Abstract
Apple scab disease caused by the fungus Venturia inaequalis is a devastating disease that seriously affects quality and yield of apples. In order to understand the mechanisms involved in scab resistance, we performed gas chromatography-mass spectrometry based metabolomics analysis of the cell culture of scab resistant cultivar ‘Florina’ and scab susceptible cultivar ‘Vista Bella’ both prior -to and -following treatment with V. inaequalis elicitor (VIE). A total 21 metabolites were identified to be altered significantly in ‘Florina’ cell cultures upon VIE-treatment. Among 21 metabolites, formation of three new specialized metabolites aucuparin, noraucuparin and eriobofuran were observed only in resistant cultivar ‘Florina’ after the elicitor treatment. The score plots of principal component analysis (PCA) exhibited clear discrimination between untreated and VIE-treated samples. The alteration in metabolite levels correlated well with the changes in the transcript levels of selected secondary metabolite biosynthesis genes. Aucuparin, noraucuparin and eriobofuran isolated from the ‘Florina’ cultures showed significant inhibitory effect on the conidial germination of V. inaequalis. The results expand our understanding of the metabolic basis of scab-resistance in apple and therefore are of interest in apple breeding programs to fortify scab resistance potential of commercially grown apple cultivars.
Similar content being viewed by others
Introduction
Apple (Malus domestica) is the main deciduous fruit crop of the temperate region of the world1. Apple fruits are rich in antioxidants and health protective polyphenols2,3. The major categories of polyphenols detected in apple are catechins, epicatechin, proanthocyanidins, flavonols, dihydrochalcones, hydroxycinnamates and anthocyanins4,5,6,7. In addition, apples are also rich source of many bioactive triterpenes8. Regular consumption of apples is known to be associated with the reduced risk of heart disease and cancer9. Apples and apple-derived products possess several health beneficial activities, such as anticancer, anti-diabetic, and cholesterol-lowering properties10.
The apple scab disease is caused by the ascomycetous fungus V. inaequalis. Scab is one of the most devastating diseases of apple in terms of economic damage11,12. The pathogenic phase of V. inaequalis infection initiates with the germination of ascospores (sexual spores), which serve as the primary source of infection on fallen leaves. The conidiospores (asexual spores) serve as a secondary infection source. In a single apple-growing season, multiple cycles of conidiospore production and secondary infections occurs13. The control of the disease relies mainly on multiple fungicides spray14, which exhibit toxic effect to human health and may even contaminate the environment. Moreover, extended use of scab controlling fungicides may result in selection pressure on the V. inaequalis, thereby inducing emergence of new resistant races15. Apple plants have natural defense systems against a wide range of pathogens. The genus Malus has coevolved with the scab fungus V. inaequalis, and many apple accessions harbor scab-resistance genes that are currently used for introgression from wild relatives to commercial cultivars through breeding programs12,16. In recent past, a number of scab-resistance genes (Rvi1 – Rvi17) have been isolated and mapped from various scab-resistant cultivars across the globe12. Notably, until now Rvi15 is the only gene available to provide complete resistance against all the available races of V. inaequalis17,18. Apple breeding programs usually take longer time and generally involve a series of back crossings between resistant and susceptible cultivars to recover near-isogenic lines expressing the desired commercial trait. Furthermore, self-incompatibility in apples make breeding process even more problematic as the elite varieties sometimes might not be recovered by crossing. As a result, strategies able to directly transfer traits to elite cultivated varieties through cis-genic approach would accelerate the breeding process19.
