Abstract
The role of angiopoietin-like 3 (ANGPTL3) in blood lipid levels, cardiovascular disease risk, and glucose metabolism has received wide attention. This study aimed to examine whether rs11207997 in ANGPTL3 is associated with a 10-year risk of diabetes mellitus (DM) and if the association is modified by the consumption of certain food groups or nutrients. A prospective cohort study was designed using the Ansan–Ansung data of the Korean Genome and Epidemiology Study (n = 7,358; age ≥40 years at baseline). Participants with the T allele of rs11207997, particularly TT homozygotes, had lower triglyceride (TG) and total cholesterol levels than those with CC. There was no association with fasting blood glucose or other biochemical parameters. ANGPTL3 mRNA was positively associated with circulating TG levels and blood pressure (all p < 0.05). Cox proportional hazard models showed that the rs11207997 T allele is associated with a lower risk of DM after adjusting for covariates (hazard ratio: 0.90, 95% confidence interval: 0.812–0.998, p = 0.046). Furthermore, the association between rs11207997 and the risk of DM was modified by dietary factors. These associations were no longer statistically significant when additionally adjusted for baseline TG, a potential mediator. Our data suggest that genetic variation of rs11207997 in the ANGPTL3 gene is associated with risk of DM, possibly through contributing to a lifelong set point of TG.
Similar content being viewed by others
Introduction
Angiopoietin-like proteins (ANGPTLs) represents a family of proteins that are involved in various physiological and pathological processes1 including lipid and glucose metabolism2, inflammation3, hematopoietic stem cell activity4, and cancer cell invasion5. Among the eight ANGPTLs discovered to date1, ANGPTL3 plays an important role in the regulation of plasma triglyceride6 and cholesterol levels, mainly via reversible inhibition of lipoprotein lipase (LPL) activity6,7,8. Recently, ANGPTL3 has been considered an emerging target of novel drug therapy for cardiovascular disease (CVD)6,7,8. Deficiency of ANGPTL3 is associated with hypolipidemia and reduced risk of CVD in Caucasians9,10,11. Although the precise mechanism is not yet fully understood, ANGPTL3 is thought to inhibit the activity of LPL and endothelial lipase, thereby increasing blood lipid levels12. ANGPTL3 levels are also associated with insulin sensitivity and glucose metabolism12,13. High levels of hepatic ANGPLT3 mRNA and protein have been observed in insulin-resistant and insulin-deficient mice13. Reportedly, loss-of-function mutations of ANGPTL3 in Italians result in lower levels of plasma insulin and glucose as well as homeostatic model assessment of insulin resistance (HOMA-IR)12.
However, the results of studies examining the involvement of ANGPTL3 in metabolic regulation are inconsistent, possibly due to differences in age and race of the study participants. For example, it has been shown that circulating ANGPTL3 levels are associated with fasting insulin levels and HOMA-IR but not with triglyceride (TG) or cholesterol levels in Korean boys and girls14, whereas the association has not been investigated in Korean adults. In Chinese boys and girls, rs1748195 polymorphisms in the ANGPTL3 gene are not associated with blood lipid levels, including TG, total cholesterol (TC), high-density lipoprotein cholesterol (HDLC), or low-density lipoprotein cholesterol (LDLC)15. In contrast, among the participants of the Healthy Lifestyle in Europe by Nutrition in Adolescence study, both adolescents and adults with the T minor allele of rs11207997 in ANGPTL3 had lower HDLC and apolipoprotein A-1 levels than those carrying two C alleles16. However, no further investigations regarding the influence of the genetic variation of rs11207997 in ANGPTL3 on glucose metabolism and clinical endpoints including diabetes mellitus (DM) risk have been reported.
Based on the reported associations between ANGPLT3 and metabolic profiles, we hypothesized that rs11207997 in ANGPTL3 is associated with life-long metabolic exposure to DM. In this study, using data from the Ansan–Ansung (urban–rural) cohort with a median follow-up of 10 years, we prospectively examined the associations of rs11207997 with DM risk among 7,358 Korean older adults. We also investigated whether the association is modified by the consumption of certain food groups previously reported to significantly affect metabolic status and the risk of DM.
