the general shutdown of NK cell effector functions. Opposite, activating KIR proteins have short cytoplasmic tails (“S” KIR genes) and their signal promotes NK cell activity20. The immune-modulating effects depend on the balance of the number and type of receptors exposed on the cell surface21. KIR genes are organized in a highly polymorphic multigene family and their combination define two main groups of haplotypes. A haplotypes are characterized by a predominance of genes encoding inhibitory receptors. B haplotypes are more heterogeneous and generally contain several activating genes22. A recent study provides evidence of the role of specific variants of KIRs on the response to anti-EGFR treatment in solid tumors including RAS wild type advanced colorectal cancer. Due to the strong immunomodulatory activity of cetuximab we hypothesized that both KIR and FcγRIIIa receptors might influence the response of mCRC patients to cetuximab independently of RAS mutation.
This prospective multicenter clinical study aimed to determine whether polymorphisms in CD16/FcγRIIIa and/or KIR genes could enhance the clinical impact of the FcγRIIa polymorphism in mCRC patients with KRAS mutated who are treated with cetuximab.
Results
Patient characteristics and clinical outcome
Table 1 summarizes the baseline demographic and disease characteristics of the participants. Baseline blood levels were: median 46.50 ng/μL (0–2930) for CEA, median 330 UI/L (150–5882) for LDH and 2.10 mg/L (1–4) for β2-microglobulin. The median time of follow-up was 6.4 months (range: 3.8–10.2 months).
Overall, 68 patients (97.1%) experienced disease progression. Fifty-six (80.0%) patients died during the study period.
Median OS was 6.71 months with a 95% CI of (5.4–8.1) and median PFS was 2.53 with a 95% CI of (2.4–2.7).
Frequency of V158F FcγRIIIa allele and presence of KIR genes
Twenty-eight out of 70 patients (39.4%) were homozygous for FcγRIIIa-158F allele, 36 patients (52.1%) were heterozygous (V/F) and 6 (8.5%) were homozygous for FcγRIIIa-158V allele (Table 2). The minor allele frequency of this polymorphism was 34% in our series, which was in concordance with the frequency described in the Spanish population23.
With regard to KIR genes (Table 2), some of the gene variants were present in more than 90% of the patients (KIR2DL1, KIR2DL3, KIR2DL4, KIR3DL1, KIR3DL2, KIR3DL3 and KIR2DP1). Owing to this high frequency, these genes were excluded from the survival analysis. The remaining KIR genes and pseudogene (KIR3DP1) were included in the survival analysis.
Survival analysis
Regarding clinical and biochemical parameters only the number of metastatic sites was significantly correlated by univariate analysis (Table 3).
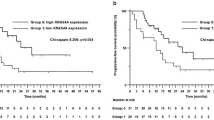
Cox regression analysis showed that only KIR2DS4 polymorphism was significantly associated with OS (Table 4). KIR2DS4 encodes two different proteins, a full length variant (KIR2DS4f) and a truncated protein with a 22-bp deletion in exon 5 (KIR2DS4d) resulting in a protein unable to attach to the cytoplasmic membrane. The genotype distribution in the study population was 50 (71.4%) homozygotes for the KIR2DS4d, four (5.7%) homozygotes for the KIR2DS4f, 10 (14.28%) heterozygotes (f/d) and six (8.6%) patients did not have any copy of the KIR2DS4 gene. As both homozygotes for the deleted variant and patients with no copy of the gene have a non-functional receptor (NFR), these patients were grouped together. A significantly lower OS was observed for patients with at least one KIR2DS4f allele compared with individuals without a functional receptor (median 4.8 months vs. 7.4 months; HR 2.27; 95% CI, 1.08–4.77; P = 0.026) (Fig. 1). Since the number of homozygotes for KIR2DS4f was very small (n = 4), it was not possible to determine the expected cumulative effect of this genotype.
Kaplan–Meier curve for overall survival according to the status of the KIR2DS4 gene. Heterozygous individuals (full variant [f] and deleted variant [d]; f/d) were compared with all patients with the non-functional receptor (homozygous deleted variant and without any copy of the gene; NFR). All patients were carriers of FcγRIIa H131 polymorphism. The median overall survival among heterozygotes for the KIR2DS4f allele was 4.8 months (95% CI, 3.45–6.08) and 7.4 months (95% CI, 5.97–8.82) for patients with NFR (P = 0.026; log-rank test).
