Abstract
The objective was to delineate hepatitis C virus (HCV) epidemiology in countries of Central Asia (CA), specifically Kazakhstan, Kyrgyzstan, Tajikistan, Turkmenistan, and Uzbekistan. A systematic review was conducted guided by the Cochrane Collaboration Handbook, and reported using PRISMA guidelines. Meta-analyses were performed using DerSimonian-Laird random-effects models with inverse variance weighting. Random-effects meta-regression analyses were performed on general population studies. The systematic review identified a total of 208 HCV prevalence measures. No incidence or Turkmenistan studies were identified. Meta-analyses estimated HCV prevalence among the general population at 0.7% (95%CI: 0.7–0.8%) in Kazakhstan, 2.0% (95%CI: 1.7–2.4%) in Kyrgyzstan, 2.6% (95%CI: 1.7–3.6%) in Tajikistan, and 9.6 (95%CI: 5.8–14.2%) in Uzbekistan. Across CA, the pooled mean prevalence was 13.5% (95%CI: 10.9–16.4%) among non-specific clinical populations, 31.6% (95%CI: 25.8–37.7%) among populations with liver-related conditions, and 51.3% (95%CI: 46.9–55.6%) among people who inject drugs. Genotypes 1 (52.6%) and 3 (38.0%) were most frequent. Evidence was found for statistically-significant differences in prevalence by country, but not for a temporal decline in prevalence. CA is one of the most affected regions by HCV infection with Uzbekistan enduring one of the highest prevalence levels worldwide. Ongoing HCV transmission seems to be driven by injecting drug use and healthcare exposures.
Similar content being viewed by others
Introduction
With approximately 71 million people chronically infected worldwide, hepatitis C virus (HCV) related morbidities place a strain on healthcare systems globally1. Since the recent development of direct-acting antivirals (DAA), a breakthrough treatment which provides opportunities to reduce HCV infection and disease burden2,3, the World Health Organization (WHO) has set a target for the elimination of HCV as a public health concern by 20304,5. As such, an understanding of HCV epidemiology and risk factors for HCV infection worldwide is essential for developing targeted and cost-effective preventative and treatment interventions, to achieve the global target and eliminate HCV.
Geographically, for the purpose of this study, Central Asia (CA) encompasses five countries: Kazakhstan, Kyrgyzstan, Tajikistan, Turkmenistan, and Uzbekistan. Since independence from the Soviet Union, these countries have been undergoing difficult political, social, and economic transition6,7. The public health and healthcare infrastructure has deteriorated, resulting in a decline in life expectancy, a rising burden of diseases, and re-emergence of infectious diseases7,8. Though the region is perceived to have one of the highest HCV prevalence levels worldwide9,10, HCV epidemiology and the drivers of HCV transmission remain poorly characterized.
Our objective was to delineate HCV epidemiology in CA by (1) performing a systematic review of all available records of HCV antibody incidence and/or antibody prevalence among the different population categories, (2) pooling all HCV antibody prevalence measures in the general population to estimate the country-specific population-level HCV prevalence, (3) estimating the number of HCV infected persons across countries of CA, (4) performing a secondary systematic review of all evidence on HCV genotype information, and (5) identifying sources of between-study heterogeneity and estimate their contribution to the variability in HCV prevalence among the general population.
Materials and Methods
The methodology in this study is informed and adapted from that of the systematic reviews of the Middle East and North Africa (MENA) HCV Epidemiology Synthesis Project11,12,13,14,15,16,17,18,19. This methodology is summarized in the ensuing subsections, and additional information is available in respective publications from this project11,12,13,14,15,16,17,18,19.
Sources of data and search strategy
Literature on HCV antibody incidence and/or antibody prevalence was systematically reviewed guided by the Cochrane Collaboration Handbook20. Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines were used in reporting our results21 (Table S1). The data sources used in this study included international PubMed and EMBASE databases (up to 9th April, 2018), a Russian scientific database—Scientific Electronic Library (eLibrary.ru) (up to 9th April, 2018), and country-level reports. The search criteria was broad with no language restrictions (Fig. S1). Articles published after 1989 were included in this review, since this was the year in which HCV was first identified22,23.
Selection of studies
Duplicate publications were found and removed using the reference manager software, Endnote. Screening of the remaining unique records’ titles and abstracts were performed individually by WB and FZ. Articles that were considered relevant or potentially relevant underwent full-text screening, using our inclusion and exclusion criteria. The references of all full-text articles and literature reviews also underwent screening to find any further relevant articles that may have been overlooked.
Inclusion and exclusion criteria
The inclusion and exclusion criteria used were adapted from that of the MENA HCV Epidemiology Synthesis Project systematic reviews11,12,13,14,15,16,17,18,19. The inclusion criteria consisted of any document reporting HCV antibody incidence and/or antibody prevalence in populations from Kazakhstan, Kyrgyzstan, Tajikistan, Turkmenistan, and Uzbekistan, based on primary data, and of any language. The exclusion criteria included studies conducted before 1989, studies that referred to HCV as non-A non-B hepatitis, case series, case reports, commentaries, editorials, letters to editors, and literature reviews. All records underwent a secondary independent screening for data on HCV genotypes, regardless of whether they reported HCV antibody incidence and/or antibody prevalence.
In the following subsections, the term ‘report’ is used to refer to any document with an outcome measure of interest, while a ‘study’ refers to stratifications of a specific outcome measure. As such, a single report may contribute multiple studies, and multiple reports of the same study (outcome measure) were recognized as duplicates and considered as one study.
Extraction and analyses of data
Data from all reports considered relevant were extracted by WB and FZ. Data from all reports were subsequently double extracted by SM to ensure consistency and minimize errors in extracted information. Extracted information included study details (author, year of publication, title, and journal), location of study, year(s) of data collection, study design, sampling method, risk population, number of participants included in the study, number of participants tested, type and name of serological test used to test for HCV, and the primary outcome (HCV incidence or/and HCV prevalence). Rounding HCV prevalence measures to two decimal places was conducted if they were below 1%, while the remaining measures were rounded to only one decimal place. When available, HCV ribonucleic acid (RNA) data were also extracted. All studies identified in the secondary independent screening for genotype information were also extracted into a separate extraction file. Risk factors for HCV infection were extracted if they were statistically-significant through multivariable meta-regression. Extracted data were classified into population categories according to exposure risk to HCV infection, as presented in Fig. 1. The classification scheme was based on existing literature10,24,25, and earlier reviews of HCV prevalence11,12,13,14,15,16,17,18,19.