Results
Characteristics of study participants
After a median follow-up of 9.8 years, 1,168 participants (15.9%) were diagnosed with DM (Table 1). Participants who developed DM within 10 years were more likely to be smokers, possibly due to the higher proportion of male individuals, were older, and had a higher mean body mass index (BMI), systolic blood pressure (SBP), diastolic blood pressure (DBP), TG, TC, LDLC, fasting blood glucose (FBG), hemoglobin A1c (HbA1c), HOMA-IR and lower HDLC at baseline than non-DM participants. The genotype distribution for rs11207997 (ANGPTL3) polymorphisms was as per the Hardy–Weinberg equilibrium. The minor allele (T) frequency was approximately 0.221 in all participants. The genetic variants of rs11207997 in ANGPTL3 were significantly associated with circulating TG and TC levels (Table 2). Participants with the T minor allele had lower TG and TC levels (CC: 154.6 ± 1.2, CT: 147.8 ± 1.5, TT: 140.5 ± 3.5 mg/dL for TG, p < 0.001; CC: 190.2 ± 0.5, CT: 189.7 ± 0.7, TT: 185.5 ± 1.7 mg/dL for TC, p = 0.029).
ANGPTL3 mRNA expression in LCLs is associated with circulating TG levels and BP
A linear regression analysis was performed in a subset of 62 healthy participants (24 males and 38 females, mean age: 54.5 ± 1.2 years) to determine if ANGPTL3 mRNA expression is associated with metabolic parameters and BP. Upregulated ANGPTL3 expression in lymphoblastoid cell lines (LCLs) was positively correlated with circulating TG levels (r = 0.454, p = 0.001), SBP (r = 0.294, p = 0.033), and DBP (r = 0.425, p = 0.002) after adjusting for age, sex, location, BMI, total energy intake, physical activity, education level, and drinking status (Fig. 1). However, ANGPTL3 mRNA expression was not correlated with glycemic parameters (i.e., FBG, HOMA-IR, and HbA1c) or other circulating lipid levels (i.e., TC, LDLC, and HDLC) (data not shown).
Relationship between ANGPTL3 mRNA expression (-ΔCt) and circulating triglyceride levels (mg/dL) or blood pressure (mmHg). r: correlation co-efficient; P: p-value. Data were tested by partial correlation analyses adjusted for age, sex, location, body mass index, total energy intake, physical activity, education level, and drinking status §log-transformed for the analysis.
Variants of rs11207997 are associated with the risk of DM
To investigate the effect of rs11207997 genotype on the risk of developing DM, Cox proportional regression was performed with adjustments for potential covariates (i.e., age, sex, location, total energy intake, physical activity, education level, and smoking and drinking status). The presence of the T minor allele was associated with a 10% lower risk of DM (hazard ratio (HR) = 0.901, 95% confidence interval (CI) = 0.812–0.998, p = 0.046; Table 3). When we evaluated the contribution of circulating TG levels, a potential intermediate variable, in the association between rs11207997 and DM, by additionally adjusting for them in the models, the association was attenuated and no longer statistically significant (HR = 0.936, 95% CI = 0.845–1.036, p = 0.200).
Association between rs11207997 genotype and the risk of DM is modulated by food consumption patterns
Table 4 presents the association between rs11207997 genotype and the risk of DM according to food group consumption quartiles (Q1–Q4). The median values of fruit intake quartiles were 3.6, 9.6, 16.8, and 28.6 servings/week, respectively. Among the participants in the Q2 group, T minor allele carriers had a lower HR for DM than CC homozygotes (HR: 0.795, p = 0.032). Regarding vegetable intake, the mean intake of participants in the Q2 group (39.2 servings/week; range: 32.6–45.3 servings/week) was slightly lower than that recommended for Koreans (≥6 servings/d17 or ≥42 servings/week). Among participants in this group, HR for DM was lower when carrying the T allele (HR: 0.793, p = 0.030). However, an association between genotype and the risk of DM was not observed when intake was above or below Q2 for vegetables. Regarding unprocessed meat consumption, T minor allele carriers in the Q4 group had a lower HR for DM than C major allele homozygotes (HR: 0.808, p = 0.042). Unprocessed meat consumption in the study participants was generally low, and the Q4 group consumed approximately 1 serving/d. As for sodium intake, the mean intake in the Q1 group (1600 mg/d) was greater than the recommended intake for Koreans (1500 mg/d)17. The maximum intake of participants in the Q1 group exceeded the “goal” for sodium intake (≤2000 mg/d17). However, the T allele in the Q2 group, but not in other quartiles, was associated with a lower HR for DM (mean: 2500 mg/d, range: 2100–2830 mg/d, HR: 0.804, p = 0.036). Among the participants consuming 0.06–0.99 servings/week of milk (Q2), the T minor allele carriers had a significantly lower HR for DM (HR: 0.800, p = 0.300).