Multivariate logistic regression model, adjusted for baseline characteristics, showed that 3 or more metastatic sites and KIR2DS4 polymorphism were independent predictors of overall survival (OS) in KRAS mutated mCRC patients carrying FcγRIIa H131 allele and receiving cetuximab (HR 3.03, P = 0.007 and HR 2.17, P = 0.045, respectively) (Table 5).
Logistic regression and disease control rate (DCR) analysis were also performed; however, no significant result was obtained (data not shown).
Discussion
Our results support with new evidences the immunomodulatory activity of cetuximab in mCRC patients regardless KRAS status. In particular, subjects carrying the non-functional receptor KIR2DS4 showed longer OS than carriers of the full-length variant.
We have previously described higher disease control rate (DCR) in KRAS mutated patients carrying FcγRIIa H131 allele; however, only a tendency was observed for patients with the FcγRIIIa V-containing genotype. Although this polymorphism is the best-studied biomarker for ADCC its clinical value is not fully confirmed. An analysis in head and neck squamous cell carcinoma, no predictive value for FcγRIIIa F158V polymorphism was detected for cetuximab efficacy24; and a study in refractory mCRC patients treated with anti-EGFR antibodies did not found significant associations between FcγRIIIa polymorphism and clinical outcome15. However, two different studies in mCRC patients with KRAS wild-type found a significant difference in outcomes among patients carrying different genotypes of this polymorphism18,25. We have evaluated this polymorphism in our cohort of KRAS mutated mCRC patients treated with cetuximab, similarly to Paez et al.15 we have observed no effect of FcγRIIIa V158F on OS or PFS.
Despite the central role of NK-cells in ADCC and the essential role of KIR receptors to modulate NK cell function, the impact of the KIRs on the mCRC clinical outcome have been essentially unexplored. A recent study demonstrated the use of genotyping KIR to predict overall survival to treatment with FOLFIRI in mCRC patients. The authors showed the absence of KIR2DS4 and 3DL1 increased complete response26. Apart from this study, there are few more analyses assessing the clinical impact of KIRs in cancer. Siebert et al.27 found higher level of ADCC and superior event-free survival in Neuroblastoma patients with haplotype B (combination of KIR genes including activating receptor genes) compared to inhibitory haplotype A (a fixed set of gene encoding for inhibitory receptors, except 2DS4). Also in Neuroblastoma patients, Forlenza et al.28 evaluated whether combinations of KIR3DL1 and HLA ligands could influence patient outcome after treatment with anti-GD2 monoclonal antibody. They evaluated a total of 245 patients and found that single KIR3DL1 did not impact survival but after assessing combinations of KIR3DL1 and HLA-B they found that when receptor and ligand did not interact at the cell surface exhibited the greatest OS and PFS. Similar results were observed by Boudreau et al.29 when investigated the impact of KIR3DL1/HLA-B combinations in 1,328 patients with acute myelogenous leukemia who underwent hematopoietic cell transplantation (HCT). Patients with weak or non-inhibiting KIR3DL1/HLA-B partnership experienced higher DFS after HCT. Recently, it has been published other study in patients with chronic myeloid leukemia treated with tyrosine kinase inhibitors (TKIs)30. The authors hypothesized that KIR and HLA polymorphisms may influence response to TKIs. They did not find association between single KIR genes or the presence of a single functional combination of an inhibitory KIR-HLA genotype with achievement of complete molecular response. However, specific alleles of KIR2DL4, KIR3DL1 and KIR2DS4 were associated with response. A study in non-small cell lung cancer showed patients with absence of KIR2DS4 had longer OS than patients who were positive31. A third study demonstrated that donor’s full-length KIR2DS4 allele is associated with lower OS rates and higher relapse incidence in patients with hematological malignancies32. Finally, a very recently study showed a protective impact of deleted KIR2DS4 on genetic predisposition to head and neck squamous cell carcinoma (HNSCC)33.