Quantitative assessment
HCV prevalence reports with a minimum of 50 participants were categorized and reported in our reporting tables by risk population. Meta-analyses of HCV prevalence measures were performed by risk population and country for all studies with at least 25 participants. In reports where HCV prevalence was reported for mixed-country samples, the study was included only in meta-analyses for CA as a region. In reports that included prevalence measures but no reported sample size, a sample size of 300 was imputed and the study was included in the review and meta-analyses. This sample size was deemed reasonable and conservative, given that the median sample size of included studies with a reported sample size was 348.
HCV prevalence for the total sample size was replaced with stratified prevalence whenever a minimum of 25 participants were available for each stratum. Stratifications were included based on a predefined order, where nationality was prioritized, then sex, year, region, and finally age. To avoid duplication one final stratification for each study was included.
Freeman-Tukey type arcsine square-root transformation was used to stabilize the variance of HCV prevalence measures26. DerSimonian-Laird random-effects model was used to pool HCV prevalence (with inverse variance weighting). This model assumes a normal distribution of true effect sizes (that is HCV prevalence) across studies, and takes into account true heterogeneity as well as random chance effects across studies27.
Heterogeneity measures were also assessed. All forest plots were visually assessed and the Cochran’s Q test was performed, with a p-value of <0.10 indicating statistically strong evidence27,28. The I² measure and its confidence interval were assessed27. The prediction interval was also calculated to estimate the range in which HCV prevalence of 95% of future studies will fall27,29.
The number of HCV antibody-positive persons in each country was determined by multiplying the country-specific pooled mean HCV antibody prevalence estimate by the population size in each country. This was subsequently multiplied by the pooled mean fraction of HCV RNA positivity in antibody-positive persons (also commonly referred to as the “viremic rate”30,31), to derive the number of HCV chronically-infected persons. The United Nations World Population Prospects database32 was used to obtain the population size of each country.
Since potential issues have been identified with the Freeman-Tukey type arcsine square-root transformation33, a sensitivity analyses was performed to confirm the validity of our results in which the generalized linear mixed models (GLMM) method was used to perform meta-analyses.
A proportion of the general population data were on blood donors, a population typically including only healthy adults. Sensitivity analysis was performed to determine whether excluding blood donors could impact the pooled mean HCV prevalence estimate in the general population. This sensitivity analysis was done for each country separately, and for CA as a whole.
Based on established methodology20, univariable and multivariable random-effects meta-egressions were performed to assess country-level associations with HCV prevalence and the sources of between-study heterogeneity in the general population. Variables included in the univariable models included country, general population subpopulations, study site, sample size (<100 or ≥100), sampling method (probability-based or non-probability-based), year of publication, and year of data collection. Variables with a p-value of <0.1 were included in the multivariable model. Variables were deemed significant in the final multivariable meta-regression if they had a p-value of <0.05.
For each country and the whole CA, the frequency of each genotype was calculated. Individuals who were positive for mixed genotypes contributed separately to the number of each of the identified genotypes. The Shannon Diversity Index (H) was determined to assess the diversity of genotypes, with a higher score (out of 1.95) indicating more diversity34.
The meta package35 on R version 3.4.336 was used to perform the meta-analyses. The metan command on STATA 1337 was used to perform meta-regressions.
Qualitative analysis
Using the Cochrane approach to surmise risk of bias (ROB), the quality of HCV incidence and/or prevalence measures was evaluated. Based on three quality domains, studies were classified into either low or high ROB. These domains included HCV ascertainment (biological assay or otherwise), sampling method (probability-based or non-probability-based), and response rate (≥80% of the target sample size was reached or otherwise).
Studies with information missing for any of the three domains were classified with unclear ROB for that specific domain. Studies in which the reported HCV measures were acquired from patients’ medical records, or from individuals voluntarily visiting facilities where routine blood screening is performed, were considered as having low ROB on strictly the response rate domain.
Studies with at least 100 participants were classified as having high precision, as informed by previous studies11,12,13,14,15,16,17,18,19.
Results
Search results
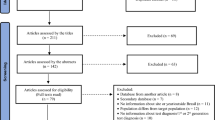
Figure 2, adapting the PRISMA flow diagram21, shows the process by which studies were selected into this systematic review. A total of 771 citations were identified: 95 from PubMed, 129 from Embase, and 547 from the Scientific Electronic Library (eLibrary.ru). A total of 99 unique reports underwent full-text screening, after duplicates were removed and titles and abstracts were screened. From these, 69 reports were removed, the reasons for which are stated in Fig. 2. Eighteen reports were added to the systematic review from gray literature/unpublished data, and from screening of references of full-text articles and reviews. Finally, 47 reports qualified for inclusion in this systematic review, yielding no incidence measure and 208 prevalence measures.
Flow chart of the process by which articles were selected for inclusion in this systematic review of hepatitis C virus (HCV) incidence and prevalence in Central Asia, adapted from the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) 2009 guidelines21.
In the secondary systematic review, all 771 citations were screened for HCV genotype information. After duplicates were removed and titles and abstracts of all unique reports were screened, 35 reports underwent full-text screening. Finally, 6 reports qualified for inclusion in this secondary systematic review (Fig. S2).
HCV antibody prevalence overview
We present here a synthesis of HCV prevalence in each country of CA. The 208 HCV prevalence measures included 67 measures from Kazakhstan, 96 from Kyrgyzstan, 20 from Tajikistan, 23 from Uzbekistan, and 2 from mixed-country samples. No study was identified from Turkmenistan (Fig. S8A).
Overall
In CA, HCV prevalence ranged from 0.5–13.1% among the general population, with a median of 2.0%. This included blood donors (number of studies (n) = 9), with HCV prevalence ranging from 0.9–7.3%, with a median of 1.5%; 0.5–6.0% among pregnant women (n = 9), with a median of 1.5%; and 0.7–13.1 among other general populations (n = 19), with a median of 2.0% (Table 1).
HCV prevalence ranged from 0.0–50.0% among populations at intermediate risk, with a median of 13.2%. These included prisoners, with HCV prevalence ranging from 7.0–50.0%, with a median of 32.0%; 0.0–28.0% among sex workers (male, female, unspecified), with a median of 11.0%; and 2.0–6.2% among HCW, with a median of 2.7% (Table S2).
HCV prevalence ranged from 4.0–40.3% among non-specific clinical populations, with a median of 8.5%. These included hospitalized populations with HCV prevalence ranging from 5.9–33.3%, with a median of 8.0%; and HIV patients with HCV prevalence ranging from 10.5–40.3%, with a median of 21.8% (Table 2).
HCV prevalence ranged from 16.6–46.0% among populations with liver-related conditions, with a median of 26.8; and 17.0–90.2% among PWID, with a median of 51.0% (Table 3).