Discussion
The present study demonstrates that the minor allele of rs11207997 in ANGPTL3 is associated with a lower incidence of DM, and the genetic effect on the risk of DM can be modified by dietary factors, such as the intake of fruits, vegetables, unprocessed meat, sodium, and milk, in older Korean adults. Compared with CC homozygotes, participants with the T allele of rs11207997 had lower TG and TC levels at baseline, and lower risk of DM. This genotype effect on DM risk is possibly mediated through chronically reduced levels of circulating TG. The inverse association between T minor allele of rs11207997 and DM was restricted to those with Q2 intakes of fruits, vegetables, milk, or sodium and those with relatively high (Q4) intakes of unprocessed meat.
The specific role of rs11207997 in ANGPTL3 function and the risk of DM has not yet been elucidated. In our participants, the protective effect of the T minor allele of rs11207997 on DM-related biochemical markers (i.e., FBG, HbA1c, and HOMA-IR) was not present at baseline, but T minor allele carriers had a lower risk of DM in our 10-year follow-up. The function of rs11207997 on glucose metabolism may be partly explained by the report by Wang et al.18. Mice lacking ANGPTL3 had a lower uptake of VLDL-TG, but fat mass was preserved by a 10-fold increase in glucose uptake and de novo synthesis of white adipose tissue18. That is, the genetic variants of rs11207997 in ANGPTL3, which regulates lipid metabolism (TG and TC), may consecutively affect glucose uptake and fat accumulation in adipose tissue, resulting in future alteration in glycemic status, insulin sensitivity, and DM development. In our participants, circulating TG and TC levels at baseline were lower in participants carrying the T allele and TG was positively associated with ANGPTL3 mRNA levels, regardless of genotype, and the genotypic associations were attenuated when additionally adjusted for TG. Further research is required to clarify the underlying mechanism of the genetic variants of rs11207997 on DM-related metabolic status.
In our Korean older adult population, the effect of the genetic variants of rs11207997 on the risk of DM was also modulated by the amounts of intakes of certain types of food. Diet is associated with CVD risk, possibly through its effect on lipid and glucose metabolic profiles, systemic inflammation19, and DM20. We observed lower HRs for DM in T minor allele carriers of rs11207997 at or near the recommended intakes for fruits, vegetables, and sodium. Regarding fruit and vegetable intakes, the effect of genetic variants on the risk of DM was observed in participants in the Q2 group, which encompasses the recommended intakes for Koreans, whereas the risk of DM in those with intakes above or below the recommendations was not associated with genotype. An association between genotype and the risk of DM was evident in participants with sodium intakes slightly above the recommendations of the World Health Organization and Dietary Reference Intakes for Koreans (<2000 mg/d), which is well below the mean sodium intake of Koreans (~4800 mg/d21). Caution is required in the interpretation of data regarding sodium intake. Analyzed intake data may not accurately reflect the participants’ actual intake because the Standard Tables of Food Composition of Korea22 was utilized to assess sodium intake, whereas sodium use may vary greatly depending on the habits of those who prepare and cook food. Thus, food intake may affect the genetic risk of DM, but the results of this study may not apply to populations with lower sodium intake.
No specific dietary recommendations for Koreans on milk alone or unprocessed meat are available. Milk intake in our population was very low (mean intake: 1.0 serving/week), compared with the Korean recommendation of 1–2 servings of dairy products/day. Calcium intake may not be the critical factor for the interaction effect between milk intake and genotype on the risk of DM in the present study. Milk is a good source of calcium and has been reported to affect metabolic status in association with certain single nucleotide polymorphisms (SNPs)23. However, approximately 14 mg of Ca/d is provided from milk when intake is within Q2 (mean intake: 0.5 servings/week), from which a significant association between rs11207997 genotype and risk of DM was observed. It seems more likely that the interaction effect between milk intake and genotype on the risk of DM is due to other factors in milk (i.e., lactose, protein, fat, phosphorus, potassium, and other nutrients) rather than calcium alone24. In contrast, we found that the T allele of rs11207997 decreases the HR for DM when unprocessed meat consumption is relatively high (Q4). Beef, pork, chicken, and processed meat consumption were not associated with metabolic syndrome risk in Koreans using Korea National Health and Nutrition Examination Survey (KNHANES) data25, but the associations may have been masked by genotype. Meat is a good source of cholesterol and saturated fat, in addition to protein, vitamins, and minerals. Although no dietary guidelines are provided for unprocessed meat alone, the mean unprocessed meat intake in the Q4 group was approximately one quarter of the current recommendations for meat, fish, eggs, and legume intakes for Koreans (4 servings/day17). Therefore, the results of the present study may not agree with results in populations with higher milk or meat consumption.