In our study, a significant survival advantage was observed for patients without functional KIR2DS4, suggesting a novel favorable and independent prognostic biomarker for overall survival in mCRC patients KRAS mutated and treated with cetuximab. KIR2DS4 encodes two different allele variants, full-length (KIR2DS4f) or deleted (KIR2DS4d). A deleted KIR2DS4 is a truncated protein without transmembrane and cytoplasmic domains compared to full-length KIR2DS4 protein. Therefore, this truncated protein is not anchored to cell membrane becoming a soluble variant21. The role of this soluble protein remains unclear but it has been suggested that the presence of this truncated protein could minimize the detrimental effects of the KIR2DS4f 33. Its function could potentially be a decoy that absorbs the available soluble HLA and thus potentially interferes with NK cell function or may be related to an alternative receptor ligand34,35. It is important to highlight that KIR2DS4 is the only activating gene within the KIR A haplotype. Therefore individuals with haplotype A being homozygous for the deleted KIR2DS4 will carry only inhibiting KIRs which has been associated with a protective role against some types of cancer such as HNSCC33. Based on our results and those of the above-mentioned studies, we can assume that the lack of functional KIR2DS4 has a positive impact on clinical outcome of cancer patients treated with anti-IgG1 therapies. In our cohort, this influence is stronger when NFR-KIR2DS4 and FcγRIIa H131 allele are combined, this effect was also observed in neuroblastoma patients treated with anti-GD2 IgG1 Ab27.
An important factor to consider was the possibility that patients with the G13D mutation skewed our analysis. However, we found no significant differences in OS (P = 0.22) or PFS (P = 0.70) in G13D patients compared to the rest of patients. It is in agreement with Segelov et al.36 who did not find statistically significant improvement in disease control at 6 months in patients with G13D mutation chemotherapy-refractory mCRC treated with cetuximab.
One of the limitations of our study that warrant consideration is that it has been performed in a limited cohort. To address whether the combination of FcγRIIa and KIR2DS4 could be a suitable predictive marker for KRAS mutated patients, it should be confirmed in large-scale prospective studies.
In conclusion, the use of FcγRIIa H131R and KIR2DS4 to identify the subset of KRAS mutated patients who might benefit from cetuximab therapy could improve the current management strategies available for these patients. Moreover, these results could explain the observed variability in efficacy of cetuximab in KRAS mutated mCRC patients and confirm the important role of ADCC-mediated toxicity to tumor cells by cetuximab.
Methods
Patients and trial design
From September 2011 through December 2013, 70 mCRC patients with KRAS mutations were prospectively enrolled in this multicenter Phase II clinical trial (registration number: NCT01450319, 07/10/2011; EudraCT Number: 2010-023580-18). Sample size was calculated using a one-sided test with a significance level of α = 0.05, assuming a recruitment period of 14 months and 12 months of follow-up and considering a minimum frequency of 30% for the KIR and FcγRIIIa-F158V polymorphisms. In order to detect an improvement of overall survival compared to historical controls with at least 80% power, it was estimated a sample size of 70 patients assuming an exponential distribution for OS, a one-sided null hypothesis H0: Median OS <= 5 months and, an actual median OS of 7 months. Primary endpoint was OS, and the secondary endpoint was progression-free survival.
This study was approved by the Institutional Ethical Committee at the University Hospital Fundacion Jimenez Diaz (authorization number EC 02-12 IIS-FJD) and accepted by the other 12 participating hospitals located across Spain. All of them were general hospitals and publically funded excepting “Clínica Universitaria of Navarra” that was private. Written informed consent was obtained from all patients before enrollment. All methods were carried out in accordance with the approved guidelines and regulations.
The patients included in this study were eligible taking into account the inclusion criteria: (A) histologically confirmed mCRC with confirmed KRAS mutation; (B) positive EGFR expression; (C) carrier of at least one histidine at position 131 in FcγRIIa; (D) aged 18 years or older; (E) Eastern Cooperative Oncology Group (ECOG) performance status of 0–2. All patients had adequate bone marrow, renal and liver functions and were refractory to at least two lines of treatment including standard chemotherapy.
Major exclusion criteria were previous anti-EGFR monoclonal antibody-based therapy, presence of brain metastases or evidence of toxicity greater than grade 1 caused by previous treatment.
After enrollment, cetuximab was administered as an intravenous dose of 500 mg/m2 of body surface area, every two weeks until unacceptable toxicity, disease progression, death or revocation of the informed consent.