Country-level
In Kazakhstan, HCV prevalence ranged from 0.7–5.1% among the general population, with a median of 0.9%; and 2.0–50.0% among populations at intermediate risk, with a median of 29.0%. Only one study was identified among non-specific clinical populations, with an HCV prevalence of 40.3% in HIV patients38. HCV prevalence ranged from 23.8–40.4% in populations with liver-related conditions, with a median of 26.6%; and 43.3–90.2% among PWID, with a median of 60.3%.
In Kyrgyzstan, HCV prevalence ranged from 0.8–5.0% among the general population, with a median of 2.0%; 0.0–35.0% among populations at intermediate risk, with a median of 7.0%; and 4.0–33.3% among non-specific clinical populations, with a median of 8.0%. No studies were identified among populations with liver-related conditions. HCV prevalence ranged from 17.0–60.4% among PWID, with a median of 46.4%.
In Tajikistan, HCV prevalence ranged from 0.5–7.3% among the general population, with a median of 3.9%. Only two studies were conducted among populations at intermediate risk39, with HCV prevalence of 4.2% among sex workers (male, female, unspecified)40, and 6.2% among HCW41. Only one study was conducted on non-specific clinical populations, with an HCV prevalence of 32.1% in HIV patients41. Only two studies were conducted on populations with liver-related conditions, reporting an HCV prevalence of 46.0%42 and 36.0%39. HCV prevalence ranged from 24.9–67.1% among PWID, with a median of 32.6%.
No studies were identified from Turkmenistan.
In Uzbekistan, HCV prevalence among the general population ranged from 6.4–13.1%, with a median of 6.5%; 9.2–18.8% among populations at intermediate risk, with a median of 11.9%; 16.5–29.2% among non-specific clinical populations, with a median of 26.9%; 16.6–41.9% among populations with liver-related conditions, with a median of 23.4%; and 20.9–63.8% among PWID, with a median of 51.7%.
Pooled mean HCV prevalence estimates and estimated number of HCV infected persons
The national population-level HCV prevalence for each country, based on pooling the general population measures, were estimated at: 0.7% (95%CI: 0.7–0.8%) in Kazakhstan, 2.0% (95%CI: 1.7–2.4%) in Kyrgyzstan, 2.6% (95%CI: 1.7–3.6%) in Tajikistan, and 9.6% (95%CI: 5.8–14.2%) in Uzbekistan. For all countries combined, the pooled mean HCV prevalence was estimated at 2.2% (95%CI: 1.9–2.6%). Figure S8B maps the pooled mean HCV prevalence estimates for CA.
Across CA, the estimated pooled mean HCV prevalence was 14.6% (95%CI: 12.8–16.5%) among populations at intermediate risk; 13.5% (95%CI: 10.9–16.4%) among non-specific clinical populations; 31.6% (95%CI: 5.8–37.7%) among populations with liver-related conditions; and 51.3% (95%CI: 46.9–55.6%) among PWID. The results of pooling these populations for each country separately can be found in Table 4.
Forest plots for the meta-analyses can be found in the Supplementary Material (Figs S3–S7). In the majority of meta-analyses, statistically significant heterogeneity was observed (Cochrane’s Q statistic’s p-value was always <0.0001; Table 4). Most of the variation across studies was due to variation in effect size (HCV prevalence) rather than chance (I2 > 59.0%). The prediction intervals ranged from narrow to wide for the different meta-analyses. Collectively, the heterogeneity measures indicated high heterogeneity in HCV prevalence in each country and risk population category.
Too few studies reported HCV RNA viremic rate in the general population to warrant calculation of the pooled mean viremic rate for CA. Accordingly, the pooled mean viremic rate of 67.6% for MENA was used in calculating chronic-infection prevalence and the number of chronically-infected persons. This choice is justified by the fact that this measure is a biological measure that (in principle) should be largely independent of the region31, and given that CA and MENA countries are both developing countries. The highest number of chronically-infected persons was found in Uzbekistan at 2.1 million, followed by Tajikistan at 160,068, Kazakhstan at 87,087, and Kyrgyzstan at 82,917.
In sensitivity analyses, the GLMM meta-analyses confirmed similar results for all risk populations (Table S3). Also in sensitivity analyses, after blood donor data were excluded, population-level HCV prevalence was overall similar across countries, and in CA as a whole (Table S4).
Meta-regressions and sources of heterogeneity
The results of the meta-regression for the general population is presented in Table 5. In the univariable meta-regression analyses, country, study site, sample size, and year of data collection were significant predictors (p-value < 0.1), and therefore were included in the final multivariable analysis. Notably, sampling method (probability-based versus non-probability-based) had no effect on observed HCV prevalence.
Study site and year of data collection lost significance (p-value > 0.05) in the multivariable analysis—only country and sample size remained statistically significant. Relative to Kazakhstan, the prevalence in Kyrgyzstan, Tajikistan, and Uzbekistan was higher with an adjusted odds ratio (AOR) of 2.0 (95%CI: 1.1–3.4), 2.8 (95%CI: 1.4–5.6), and 10.0 (95%CI: 4.6–21.7), respectively. Sample size (>100) was associated with lower HCV prevalence, with an AOR of 0.4 (95%CI: 0.1–1.0). Notably, the AOR for year of data collection was 1.0 (95%CI: 1.0–1.1)—there was thus no evidence for declines in HCV prevalence with time. The model explained 51.4% of the variability in HCV prevalence.
HCV RNA prevalence
Our search identified only four HCV RNA measures, all of which were reported among HCV antibody-positive individuals: 39.2% in a study on a general population43, 100% in a study on HIV patients38, 100% in a study on chronic hepatitis patients42, and 70.5% in a study on liver cirrhosis patients42.
HCV genotypes
HCV genotype information was available in six studies with a total of 382 HCV RNA positive individuals (Table S5). Only 0.5% of individuals were infected with multiple genotypes, while the remaining majority were infected with a single genotype. No genotype information was available for Kyrgyzstan and Turkmenistan.
The highest proportions of infections for each HCV genotype in CA as a whole were for genotype 1 at 52.6% and genotype 3 at 38.0%, followed by genotype 2 at 9.4%. Genotypes 4, 5, 6, and 7 were not identified. Genotype diversity tended towards being low, but varied across CA, with the highest diversity observed in Kazakhstan (H = 1.04 out of 1.95; 53.7%), followed by Uzbekistan (H = 0.85 out of 1.95; 43.6%), and Tajikistan (H = 0.54 out of 1.95; 27.5%). Collectively in CA, genotype diversity was rather low (H = 0.93 out of 1.95; 47.7%).