The food intake of participants in our study, reported from a food frequency questionnaire (FFQ) of 103 items, was similar to that reported in previous reports that used different methods to assess food intake21,23,26. Mean fruit intake reported from 24-hour recalls of the KNHANES 2013, a nationally representative database of non-institutionalized Koreans, was 219 g/d for adults 50–64 years old and 147 g/d for adults ≥65 years old27. This equals approximately 1–2 servings/day or 7–14 servings/week. (One serving of fruit is defined by the energy content, 50 kcal, which equals 150 g for watermelon and strawberries, and 100 g for most other fresh fruit and all fruit juices17.) The median fruit intake in our study population was similar (13 servings/week). Regarding mean vegetable consumption in adults 50–64 and ≥65 years old was 374 g/d and 310 g/d, respectively, when assessed by a 24-hour recall in the KNHANES26, which equals approximately 31.5–49 servings/week. (One vegetable serving size is 50 kcal of vegetables17, which is approximately 70 g for most vegetables and ≤40 g for burdock, seaweeds, and mushrooms in the Korean diet.) Similarly, the median vegetable intake in our study population was 45.3 servings/week. Milk intake in our population was slightly lower than those of the Health Examinees participants. We were unable to find reports on unprocessed meat consumption in different Korean cohorts. Still, there is little evidence that food consumption of our study population could have deviated from that of normal Korean older adults.
Our study has some limitations. First, the 10-year follow-up in adults ≥40 years old may not be adequate to assess the long-term effect of exposure. The Korea Centers for Disease Control and Prevention (KCDC) is continuing to collect follow-up data from these adults, which may be analyzed in the future. In contrast, because the onset of type 2 DM commonly occurs around 40 years of age, the interaction between food intake and genetics needs to be examined earlier to prevent or alleviate the risk of DM. By excluding participants with DM at baseline, we may have underestimated the association and interaction by eliminating a large population that may potentially prevent or delay the onset of DM by altering food intake according to one’s genetic variation. However, due to the lack of younger participants at baseline in the Korean Genome and Epidemiology Study (KoGES) database, we were unable to assess the genotype association with DM incidence modified by food/nutrient intake in younger populations (<40 years). Second, though FFQs are the most commonly used measurement for average food intake, the accuracy of FFQs may be limited due to its reliance on memory. However, in our prospective cohort study, dietary intakes were assessed prior to DM diagnosis and thus the resulting measurement error is most likely independent of outcome (i.e., non-differential) and may have only attenuated the association. Third, the function of ANGPTL3 has been reported to be closely associated with ANGPTL8, which is known to promote the ability of ANGPTL3 to bind and inhibit LPL in vitro28. However, we were unable to investigate the interaction between ANGPTL3 rs11207997 and ANGPTL8 due to limited data. Despite these limitations, this study evaluated associations with clinical endpoint (vs. intermediate biochemical endpoints) and disease incidence (vs. prevalence) using a prospective cohort design. The prospective nature of this 10-year follow-up study reduces disease-related recall bias and further supports a possible causal effect.
In summary, we found that among older Koreans, T allele carriers of rs11207997 in ANGPTL3 had a lower risk of DM, possibly through a lifelong set point of circulating TG. In addition, among those who habitually consume fruits, vegetables, or sodium at or near the recommended intakes, adults with the T allele have lower HRs for DM than CC carriers. The association of the genetic variants of rs11207997 with the risk of DM may be masked when food or sodium intakes are far from the recommended intakes.
Methods
Study population
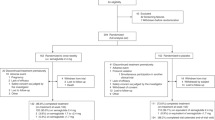
Data for this study were obtained from the Ansan–Ansung cohort, which was part of the KoGES. Detailed information about the KoGES has been published in a previous study29. Briefly, the baseline survey of the Ansan–Ansung cohort was performed in 2001–2002 with a total of 10,030 participants aged 40–69 years, and follow-up survey data were collected biennially. Person-years for each participant was estimated from the baseline examination until the date of the first DM diagnosis at the hospital, the date of the last contact, or the date of the last follow-up visit (November 2012), whichever happened first. Each investigation data record included demographic information, lifestyle characteristics, medical anthropometric and biochemical measurements, history, and disease incidence data. Our study focused on 8,841 participants with finalized DNA genotyping and quality control data (Fig. 2). Among these 8,841 participants, individuals with cancer (n = 104), CVD (n = 243), DM (n = 1,060), and high TG levels (>600 mg/dL) (n = 76) at baseline were excluded from the study, leaving a total of 7,358 participants (3,427 men and 3,931 women) whose data were included in the final analysis. Informed consent was provided by all participants. The study protocol was approved by the Institutional Review Board of the KCDC (KBP-2016-062) and the Institutional Review Board at Korea University (KU-IRB-16-EX-272-A-1) and all methods were carried out in accordance with the approved protocol and the relevant guidelines. This study is not a clinical trial.