Baseline measurements
Peripheral blood was collected in 5-ml tubes with EDTA. Blood levels of carcinoembryonic antigen (CEA), lactate dehydrogenase (LDH) and β2-microglobulin levels were evaluated before initiation of cetuximab therapy as they have been described as prognostic factors in colorectal cancer37,38,39. Patients were grouped according to the upper limit of the normal range (ULN) for each measure (>5 mcg/L for CEA, 333 UI/L for LDH and 3 mg/L for β2-microglobulin).
FcγRIIIa V158F polymorphism and KIRs genotyping
Genomic DNA extraction was performed using the QIAamp DNA Blood Mini Kit (Qiagen). The purified DNA was quantified with a NanoDrop 3.0 spectrophotometer (Nucliber).
The FcγRIIIa genotype was determined using the TaqMan Allelic Discrimination Assay (assay code C_25815666_10, Applied Biosystems) according to the manufacturer’s instructions. After thermal cycling, allelic determination was performed using the 7500 Fast real-time PCR instrument (Applied Biosystems). Samples of known genotype were included as the positive control.
The 17 KIR genes were determined using the KIR genotyping SSP Kit (Applied Biosystems) according to the manufacturer’s protocol. After thermal cycling the genotype-specific PCR products were resolved using 2% agarose gels and interpreted according to the manufacturer’s instructions.
Statistical Methods
Overall survival was defined as the time from enrollment until death from any cause. Progression-free survival was calculated as the time from enrollment until disease progression, or death due to any cause. In the absence of confirmation of disease progression or death, the participant was censored at the last contact, known to be alive and progression free. Those patients who started a new treatment (different from cetuximab) were censored at the date of starting the new treatment.
Univariate analysis was performed to assess the effect of genetic markers and clinical variables on the prediction of outcome. Only variables that were statistically significant by univariate analysis were considered as covariates in the multivariate Cox regression model. The survival probability was estimated using the Kaplan-Meier method, and the log-rank test was used to test the differences between the subgroups.
Data analysis was performed using the SPSS statistics version 20.0 software package.
Change history
17 May 2019
A correction to this article has been published and is linked from the HTML and PDF versions of this paper. The error has been fixed in the paper.
References
Ferlay, J. et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. International Agency for Research on Cancer, Lyon (France); 2013 [accessed 2018 January 31]. http://globocan.iarc.fr.
GLOBOCAN 2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012. ARCI: OMS; [accessed 2018 January 31] http://globocan.iarc.fr/Default.aspx.
Cartwright, T. H. Treatment decisions after diagnosis of metastatic colorectal cancer. Clin. Colorectal Cancer. 11, 155–166 (2012).
Attar, B. M., Atten, M. J. & Holian, O. MAPK activity is down-regulated in human colon adenocarcinoma:correlation with PKC activity. Anticancer Res. 16, 395–399 (1996).
Di Fiore, F., Sesboüé, R., Michel, P., Sabourin, J. C. & Frebourg, T. Molecular determinants of anti-EGFR sensitivity and resistance in metastatic colorectal cancer. Br. J. Cancer. 103, 1765–1772 (2010).
Karapetis, C. S. et al. K-ras mutations and benefit from cetuximab in ad-vanced colorectal cancer. N. Engl. J. Med. 359, 1757–1765 (2008).
Qiu, L. X. et al. Predictive and prognostic value of KRAS mutations in met-astatic colorectal cancer patients treated with cetuximab: a meta-analysis of 22 studies. Eur. J. Cancer. 46, 2781–2787 (2010).
Rodríguez, J. et al. Fc gamma receptor polymorphisms as predictive mark-ers of Cetuximab efficacy in epidermal growth factor receptor downstream-mutated metastatic colorectal cancer. Eur. J. Cancer. 48, 1774–1780 (2012).
Ferris, R. L. et al. Rationale for combination of therapeutic antibodies tar-geting tumor cells and immune checkpoint receptors: Harnessing innate and adaptive immunity through IgG1 isotype immune effector stimulation. Cancer Treat. Rev. 63, 48–60 (2017).
Trivedi, S. et al. Anti-EGFR Targeted Monoclonal Antibody Isotype Influ-ences Antitumor Cellular Immunity in Head and Neck Cancer Patients. Clin. Cancer Res. 22, 5229–5237 (2016).