HCV risk factors
Only two studies reported statistically-significant risk factors for HCV infection after controlling for confounders. In Tajikistan, among PWID, daily injection, history of incarceration, and living/working outside of Tajikistan in the past 10 years, were associated with HCV infection44. In Kazakhstan, among a general population, tattooing and (unexpectedly) towel sharing were reported as associated with HCV infection45.
Quality assessment
Table S6 summarizes the results of the ROB assessment performed on HCV prevalence measures. The majority of measures were of high precision (94.7%), with a sample size ≥100. Most measures were of low risk of bias in the HCV ascertainment domain, with 99% being based on biological assays, and 1% being based on self-reporting. Though most of the studies reported the name of the biological assay used to assess HCV antibody prevalence, the majority of studies (90%) did not explicitly report the generation of the assay. Among studies reporting the generation of the used assay, all used the more sensitive and specific 3rd generation enzyme-linked immunosorbent assays (ELISA) tests. The majority of studies employed non-probability-based sampling, and were characterized by a high response rate.
To summarize, 100% of studies had low ROB based on at least one ROB domain, 65.0% of studies had low ROB based on at least two ROB domains, and 13.4% of studies had low ROB based on all three ROB domains. No study had high ROB based on two or three ROB domains. In all, the quality assessment indicates reasonable though not optimal study quality.
Discussion
We presented, to our knowledge, the first systematic review and synthesis of HCV epidemiology in CA, a region perceived to be heavily affected by this infection9,10. Our results indicated that HCV antibody prevalence varies across countries of CA, ranging from 0.7% in Kazakhstan to 9.6% in Uzbekistan (Table 4 and Fig. S8). Accordingly, HCV prevalence in Uzbekistan is considerably higher than global levels, and one of the highest worldwide9,10. This finding is of concern considering that Uzbekistan is also the most populous country in CA, with 32 million inhabitants46, and a country struggling with a weakened healthcare system since the collapse of the Soviet Union6. With an estimated 2.1 million chronically-infected persons, >80% of all chronically-infected persons in CA reside in Uzbekistan. Notably, Uzbekistan has also the highest rate of HIV among all countries in this region47, and a main mode of transmission appears to be injecting drug use, a shared mode of transmission with HCV.
Remarkably, HCV prevalence does not appear to be decreasing with time in CA (Table 5), contrary to global trend48,49. This may in part be reflective of the majority of studies from this region being reported more recently, with approximately 85% of all studies included in this review being from the last decade.
High HCV antibody prevalence was observed across all risk populations (Tables 1–3 and S2), and more so for PWID, HIV patients, and prisoners, suggesting a major role for injecting drug use in infection transmission. HCV antibody prevalence was also high in populations with liver-related conditions, suggesting a major role that HCV plays in liver disease burden in CA.
Strikingly, no studies were identified among high risk clinical populations such as haemodialysis, haemophilia, and thalassemia patients—the role of healthcare in transmission remains uncertain. However, the relatively high HCV antibody prevalence in non-specific clinical populations (Table 2), and HCV epidemiology in other soviet-era-related countries9,10,49,50, suggest that healthcare could be a major mode of exposure, at least in earlier decades.
Subregional disparities in quality of healthcare services may have also contributed to the heterogeneity in HCV prevalence across CA50. For example, in Uzbekistan, it appears (anecdotally) that there is an excessive practice of medical and non-medical invasive procedures, such as blood transfusions and bloodletting, in addition to poor infection control51, inadequate blood screening43,51,52, and use of unsafe medical injections50,51, all of which are probable causes for the high HCV prevalence in this country, as has been observed in other developing countries53,54,55. Furthermore, the challenging political climate in Uzbekistan has prevented the introduction of up-to-date healthcare guidelines and effective approaches to reduce HCV transmission in healthcare settings51,52.
While no genotype information was available for Kyrgyzstan and Turkmenistan, pooled analysis on data from Kazakhstan, Tajikistan, and Uzbekistan suggest that HCV genotype 1 (53% of infections) and genotype 3 (38%) are the major circulating strains, but with strong presence of genotype 2 (9%; Table S5). While genotype 1 is common globally10,56, its major presence may reflect healthcare-related exposures, given the frequency of identifying this genotype in clinical populations in CA42,43,57,58, as well as the global association between this genotype and healthcare exposures56. The major presence of genotype 3 may be due to injecting drug use being a major driver of incidence, given the global association between this genotype and injecting drug use56, or may just reflect a sub-regional pattern—genotype 3 is the main circulating strain in each of Afghanistan13,59 and Pakistan59, both of which are neighbouring countries of CA.
The pooled mean HCV prevalence in PWID indicated that over half of this population is already exposed to HCV (Table 4), similar to global trends60,61,62. Notably, CA is geographically located along drug trafficking routes originating from Afghanistan7,63, and is believed to have one of the highest rates of injecting drug use in the world64. These factors further corroborate a major role for injecting drug use in transmission. Furthermore, with the high HCV prevalence found in prisoners (Table S2), incarceration could be influential in HCV transmission dynamics, just as in other regions62,65. The high prevalence observed among sex workers (male, female, unspecified; Table S2) may suggest also high rates of injecting drug use in these populations, as supported by HIV biobehavioral surveillance data—sexual and injecting networks could be overlapping hotspots of both HCV and HIV transmission7,64.
Despite progress in characterizing HCV epidemiology in CA, our study highlights key challenges and limitations to establishing a satisfactory understanding. Evidence varied by country, with no data identified from Turkmenistan (Fig. S8A). No data were available for high risk clinical populations, though healthcare could be a major mode of exposure, as it is in other soviet-era-related countries9,10,50, and in countries with similar stage of development, e.g. in MENA11,12,13,14,15,16,17,18,19,66. No data was identified for community-related exposures, e.g. informal healthcare, but such exposures could play a role as seen in other regions67. There was an insufficient number of studies reporting HCV RNA prevalence in CA, a measure that informs assessment of chronic-infection prevalence, as antibody prevalence reflects both current infection as well as past infection (that is persons who spontaneously cleared the infection or were treated)68.
Most available studies were descriptive—few had analytic epidemiologic designs where risk factors and modes of exposure could be ascertained. Most studies employed non-probability-based sampling, however, results of the meta-regressions indicated this had no effect on HCV prevalence in the general population, and therefore may not have limited the representativeness of reviewed data in our study. There was high heterogeneity in HCV prevalence measures (Table 4), but most heterogeneity (for the general population) was subsequently explained—differences by country were the main driver of prevalence variation (Table 5). A small-study effect was observed, with studies with a smaller sample size reporting higher HCV prevalence (Table 5), thereby potentially limiting the representativeness of reviewed data. HCV genotype data was relatively sparse, with no studies identified from Kyrgyzstan and Turkmenistan.