General characteristics
At baseline and each follow-up examination, study participants were asked to complete survey questionnaires regarding demographic and behavioral data, including age, sex, location, total energy intake, BMI, physical activity, income level, education level, cigarette smoking, and alcohol use. Total daily energy intake (kcal) was estimated by a semi-quantitative FFQ, which was validated by the KoGES30. BMI (kg/m2) was calculated as the weight in kilograms divided by the square of the body height in meters. Height was measured to the nearest 0.1 cm. Weight, with participants in light clothing, was measured to the nearest 0.1 kg. Total metabolic equivalent (MET; hours/day) was used for measuring the energy cost of physical activities, calculated by summing the MET values of each physical activity type (2.4 for light, 5.0 for moderate, and 7.5 for intense activities)31,32. Monthly household income level was categorized into lowest (<1 million Korean won), lower-middle (1–2 million), upper-middle (2–4 million), and highest (>4 million). Education level was categorized into elementary school, middle school, high school, and university, considering the highest level of education completed by the participants. Smoking and drinking status were categorized into current smokers/drinkers (defined as those who answered on the questionnaire as smoking cigarettes or drinking alcoholic beverages currently) and non-smokers/drinkers.
Dietary information
Dietary information was investigated using a semi-quantitative FFQ at the baseline survey (2001–2002) and at the second follow-up (2005–2006), which comprised questions on 103 food items. The questionnaire responses for frequency of food consumption and serving size of each food item were collected, and weighted food frequencies for each parameter were derived. To identify the effect modifier regarding associations between ANGTPL3 polymorphisms and DM, we selected dietary intake as the average of two surveys for fruit, vegetables, whole grains, unprocessed red meat, sodium, and milk. Fruit intake was the sum of the weekly consumption of persimmons, mandarin oranges, melons, bananas, pears, apples, oranges, watermelons, peaches or plums, strawberries, and grapes. Vegetable intake was the sum of the weekly consumption of tomatoes, green peppers, pepper leaves, spinach, lettuce, perilla leaves, chives or water parsley, other green vegetables, radishes, balloon flowers or deodeok (Codonopsis lanceolata), onions, cabbages, cucumbers, bean sprouts, carrots, pumpkins, bracken or sweet potato vines, kimchi (salted and fermented vegetables: cabbage kimchi, diced radish kimchi, radish kimchi, water kimchi, or other kimchi), pickled vegetables, and mushrooms. Whole grain intake was the sum of the weekly consumption of barley rice, multi-grain rice, flour made of mixed grains, and noodles made of buckwheat. Unprocessed meat intake was calculated as the sum of the weekly consumption of pork (roast pork ribs and sirloin, pork belly, and steamed pork), beef (roast beef, beef soup, beef stew, and beef ribs), unprocessed poultry (chicken legs, chicken wings, and other chicken meat), canine meat, and organ meat. Daily sodium intake was calculated based on the Standard Tables of Food Composition of Korea22.
Anthropometric and biochemical measurements
Study data included biochemical measurements, for which detailed procedures and analysis methods have been described in a previous study33. BP was measured in both arms using a mercury sphygmomanometer. Two measurements were made with the arm above the heart level in a sitting position after at least 5 min of rest, and the average of the two was used for systolic and diastolic BP (SBP and DBP; mmHg). Blood samples for biochemical analysis were collected after fasting for at least 8 h. TG (mg/dL), TC (mg/dL), HDLC (mg/dL), FBG (mg/dL) and the oral glucose tolerance test (OGTT)−120 levels were measured using an automatic analyzer (ADVIA 1650 and 1680; Siemens, Tarrytown, NY, USA). In this study, we used the Friedewald equation for calculating LDLC (mg/dL): LDLC = TC − TG/5 − HDLC in participants with TG <400 mg/dL34. HbA1c was determined from whole blood samples. Blood glucose and insulin concentrations were measured at 60 and 120 min during the OGTT. The following equation was used to calculate HOMA-IR: HOMA-IR = [fasting glucose (mmol/L) × fasting insulin (μIU/mL)]/22.535.