Desjarlais, J. R., Lazar, G. A., Zhukovsky, E. A. & Chu, S. Y. Optimizing en-gagement of the immune system by anti-tumor antibodies: an engineer’s perspective. Drug Discov. Today. 12, 898–910 (2007).
Lee, S. C., Srivastava, R. M., López-Albaitero, A., Ferrone, S. & Ferris, R. L. Natural killer (NK):dendritic cell (DC) cross talk induced by therapeutic monoclonal antibody triggers tumor antigen-specific T cell immunity. Im-munol. Res. 50, 248–254 (2011).
Bibeau, F. et al. Impact of Fc{gamma}RIIa-Fc{gamma}RIIIa polymor-phisms and KRAS mutations on the clinical outcome of patients with meta-static colorectal cancer treated with cetuximab plus irinotecan. J. Clin. On-col. 27, 1122–1129 (2009).
Cartron, G. et al. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood. 99, 754–758 (2002).
Paez, D. et al. Immunoglobulin G fragment C receptor polymorphisms and KRAS mutations: are they useful biomarkers of clinical outcome in ad-vanced colorectal cancer treated with anti-EGFR-based therapy. Cancer Sci. 101, 2048–2053 (2010).
Negri, F. V. et al. Role of immunoglobulin G fragment C receptor polymor-phism-mediated antibody-dependent cellular cytotoxicity in colorectal cancer treated with cetuximab therapy. Pharmacogenomics J. 14, 14–19 (2014).
Lo Nigro, C. et al. Evaluation of antibody-dependent cell-mediated cytotox-icity activity and cetuximab response in KRAS wild- type metastatic colo-rectal cancer patients. World J. Gastrointest. Oncol. 8, 222–230 (2016).
Trotta, A. M. et al. Prospective Evaluation of Cetuximab-Mediated Anti-body-Dependent Cell Cytotoxicity in Metastatic Colorectal Cancer Patients Predicts Treatment Efficacy. Cancer. Immunol. Res. 4, 366–374 (2016).
Eriksson, M. et al. Inhibitory receptors alter natural killer cell interactions with target cells yet allow simultaneous killing of susceptible targets. J. Exp. Med. 190, 1005–1012 (1999).
Bakker, A. B., Phillips, J. H., Figdor, C. G. & Lanier, L. L. Killer cell inhibitory receptors for MHC class I molecules regulate lysis of melanoma cells medi-ated by NK cells, gamma delta T cells, and antigen-specific CTL. J. Immu-nol. 160, 5239–5245 (1998).
Shilling, H. G. et al. Allelic polymorphism synergizes with variable gene con-tent to individualize human KIR genotype. J. Immunol. 168, 2307–2315 (2002).
Martin, A. M., Freitas, E. M., Witt, C. S. & Christiansen, F. T. The genomic organi-zation and evolution of the natural killer immunoglobulin-like receptor (KIR) gene cluster. Immunogenetics. 51, 268–280 (2000).
Lopez-Escamez, J. A. et al. Polymorphisms of CD16A and CD32 Fcγ recep-tors and circulating immune complexes in Ménière’s disease: a case-control study. BMC Med. Genet. 12, 2 (2011).
Srivastava, R. M. et al. Cetuximab-activated natural killer and dendritic cells collaborate to trigger tumor antigen-specific T-cell immunity in head and neck cancer patients. Clin. Cancer Res. 19, 1858–1872 (2013).
Calemma, R. et al. Fc gamma receptor IIIa polymorphisms in advanced colorectal cancer patients correlated with response to anti-EGFR antibodies and clinical outcome. J. Transl. Med. 10, 232–243 (2012).
De, R. V. et al. Genetic diversity of the KIR/HLA system and outcome of patients with metastatic colorectal cancer treated with chemotherapy. PLoS One 9, e84940 (2014).
Siebert, N. et al. Neuroblastoma patients with high-affinity FCGR2A, -3A and stimulatory KIR 2DS2 treated by long-term infusion of anti-GD(2) an-tibody ch14.18/CHO show higher ADCC levels and improved event-free survival. Oncoimmunology. 5, e1235108 (2016).
Forlenza, C. J. et al. KIR3DL1 Allelic Polymorphism and HLA-B Epitopes Modulate Response to Anti-GD2 Monoclonal Antibody in Patients With Neuroblastoma. J. Clin. Oncol. 34, 2443–2451 (2016).