In spite of these limitations, a key strength of our study is that we identified a substantial number of studies, including a volume of unpublished data, in a significantly affected, but poorly understood region, thereby facilitating a synthesis of evidence and identification of knowledge gaps. A priority in addressing these gaps is to carry out nationally-representative probability-based and population-based surveys in each of these countries. Such surveys can yield a precise estimate of HCV prevalence, delineation of spatial variability in infection exposure, identification of modes of transmission, and assessment of HCV knowledge and attitudes, as has been done in recent years in other countries, e.g. in Egypt15,69,70,71,72,73,74 and Pakistan11,75,76,77.
Conclusion
In context of inadequate and underfunded healthcare systems8,52, CA is one of the most affected regions by HCV infection. Uzbekistan, in particular, appears to be enduring one of the highest prevalence levels worldwide. HCV transmission appears to be driven by injecting drug use and healthcare exposures, with no evidence for declines in prevalence in recent years. Genotypes 1 and 3 are the most frequently-circulating strains, with some presence for genotype 2.
Our findings inform HCV response for public health planning, health service provision, development of HCV policy guidelines, and implementation of HCV programming to reduce transmission and associated disease burden. Achieving HCV elimination in CA by 2030 can only be accomplished by aggressive action and commitment, given the extent of challenges. There is an urgent need for expansion of affordable HCV testing and treatment for key populations, and targeted control based on settings of exposure. In context of this region being heavily affected by injecting drug use and the global opioid epidemic, harm reduction services must incorporate HCV services and be accessible to all PWID, by being expanded to all relevant settings, such as prisons. Nationally-representative probability-based population-based surveys must be conducted to precisely delineate HCV epidemiology in these countries and address the knowledge gaps, as identified in this study. Improving infection control in healthcare facilities is also warranted, such as through updating (otherwise outdated) clinical guidelines for healthcare workers52, and adopting safety-engineered syringes as recommended by WHO28,29.
References
WHO. Global Hepatitis Report, 2017. Online at: http://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/ (2017).
Brown, R. S. & Gaglio, P. J. Scope of worldwide hepatitis C problem. Liver transplantation. 9(11), S10–S3 (2003).
Ayoub, H. & Abu‐Raddad, L. J. Impact of treatment on hepatitis C virus transmission and incidence in Egypt: A case for treatment as prevention. Journal of viral hepatitis. 24(6), 486–95 (2017).
World Health Organization. Global health sector strategy on viral hepatitis 2016–2021. Towards ending viral hepatitis. Online at: http://www.who.int/hepatitis/strategy2016-2021/ghss-hep/en/ (2016).
World Health Organization. Combating hepatitis B and C to reach elimination by 2030: advocacy brief. Online at: http://apps.who.int/iris/handle/10665/206453 (2016).
Batsaikhan, U. & Dabrowski, M. Central Asia—twenty-five years after the breakup of the USSR. Russian. Journal of Economics. 3(3), 296–320 (2017).
Thorne, C., Ferencic, N., Malyuta, R., Mimica, J. & Niemiec, T. Central Asia: hotspot in the worldwide HIV epidemic. The Lancet Infectious Diseases. 10(7), 479–88 (2010).
McKee, M., Healy, J. & Falkingham, J. Health care in central Asia. Open University Press Buckingham (2002).
Mohd Hanafiah, K., Groeger, J., Flaxman, A. D. & Wiersma, S. T. Global epidemiology of hepatitis C virus infection: New estimates of age‐specific antibody to HCV seroprevalence. Hepatology. 57(4), 1333–42 (2013).
Gower, E., Estes, C., Blach, S., Razavi-Shearer, K. & Razavi, H. Global epidemiology and genotype distribution of the hepatitis C virus infection. Journal of hepatology. 61(1), S45–S57 (2014).
Al-Kanaani, Z., Mahmud, S., Kouyoumjian, S. & Abu-Raddad, L. J. The epidemiology of hepatitis C virus in Pakistan: systematic review and meta-analyses Royal Society of Open. Science. 5(4), 180257 (2008).
Mahmud, S., Akbarzadeh, V. & Abu-Raddad, L. J. The epidemiology of hepatitis C virus in Iran: systematic review and meta-analyses. Scientific reports. 8(1), 150 (2018).
Chemaitelly, H., Mahmud, S., Rahmani, A. M. & Abu-Raddad, L. J. The epidemiology of hepatitis C virus in Afghanistan: systematic review and meta-analysis. International Journal of Infectious Diseases. 40, 54–63 (2015).
Mohamoud, Y. A., Mumtaz, G. R., Riome, S., Miller, D. & Abu-Raddad, L. J. The epidemiology of hepatitis C virus in Egypt: a systematic review and data synthesis. BMC infectious diseases. 13(1), 288 (2013).
Kouyoumjian, S., Chemaitelly, H. & Abu-Raddad, L. J. Characterizing hepatitis C virus epidemiology in Egypt: systematic reviews, meta-analyses, and meta-regressions. Scientific Reports. 8(1), 1661 (2018).
Chemaitelly, H., Chaabna, K. & Abu-Raddad, L. J. The epidemiology of hepatitis C virus in the Fertile Crescent: systematic review and meta-analysis. PloS one. 10(8), e0135281 (2015).
Fadlalla, F. A., Mohamoud, Y. A., Mumtaz, G. R. & Abu-Raddad, L. J. The epidemiology of hepatitis C virus in the maghreb region: systematic review and meta-analyses. PloS one. 10(3), e0121873 (2015).
Mohamoud, Y. A., Riome, S. & Abu-Raddad, L. J. Epidemiology of hepatitis C virus in the Arabian Gulf countries: Systematic review and meta-analysis of prevalence. International Journal of Infectious Diseases. 46, 116–25 (2016).
Chaabna, K., Kouyoumjian, S. P. & Abu-Raddad, L. J. Hepatitis C virus epidemiology in Djibouti, Somalia, Sudan, and Yemen: systematic review and meta-analysis. PloS one. 11(2), e0149966 (2016).
Higgins, J. P. & Green, S. Cochrane handbook for systematic reviews of interventions: John Wiley & Sons (2011).
Moher, D., Liberati, A., Tetzlaff, J., Altman, D. G. & Group, P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS medicine 6(7), e1000097 (2009).
Choo, Q.-L. et al. Isolation of a cDNA Clone Derived from a Blood-Borne Non-A, Non-B Viral Hepititis Genome. Science. 244(4902), 359 (1989).