Genotyping information
Detailed information regarding DNA preparation, genotyping, and quality control in the KoGES has been reported elsewhere33. Briefly, genomic DNA was isolated from the peripheral blood of all participants and genotyped on the Affymetrix Genome-Wide Human SNP Array 5.0 (Affymetrix, Inc., Santa Clara, CA, USA). Bayesian robust linear modeling using the Mahalanobis distance genotyping algorithm was used for genotyping accuracy. A total of 352,228 SNPs were available after quality control with a high missing gene call rate (>5%), low minor allele frequency (<0.01), considerable deviation from the Hardy–Weinberg equilibrium (HWE; P < 1 × 10−6), and sex mismatch. After rejecting an additional 48,003 SNPs outside the HWE (P < 1 × 10−5), a subset of 304,225 SNPs was processed by the EIGENSTRAT software package36. To test its association with DM, we selected a gene variant of ANGPTL3 (rs11207997).
Definition of DM
DM was defined by self-reported information from a biennial questionnaire implemented by trained technicians. Participants were defined to have DM if (i) they had been diagnosed with DM by a physician, (ii) their FBG level was ≥126 mg/dL, (iii) their OGTT−120 glucose level was ≥200 mg/dL, or (iv) they were taking diabetes medication.
RNA extraction and semi-quantitative reverse transcription
Total RNA was isolated from LCLs using the RibospinTM Kit (Gene All, Korea), according to the manufacturer’s protocol. To demonstrate the relationship between the mRNA expression of ANGPTL3 genetic variants and cardiometabolic parameters, LCLs of 62 healthy subjects (24 men and 38 women) were collected from the Ansan–Ansung cohort of the KoGES (Institutional Review Board no. KBP-2016-062, KU-IRB-16-EX-272-A-1). We obtained cDNA from 1 μg of RNA using oligo-dT and superscript II reverse transcriptase (Invitrogen, USA). One microgram of cDNA was subjected to quantitative real-time polymerase chain reaction (PCR) amplification using the SYBR Green PCR kit (Qiagen, USA). The primer sequences were as follows: ANGPTL3 (sense, 5′-TCT CCA GAG CCA AAA TCA AGA T-3′; antisense, 5′-TTT CAC TGG TTT GCA GCG AT-3′) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; sense, 5′-TCC ACC ACC CTG TTG CTG TA-3′; antisense, 5′-ACC ACA GTC CAT GCC ATC AC-3′). The PCR program was conducted under the following conditions: 15 min at 95 °C, followed by 40 thermal cycles at 94 °C for 30 s, 60 °C for 20 s, and 72 °C for 30 s. Semi-quantitative reverse transcription-PCR and quantification of gene expression were performed using QuantStudio™ 6 Flex Real-Time PCR System (Applied Bio systems, Foster City, CA). Data were analyzed using the comparative cycle threshold method, and the GAPDH expression value was used as a reference for normalization.
Statistical analyses
Data are represented as mean ± standard error for continuous variables and percentages and the number of counts for categorical variables. Main general characteristics and biochemical parameters were compared according to DM using Student’s t-test and chi-square tests for continuous and categorical variables. Data for the statistical analyses of TG were used after log-transformation because TG was not normally distributed in this population. Differences of clinical parameters according to ANGPTL3 genotype were obtained using analysis of variation and the generalized linear regression model with Bonferroni correction after adjusting for age, sex, location, total energy intake, BMI, physical activity, education level, and drinking status. The relationships between the ANGPTL3 mRNA level and TG, TC, HDLC, LDLC, SBP, and DBP were tested by partial correlation analyses with adjustments for potential covariates. Results are presented as estimated correlation co-efficient (r) with p-values for continuous variables. The Cox proportional hazards model was used to estimate the genetic effect of ANGPTL3 on the risk of developing DM. The Cox regression model was adjusted for age, sex, location, total energy intake, BMI, physical activity, education level, smoking and drinking status. Results are shown as HRs with 95% CI and corresponding p-values. Because circulating levels of TG at baseline could be an intermediate variable through which rs11207997 is associated with DM, we adjusted for them only in the secondary analyses. We also performed stratification analyses in the Cox hazard model by dietary risk factors in evaluating the associations of rs11207997 (ANGPTL3) polymorphism with DM incidence. All statistical analyses were performed using Stata SE 13.0 (Stata Corp., Carolina, USA).