Boudreau, J. E. et al. KIR3DL1/HL A-B Subtypes Govern Acute Mye-logenous Leukemia Relapse After Hematopoietic Cell Transplantation. J Clin Oncol. 35, 2268–2278 (2017).
Ureshino, H. et al. Allelic Polymorphisms of KIRs and HLAs Predict Fa-vorable Responses to Tyrosine Kinase Inhibitors in CML. Cancer Immunol. Res. 6, 745–754 (2018).
He, Y., Bunn, P. A., Zhou, C. & Chan, D. KIR 2D (L1, L3, L4, S4) and KIR 3DL1 protein expression in non-small cell lung cancer. Oncotarget. 7, 82104–82111 (2016).
Burek Kamenaric, M. et al. The impact of KIR2DS4 gene on clinical out-come after hematopoietic stem cell transplantation. Hum. Immunol. 78, 95–102 (2017).
Barani, S., Khademi, B., Ashouri, E. & Ghaderi, A. KIR2DS1, 2DS5, 3DS1 and KIR2DL5 are associated with the risk of head and neck squamous cell carcinoma in Iranians. Hum. Immunol. 79, 218–223 (2018).
Maxwell, L. D., Wallace, A., Middleton, D. & Curran, M. D. A common KIR2DS4 deletion variant in the human that predicts a soluble KIR mole-cule analogous to the KIR1D molecule observed in the rhesus monkey. Tis-sue Antigens. 60, 254–258 (2002).
Katz, G. et al. MHC class I-independent recognition of NK-activating re-ceptor KIR2DS4. J. Immunol. 173, 1819–1825 (2004).
Segelov, E. et al. ICECREAM:randomised phase II study of cetuximab alone or in combination with irinotecan in patients with metastatic colorectal cancer with either KRAS, NRAS, BRAF and PI3KCA wild type, or G13D mutated tumours. BMC Cancer. 16, 339 (2016).
Giessen, C. et al. Evaluation of prognostic factors in liver-limited metastatic colorectal cancer: a preplanned analysis of the FIRE-1 trial. Br. J. Cancer. 109, 1428–1436 (2013).
Eker, B. et al. Factors affecting prognosis in metastatic colorectal cancer patients. Asian Pac. J. Cancer Prev. 16, 3015–3021 (2015).
Shrout, J. et al. beta(2)microglobulin mRNA expression levels are prognostic for lymph node metastasis in colorectal cancer patients. Br. J. Cancer. 98, 1999–2005 (2008).
Acknowledgements
The authors would like to thank all patients that took part in this clinical trial. We also want to thank all medical oncologists that carried out the recruitment and follow up of the patients and scientific people involved in the laboratory work of this study. We also thank Beatriz Quirós, Account Manager of Nature Publishing Group Iberoamérica for her corrections and improvements in the final manuscript. This clinical trial was approved and supported by Merck S.L., an affiliate of Merck KGaA, Darmstadt. Germany [research project number 2010-023580-18, date: 05-06-2014].
Author information
Authors and Affiliations
Contributions
J.G.-F. conceived and designed the research study. M.R.-R., R.R., R.V.-B. y A.P.-O. developed the methodology for KRAS status and EGFR expression. F.R. interpreted these analyses. R.G.-C., P.G.-A., E.A., E.E., R.L.-L., A.C., M.V., C.N., J.M.V., C.G.-P., J.R., I.H. and J.G.-F. collected the clinical samples. A.B.-P., A.C. and T.G.-P. analyzed and interpreted the genotyping data. A.B.-P., A.C., T.G.-P., R.G.-C., P.G.-A., E.A., E.E., R.L., A.C., M.V., C.N., J.M.V., C.G., J.R., I.H., J.L.G., J.M.-U., L. del P.-N. and J.G.-F. contributed to preparing and reviewing the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Borrero-Palacios, A., Cebrián, A., Gómez del Pulgar, M.T. et al. Combination of KIR2DS4 and FcγRIIa polymorphisms predicts the response to cetuximab in KRAS mutant metastatic colorectal cancer. Sci Rep 9, 2589 (2019). https://doi.org/10.1038/s41598-019-39291-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-39291-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.