Kuo, G. et al. IAn assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 244(4902), 362–4 (1989).
Nelson, P. et al. IThe epidemiology of viral hepatitis among people who inject drugs: results of global systematic reviews. Lancet. 378(9791), 571 (2011).
Lavanchy D. Evolving epidemiology of hepatitis C virus. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 17(2):107–15. Epub 2010/11/26, https://doi.org/10.1111/j.1469-0691.2010.03432.x. PubMed PMID: 21091831.(2011)
Freeman, M. F. & Tukey, J. W. Transformations related to the angular and the square root. The Annals of Mathematical Statistics. 607–11. (1950)
Borenstein, M., Hedges, L. V., Higgins, J. P. T. & Rothstein, H. R. Front Matter, in Introduction to Meta-Analysis. Chichester, UK: John Wiley & Sons, Ltd (2009).
Higgins, J. P., Thompson, S. G., Deeks, J. J. & Altman, D. G. Measuring inconsistency in meta-analyses. BMJ: British Medical Journal. 327(7414), 557 (2003).
Higgins, J., Thompson, S. G. & Spiegelhalter, D. J. A re‐evaluation of random‐effects meta‐analysis. Journal of the Royal Statistical Society: Series A (Statistics in Society). 172(1), 137–59 (2009).
Harfouche, M. et al. IHepatitis C virus viremic rate in the Middle East and North Africa: Systematic synthesis, meta-analyses, and meta-regressions. PloS one. 12(10), e0187177 (2017).
Ayoub, H. H., Chemaitelly, H., Omori, R. & Abu-Raddad, L. J. Hepatitis C virus infection spontaneous clearance: Has it been underestimated? International Journal of Infectious Diseases. 75, 60–6 (2018).
United Nations, Department of Economic and Social Affairs, Population Division. World Population Prospects: The 2017 Revision. Available at: https://population.un.org/wpp/DataQuery/ (2017).
Schwarzer G., Abu-Raddad L. J., Chemaitelly H. & Rucker G. Seriously misleading result using inverse of Freeman-Tukey double arcsine transformation in meta-analysis of single proportions (under review).
Shannon, C. E. A mathematical theory of communication. ACM SIGMOBILE Mobile Computing and Communications Review. 5(1), 3–55 (2001).
Schwarzer, G. General Package for Meta-Analysis. Version 4.1–0. Available at: http://cran.r-project.org/web/packages/meta/meta.pdf.
Team RC. R: A language and environment for statistical computing (2013).
StataCorp. Stata Statistical Software: Release 13. College Station, TX: StataCorp LP. (2013).
Begaidarova, R. et al. IAssessment of the severity of immunodeficiency in patients of Asian ethnicity with HIV/HCV co-infection. Georgian medical news. 254, 53–6 (2016).
Mirojov, G. et al. IPrevalence, etiological factors, survival rates, lifetime and causes of death of patients with liver cirrhosis in Tajikistan. Hepatology International. 7:S529, https://doi.org/10.1007/s12072-013-9429-0. PubMed PMID: 71308897 (2013).
Алсалих Н, Сычев Д, Подопригора И. Распространенность вирусного гепатита С среди трудовых мигрантов, прибывающих в Российскую Федерацию. Электронный научно-образовательный вестник «Здоровье и образование в XXI веке». 2017;19(7).
Azimova, S. M., Dustov, A. & Tursunov, R. Chronic hepatitis “C” in Tajikistan [Russian]. Avicenna Tajik State Medical University. 2(63), 82–9 (2015).
Khan, A. et al. Epidemiological and clinical evaluation of hepatitis B, hepatitis C, and delta hepatitis viruses in Tajikistan. J Med Virol. 80(2):268–76. Epub 2007/12/22, https://doi.org/10.1002/jmv.21057. PubMed PMID: 18098133 (2008).
Kurbanov, F. et al. Hepatitis C virus molecular epidemiology in Uzbekistan. J Med Virol. 69(3):367–75. Epub 2003/01/15, https://doi.org/10.1002/jmv.10298. PubMed PMID: 12526047 (2003).
Beyrer, C. et al. Characterization of the emerging HIV type 1 and HCV epidemics among injecting drug users in Dushanbe, Tajikistan. AIDS Res Hum Retroviruses. 25(9):853–60. Epub 2009/08/20, https://doi.org/10.1089/aid.2008.0206. PubMed PMID: 19689193; PubMed Central PMCID: PMCPmc2858926 (2009).
Nurgalieva, Z. Z., Hollinger, F. B., Graham, D. Y., Zhangabylova, S. & Zhangbylov, A. Epidemiology and transmission of hepatitis B and C viruses in Kazakhstan. World Journal of Gastroenterology. 13(8):1204–7. PubMed PMID: 2007184913 (2007).
United Nationas DESA/Population Division. Revision of World Urbanization Prospects. Online at: https://population.un.org/wup/ (2018).
UNAIDS Country factsheets: Uzbekistan Online at: http://www.unaids.org/en/regionscountries/countries/uzbekistan (2017).
Kamal, S. M. Acute hepatitis C: a systematic review. The American journal of gastroenterology. 103(5), 1283 (2008).
Polaris Observatory. Online at: http://cdafound.org/polaris-hepC-dashboard/.
Batash, S., Khaykis, I., Raicht, R. F. & Bini, E. J. High prevalence of hepatitis C virus infection among immigrants from the former Soviet Union in the New York City metropolitan area: results of a community-based screening program. Am J Gastroenterol.;103(4):922–7. Epub 2008/04/10, https://doi.org/10.1111/j.1572-0241.2008.01789.x. PubMed PMID: 18397420 (2008).
Ruzibakiev, R. et al. Risk factors and seroprevalence of hepatitis B virus, hepatitis C virus, and human immunodeficiency virus infection in Uzbekistan. Intervirology. 44(6), 327–32 (2001).
Marquez, P. V. Blood services in Central Asian health systems: a clear and present danger of spreading HIV/AIDS and other infectious diseases. 2008.
Simonsen, L., Kane, A., Lloyd, J., Zaffran, M. & Kane, M. Unsafe injections in the developing world and transmission of bloodborne pathogens: a review. Bulletin of the World Health Organization. 77(10):789–800. Epub 1999/12/11. PubMed PMID: 10593026; PubMed Central PMCID: PMCPMC2557743 (1999).
Hutin, Y. J., Hauri, A. M. & Armstrong, G. L. Use of injections in healthcare settings worldwide, 2000: literature review and regional estimates. Bmj. 327(7423), 1075 (2003).