References
Santulli, G. Angiopoietin-like proteins: a comprehensive look. Front Endocrinol (Lausanne) 5, 4, https://doi.org/10.3389/fendo.2014.00004 (2014).
Kersten, S. Regulation of lipid metabolism via angiopoietin-like proteins. Biochem Soc Trans 33, 1059–1062, https://doi.org/10.1042/BST20051059 (2005).
Tabata, M. et al. Angiopoietin-like protein 2 promotes chronic adipose tissue inflammation and obesity-related systemic insulin resistance. Cell Metab 10, 178–188, https://doi.org/10.1016/j.cmet.2009.08.003 (2009).
Zhang, C. C. et al. Angiopoietin-like proteins stimulate ex vivo expansion of hematopoietic stem cells. Nat Med 12, 240–245, https://doi.org/10.1038/nm1342 (2006).
Galaup, A. et al. Angiopoietin-like 4 prevents metastasis through inhibition of vascular permeability and tumor cell motility and invasiveness. Proc Natl Acad Sci USA 103, 18721–18726, https://doi.org/10.1073/pnas.0609025103 (2006).
Nordestgaard, B. G., Nicholls, S. J., Langsted, A., Ray, K. K. & Tybjaerg-Hansen, A. Advances in lipid-lowering therapy through gene-silencing technologies. Nature reviews. Cardiology 15, 261–272, https://doi.org/10.1038/nrcardio.2018.3 (2018).
Olkkonen, V. M., Sinisalo, J. & Jauhiainen, M. New medications targeting triglyceride-rich lipoproteins: Can inhibition of ANGPTL3 or apoC-III reduce the residual cardiovascular risk? Atherosclerosis 272, 27–32, https://doi.org/10.1016/j.atherosclerosis.2018.03.019 (2018).
Li, Y. & Teng, C. Angiopoietin-like proteins 3, 4 and 8: regulating lipid metabolism and providing new hope for metabolic syndrome. J Drug Target 22, 679–687, https://doi.org/10.3109/1061186X.2014.928715 (2014).
Stitziel, N. O. et al. ANGPTL3 Deficiency and Protection Against Coronary Artery Disease. J Am Coll Cardiol 69, 2054–2063, https://doi.org/10.1016/j.jacc.2017.02.030 (2017).
Romeo, S. et al. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J Clin Invest 119, 70–79, https://doi.org/10.1172/JCI37118 (2009).
Dewey, F. E. et al. Genetic and Pharmacologic Inactivation of ANGPTL3 and Cardiovascular Disease. N Engl J Med 377, 211–221, https://doi.org/10.1056/NEJMoa1612790 (2017).
Robciuc, M. R. et al. Angptl3 deficiency is associated with increased insulin sensitivity, lipoprotein lipase activity, and decreased serum free fatty acids. Arterioscler Thromb Vasc Biol 33, 1706–1713, https://doi.org/10.1161/ATVBAHA.113.301397 (2013).
Inukai, K. et al. ANGPTL3 is increased in both insulin-deficient and -resistant diabetic states. Biochem Biophys Res Commun 317, 1075–1079, https://doi.org/10.1016/j.bbrc.2004.03.151 (2004).
Chung, H. S. et al. Circulating angiopoietin-like protein 8 (ANGPTL8) and ANGPTL3 concentrations in relation to anthropometric and metabolic profiles in Korean children: a prospective cohort study. Cardiovasc Diabetol 15, 1, https://doi.org/10.1186/s12933-015-0324-y (2016).
Shen, Y. et al. Common genetic variants associated with lipid profiles in a Chinese pediatric population. Hum Genet 132, 1275–1285, https://doi.org/10.1007/s00439-013-1332-1 (2013).
Legry, V. et al. Associations between common genetic polymorphisms in angiopoietin-like proteins 3 and 4 and lipid metabolism and adiposity in European adolescents and adults. J Clin Endocrinol Metab 94, 5070–5077, https://doi.org/10.1210/jc.2009-0769 (2009).
Ministry of Health and Welfare. 2015 Dietary Reference Intakes for Koreans. (2015).
Wang, Y. et al. Hepatic ANGPTL3 regulates adipose tissue energy homeostasis. Proc Natl Acad Sci USA 112, 11630–11635, https://doi.org/10.1073/pnas.1515374112 (2015).