Kane, A., Lloyd, J., Zaffran, M., Simonsen, L. & Kane, M. Transmission of hepatitis B, hepatitis C and human immunodeficiency viruses through unsafe injections in the developing world: model-based regional estimates. Bulletin of the World Health Organization. 77(10), 801 (1999).
Messina, J. P. et al. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 61(1), 77–87 (2015).
Viazov, S. et al. Hepatitis C virus genotypes in different regions of the former Soviet Union (Russia, Belarus, Moldova, and Uzbekistan). J Med Virol.;53(1):36–40. Epub 1997/09/23. PubMed PMID: 9298730 (1997).
Kurbanov, F. et al. Molecular epidemiology and interferon susceptibility of the natural recombinant hepatitis C virus strain RF1_2k/1b. Journal of Infectious Diseases. 198(10):1448–56, https://doi.org/10.1086/592757. PubMed PMID: 2008506636. (2008).
Mahmud, S. et al. Hepatitis C virus genotypes in the Middle East and North Africa: Distribution, diversity, and patterns. Journal of medical virology. 90(1), 131–41 (2018).
Mumtaz, G. R. et al. HIV among people who inject drugs in the Middle East and North Africa: systematic review and data synthesis. PLoS medicine. 11(6), e1001663 (2014).
Mumtaz, G. R., Weiss, H. A., Vickerman, P., Larke, N. & Abu-Raddad, L. J. Using hepatitis C prevalence to estimate HIV epidemic potential among people who inject drugs in the Middle East and North Africa. AIDS (London, England). 29(13), 1701 (2015).
Larney, S. et al. Incidence and prevalence of hepatitis C in prisons and other closed settings: results of a systematic review and meta‐analysis. Hepatology. 58(4), 1215–24 (2013).
Drugs UNOo, Asia CROfC. Illicit Drug Trends in Central Asia. 2008.
El-Bassel, N., Strathdee, S. A. & El Sadr, W. M. HIV and people who use drugs in central Asia: confronting the perfect storm. Drug & Alcohol Dependence. 132, S2–S6 (2013).
Heijnen, M., Mumtaz, G. R. & Abu-Raddad, L. J. Status of HIV and hepatitis C virus infections among prisoners in the Middle East and North Africa: review and synthesis. Journal of the International AIDS Society.19(1) (2016).
Harfouche, M. et al. Epidemiology of hepatitis C virus among hemodialysis patients in the Middle East and North Africa: systematic syntheses, meta-analyses, and meta-regressions. Epidemiology & Infection. 145(15), 3243–63 (2017).
Mahmud, S., Kouyoumjian, S. P., Al Kanaani, Z., Chemaitelly, H. & Abu-Raddad, L. J. Individual-level key associations and modes of exposure for hepatitis C virus infection in the Middle East and North Africa: a systematic synthesis. Annals of epidemiology (2018).
Chen, S. L. & Morgan, T. R. The natural history of hepatitis C virus (HCV) infection. International journal of medical sciences. ;3(2):47–52. Epub 2006/04/15. PubMed PMID: 16614742; PubMed Central PMCID: PMCPMC1415841. (2006)
El-Zanaty F, W. A. Egypt Demographic and Health Survey 2008. Cairo: Egyptian Ministry of Health, National Population Council, El-Zanaty and Associates, and ORC Macro. https://dhsprogram.com/publications/publication-fr220-dhs-final-reports.cfm (2008).
Cuadros, D. F., Branscum, A. J., Miller, F. D. & Abu‐Raddad, L. J. Spatial epidemiology of hepatitis C virus infection in Egypt: analyses and implications. Hepatology. 60(4), 1150–9 (2014).
Miller, F. D. & Abu-Raddad, L. J. Evidence of intense ongoing endemic transmission of hepatitis C virus in Egypt. Proceedings of the National Academy of Sciences. 107(33):14757–62 (2010).
Chemaitelly, H., Abu-Raddad, L. J. & Miller, F. D. An apparent lack of epidemiologic association between hepatitis C virus knowledge and the prevalence of hepatitis C infection in a national survey in Egypt. PloS one. 8(7), e69803 (2013).
Benova, L., Awad, S. F., Miller, F. D. & Abu‐Raddad, L. J. Estimation of hepatitis C virus infections resulting from vertical transmission in Egypt. Hepatology. 61(3), 834–42 (2015).
Guerra, J., Garenne, M., Mohamed, M. & Fontanet, A. HCV burden of infection in Egypt: results from a nationwide survey. Journal of viral hepatitis. 19(8), 560–7 (2012).
Umar, M. et al. Hepatitis C in Pakistan: a review of available data. Hepatitis monthly. 10(3), 205 (2010).
Qureshi, H., Bile, K., Jooma, R., Alam, S. & Afridi, H. Prevalence of hepatitis B and C viral infections in Pakistan: findings of a national survey appealing for effective prevention and control measures. Eastern Mediterranean Health Journal. 16, S15 (2010).
Benova, L., Awad, S. F. & Abu‐Raddad, L. J. Estimate of vertical transmission of Hepatitis C virus in Pakistan in 2007 and 2012 birth cohorts. Journal of viral hepatitis (2017).
Skorikova, S. V., Burkitbaev, Z. K., Savchuk, T. N. & Zhiburt, E. B. [Prevalence and incidence of infections among blood donors in Astana]. Vopr Virusol. 60(1):34–6. Epub 2015/05/30. PubMed PMID: 26021072 (2015).
El-Bassel, N. et al. Implications of mobility patterns and HIV risks for HIV prevention among migrant market vendors in Kazakhstan. Am J Public Health. 101(6):1075–81. Epub 2011/04/16. doi: 10.2105/ajph.2010.300085. PubMed PMID: 21493929; PubMed Central PMCID: PMCPmc3093276 (2011).
Джумагалиева А, Оракбай Л, Омарова М, Шуратов И. Анализ эпидемиологических параметров гепатита а в ряде регионов Казахстана. Современные проблемы науки и образования. (1–1). (2015).
Khasenova, G. The results of sentinel epidemiological surveillance among pregnant women in Kazakhstan, 2006. In Russian: Результаты дозорного эпидемиологического надзора среди беременных женщин, Казахстан, 2006 год. Almaty 2007.
Data from Kazakhstan Blood Center. 2015.
Tashtemirov, K., Imangazinov, S., Tashtemirova, O. & Egoshin, V. Donation and some problems of defect donor’s blood. Georgian medical news. 261, 80–8 (2016).
Mamaev, T. M. The results of sentinel surveillance of HIV infection among pregnant women, Kyrgyzstan, 2005. In Russian: Результаты дозорного эпидемиологического надзора за ВИЧ инфекцией среди беременных женщин, Кыргызстан, 2005. Republican AIDS Association, Osh Oblast AIDS Centre. Bishkek, 2006. Provided by the WHO Country Office Kyrgyzstan.
Djumagulova, A. Ş. IKA. The situation on viral hepatitis in the Kyrgyz Republic [Russian] (2016).
Baimakhanov, Z. et al. editors. Liver transplantation in Kazakhstan: current status. Transplant International; 2017: Wiley 111 Rivert St, Hoboken 07030–5774, NJ USA.
Bahovadinov, B., Tretyakova, A., Aripova, D. & Edalieva, C. Prevalence of markers of hemotransmitted infections among donors of Tajikistan. Vox Sanguinis.;99:272, https://doi.org/10.1111/j.1423-0410.2010.01343-2.x. PubMed PMID: 70237438 (2010).
Abdurashit, R., Shakhnoza, N. & Yakov, A. Results of sentinel surveillance of HIV infection amongst pregnant women in Tajikistan 2005–2007. Tajikistan, www.ncc.tj/images/DEN/pregnant%20_eng.pdf (2008).
Ruzibakiev R. et al. Risk factors and seroprevalence of hepatitis B virus, hepatitis C virus, and human immunodeficiency virus infection in uzbekistan. Intervirology. 44(6):327–32. Epub 2002/01/24. PubMed PMID: 11805437 (2001).
Berger, S. Infectious Diseases of Uzbekistan: GIDEON Informatics Inc (2015).
Ni, Y. Q. et al. Clinical epidemiological analysis of 3602 cases of primary liver cancer in Xinjiang. [Chinese]. Chinese Journal of Oncology. 34(5):374–7, https://doi.org/10.3760/cma.j.issn.0253-3766.2012.05.012 PubMed PMID: 2012347911 (2012).
Nersesov, A. V. et al. Epidemiological and clinical characteristics of hepatocellular carcinoma in Kazakhstan. Hepatology International. ;11 (1 Supplement 1):S388. PubMed PMID: 614580527 (2017).
Deryabina, A., Patnaik, P., Gwynn, C. & El-Sadr, W. M. Sexual transmission of HIV and possible underreporting of drug use in Kazakhstan. Topics in Antiviral Medicine. 23:474. PubMed PMID: 72119923 (2015).
Zhussupov, B. et al. Study of behaviors associated with HIV infection, STI and viral hepatitis among injecting drug users in Temirtau and Karaganda, Republic of Kazakhstan. Central Asia Office (2007).
Gilbert, L. et al. Couple-Based HIV Prevention for Injecting Drug Users in Kazakhstan: A Pilot Intervention Study. Journal of Prevention & Intervention in the Community. 38(2), 162–76, https://doi.org/10.1080/10852351003640914 (2010).
El-Bassel, N. et al. Effects of a couple-based intervention to reduce risks for HIV, HCV, and STIs among drug-involved heterosexual couples in Kazakhstan: a randomized controlled trial. J Acquir Immune Defic Syndr. 2014;67(2):196–203. Epub 2014/07/06. PubMed PMID: 24991973; PubMed Central PMCID: PMCPmc4162759, https://doi.org/10.1097/qai.0000000000000277.
El-Bassel, N. et al. HIV risks among injecting and non-injecting female partners of men who inject drugs in Almaty, Kazakhstan: implications for HIV prevention, research, and policy. Int J Drug Policy. ;25(6):1195–203. Epub 2014/02/22. PubMed PMID: 24556208, https://doi.org/10.1016/j.drugpo.2013.11.009 (2014).
El-Bassel, N. et al. HIV among injection drug users and their intimate partners in Almaty, Kazakhstan. AIDS Behav.17(7):2490-500. Epub 2013/04/25, https://doi.org/10.1007/s10461-013-0484-2. PubMed PMID: 23612942 (2013).
T Zabransky, V. Mravcik (Eds.), The Regional Report on Drug Situation in Central Asia [Регионaльный обзор о нaркоситуaции в Центрaльной Азии] (1st ed.), Prague ResAd (2013).
Soliev, A. Analysis on epidemiological situation and responses based on second generation sentinel surveillance system among injecting drug users, Tajikistan, Kazakhstan, Kyrgyzstan, 2006–2009. Presented at the regional conference: HIV infection epidemic in Central Asia: further development of epidemiological surveillance; Almaty, May 18–19 (2010).
Ganina, L. Y., Elizarieva, L. & Kaspirova, A. Report: Overview of the epidemiological situation of HIV in the Republic of Kazakhstan in 2013–2015. Republican Center on Prevention and Control of AIDS.
Rosenkranz, M. et al. Assessment of health services for people who use drugs in Central. Asia: findings of a quantitative survey in Kazakhstan and Kyrgyzstan. Harm reduction journal. 13(1), 3 (2016).
Drew, R. & Choudhri, Y. Assessment of HIV/AIDS surveillance in the Europe and Eurasia Region. USAID (2005).
Kurbanov, F. et al. Human immunodeficiency virus in Uzbekistan: epidemiological and genetic analyses. AIDS Res Hum Retroviruses. 19(9):731–8. Epub 2003/10/31, https://doi.org/10.1089/088922203769232520. PubMed PMID: 14585203 (2003).
Inogamov, Z. I. The results of sentinel surveillance of HIV infection among injecting drug users in 14 sentinel sites of the Republic of Uzbekistan, 2007. Presented at: State of the HIV epidemic in the Republic of Uzbekistan Results of the SS in, Tashkent, Uzbekistan, 18–21 August 2008 (2007).
Acknowledgements
This publication was made possible by NPRP grant 9-040-3-008 from the Qatar National Research Fund (a member of Qatar Foundation). The statements made herein are solely the responsibility of the authors. The authors are also grateful for infrastructure support provided by the Biostatistics, Epidemiology, and Biomathematics Research Core at Weill Cornell Medicine-Qatar.
Author information
Authors and Affiliations
Contributions
Welathanthrige S.P. Botheju, Fawzi Zghyer and Sarwat Mahmud conducted the systematic review of the literature, data retrieval, extraction, and analysis, and wrote the first draft of the paper. Assel Terlikbayeva and Nabila El-Bassel contributed data and participated in the drafting of this article. Laith J. Abu-Raddad conceived and led the design of the study, analyses, and drafting of the article. All authors read, amended, and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Botheju, W.S.P., Zghyer, F., Mahmud, S. et al. The epidemiology of hepatitis C virus in Central Asia: Systematic review, meta-analyses, and meta-regression analyses. Sci Rep 9, 2090 (2019). https://doi.org/10.1038/s41598-019-38853-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-38853-8
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.