Gambardella, J. & Santulli, G. Integrating diet and inflammation to calculate cardiovascular risk. Atherosclerosis 253, 258–261, https://doi.org/10.1016/j.atherosclerosis.2016.08.041 (2016).
Wellen, K. E. & Hotamisligil, G. S. Inflammation, stress, and diabetes. The Journal of clinical investigation 115, 1111–1119 (2005).
Park, Y. H. C. S. A comparison of sources of sodium and potassium intake by gender, age and regions in Koreans: Korea National Health and Nutrition Examination Survey (KNHANES) 2010-2012. Korean Journal of Community Nutrition 21, 558–573 (2016).
Rural Development Administration. Food Composition Tables. 6th revision edition. (Rural Nutrition Institute, 2001).
Kim, K., Yang, Y. J., Kim, K. & Kim, M. K. Interactions of single nucleotide polymorphisms with dietary calcium intake on the risk of metabolic syndrome. Am J Clin Nutr 95, 231–240, https://doi.org/10.3945/ajcn.111.022749 (2012).
Ballard, C. S. et al. Effect of corn silage hybrid on dry matter yield, nutrient composition, in vitro digestion, intake by dairy heifers, and milk production by dairy cows. J Dairy Sci 84, 442–452, https://doi.org/10.3168/jds.S0022-0302(01)74494-3 (2001).
Kim, Y. & Je, Y. Meat Consumption and Risk of Metabolic Syndrome: Results from the Korean Population and a Meta-Analysis of Observational Studies. Nutrients 10, https://doi.org/10.3390/nu10040390 (2018)
Lee, J. S. & Kim, J. Vegetable intake in Korea: data from the Korean National Health and Nutrition Examination Survey 1998, 2001 and 2005. Br J Nutr 103, 1499–1506, https://doi.org/10.1017/S0007114509993527 (2010).
Paik YJ, K., S.H., Oh, K.W. Energy and Nutrient Intakes from Fruit in the Korea National Health and Nutrition Examination Survey (KNHANES). 777–780 (KCDC, 2014).
Chi, X. et al. ANGPTL8 promotes the ability of ANGPTL3 to bind and inhibit lipoprotein lipase. Mol Metab 6, 1137–1149, https://doi.org/10.1016/j.molmet.2017.06.014 (2017).
Kim, Y., Han, B. G. & KoGES group. Cohort Profile: The Korean Genome and Epidemiology Study (KoGES) Consortium. Int J Epidemiol 46, e20, https://doi.org/10.1093/ije/dyv316 (2017).
Ahn, Y. et al. Validation and reproducibility of food frequency questionnaire for Korean genome epidemiologic study. Eur J Clin Nutr 61, 1435–1441, https://doi.org/10.1038/sj.ejcn.1602657 (2007).
Ainsworth, B. E. et al. Compendium of physical activities: an update of activity codes and MET intensities. Med Sci Sports Exerc 32, S498–504 (2000).
Baik, I. & Shin, C. Prospective study of alcohol consumption and metabolic syndrome. Am J Clin Nutr 87, 1455–1463, https://doi.org/10.1093/ajcn/87.5.1455 (2008).
Cho, Y. S. et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet 41, 527–534, https://doi.org/10.1038/ng.357 (2009).
Friedewald, W. T., Levy, R. I. & Fredrickson, D. S. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 18, 499–502 (1972).
Matthews, D. R. et al. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28, 412–419 (1985).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38, 904–909, https://doi.org/10.1038/ng1847 (2006).
Acknowledgements
Bioresources for the study were provided by the National Biobank of Korea and the Centers for Disease Control and Prevention, Republic of Korea (KBP-2016-062). This research was supported by the Basic Science Research Program, through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science, and Technology (NRF-2015R1A2A1A15054758). This research was also supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science & ICT (NRF-2012M3A9C4048761).
Author information
Authors and Affiliations
Contributions
M.J.S. conceived the study and acquired data. C.Y.P. and M.J.S. developed the statistical analysis plan. J.M., G.J. and J.L. analyzed the data. C.Y.P. and M.J.S. prepared the first draft of manuscript. C.Y.P., J.M., G.J., J.L., O.Y.K., H.L., H.O. and M.J.S. contributed to the writing of the manuscript. All authors approved the submission of the article. M.J.S. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Park, C.Y., Moon, J., Jo, G. et al. The association between genetic variants of angiopoietin-like 3 and risk of diabetes mellitus is modified by dietary factors in Koreans. Sci Rep 9, 766 (2019). https://doi.org/10.1038/s41598-018-36581-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-36581-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.