Several apple cultivars such as ‘Liberty’, ‘Florina’, ‘Prima’, ‘Sir Prize’ are resistant to scab infections. The disease resistance mechanism depends on the nature of genetic interactions between V. inaequalis and Malus species in accordance to the gene-for-gene concept20. To date, more than 17 ‘R’ genes have been indentified from different Malus species providing varying levels of resistance against V. inaequalis11,12. The early stages of scab infection beginning with spore germination, appressoria formation, cuticular penetration, and fungal hyphae development are similar in both resistant and susceptible apple cultivars21. Subsequently, resistant cultivars show hypersensitive response (HR) whereas susceptible cultivars exhibit sporulating lesions and disease symptoms depending on the resistance genes, pathogen race, and ontogenic resistance22. The HR in apples are exhibited by several previously characterized ‘R’ genes such as Rvi4, Rvi5, Rvi7, Rvi10, Rvi15 and Rvi1618 that produces typical “pin-point pits”. These pin-point-pits are formed as a result of rapid death and subsequent collapse of cells immediately surrounding the germinating spore penetration site23. The HR prevents colonization of fungus through programmed cell death and may involve cell to cell signaling mediated by formation of reactive oxygen species and defense responsive metabolites such as phloretin, biphenyls, dibenzofurans and phenolics24,25,26. Phenolic metabolites produced in response to scab infections in apple are known to inhibit growth of V. inaequalis, suggesting their involvement in scab resistance11. Suppression of phenylalanine ammonia lyase, the key enzyme in phenolic biosynthesis, in the scab resistant apple cultivar ‘Sir Prize’ turned it to scab susceptible27. The breakdown of phloridzin into phloretin by V. inaequalis is known to be associated with hindering fungal growth28. Earlier, it was reported that the treatment of V. inaequalis elicitor induces synthesis of salicylic acid in the cell suspensions of apple cultivar ‘Florina’29. The elicitor-induced formation of a novel phytoalexin malusfuran was reported from the cell suspensions of scab resistant cultivar ‘Liberty’28. Malusfuran was found to suppress the rate of V. inaequalis conidia germination. However, systematic understanding of the defense-responsive metabolites underlying scab-resistance in apples is still elusive. It was also reported that cell wall surrounding the scab infection site undergoes rapid lignifications to reduce the risk of secondary infections in many scab resistant cultivars30. In addition to HR, the other resistance phenotypes such as chlorotic flecking31 and necrotic flecking32 have been observed in the selected cultivars. Previously, it was reported that pathogenicity related proteins like β-1,3-glucanase, chitinase, and thaumatin-like protein were constitutively expressed in the apoplast of the resistant apple cultivar ‘Remo’ and were induced in susceptible cultivar ‘Elstar’ after V. inaequalis infection33. The transgenic apple lines of scab susceptible cultivar ‘McIntosh’ expressing either the endo (ech42) or the exo (nag70) chitinase gene of Trichoderma harzianum are shown to be scab tolerant34. Similarly cis-genic apple lines of the cultivars ‘Brookfield Baigent’, ‘Mitchgla’, ‘Novajo’, and ‘Pinova’ harboring Rvi6 gene showed enhanced scab resistance19.
Metabolomics is widely used to investigate the tolerance of plants to biotic stresses35. After pathogen infection, the plant starts synthesizing an array of defense-responsive metabolites, both primary and secondary. Alteration in the metabolic profiles of a plant in response to pathogen infection can be analyzed with metabolomics approach. This technology provides the opportunity to evaluate pathogen-induced local and systemic alterations in plant metabolite patterns without any prior assumptions. Measuring the level of metabolites prior and after pathogen infection may provide an exact status of the physiological condition of the plant tissue under consideration. The gas chromatography-mass spectrometry (GC-MS) is routinely used in plant metabolomics36, especially for facilitating the identification and quantification of the primary metabolites such as amino acids, sugars, organic acids37 and an wide array of secondary metabolites like phenolics, flavonoids, terpenes and specialized phytoalexins. Most metabolomics based studies on apples published so far were targeted analyses focusing on specific metabolites and limited to fruit quality38,39,40. Recently NMR based metabolomics were performed to identify new scab-preventive metabolites from scab resistant apples41. However, untargeted metabolite analysis provides a more comprehensive view on the differential accumulation of metabolites upon pathogen infection32.
We hypothesized that scab resistant and scab susceptible cultivars of apple would respond differentially towards scab infection in terms of metabolic re-programming and for that a non-targeted GC-MS based metabolomics approach can be used. To investigate this hypothesis, we employed non-targeted comparative GC-MS to identify scab-responsive metabolites from the VIE-treated cell suspension culture of scab resistant and susceptible apple cultivars namely ‘Florina’ and ‘Vista Bella’ respectively. Comparative metabolomics resulted in a profile of total 60 metabolites from the ‘Florina’ cell cultures after VIE treatment, out of which 21 metabolites showed strict differential accumulation in the ‘Florina’. Based on these results, selected secondary metabolites were chosen for analyzing their inhibitory effect on the V. inaequalis conidial germination. Information derived from this study will provide new insights on scab-resistant mechanisms in apple in terms of metabolites.
Results
Comparative metabolomics of VIE-treated apple cell cultures
Cell cultures of scab resistant (SR) apple cultivar ‘Florina’ and scab susceptible (SS) cultivar ‘Vista Bella’ was used for comparative metabolomics. In order to understand metabolic re-programming following VIE-treatment, metabolomics was performed on samples prepared from treated SR and SS cell cultures at 6, 12, 24, 36, 48 and 72 hours post elicitation (hpe) and control (SR0 and SS0) untreated cultures. After double derivatization, metabolites were profiled by GC-MS metabolomics analyses, comparing the levels of each metabolite at defined time points to the equivalent un-treated controls. Representative GC-MS chromatograms of ‘Vista Bella’ and ‘Florina’ cell cultures are shown in Supplementary Fig. S1. A total of 60 low-molecular weight metabolites were detected in the ‘Florina’ cell cultures, whereas 55 metabolites were detected in the ‘Vista Bella’ cultures as shown in Supplementary Table S1.
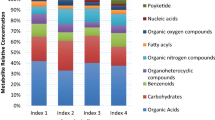
In the next step, we identified VIE-responsive metabolites that showed significant (p ≤ 0.05) differential concentrations between SR and SR0 and between SS and SS0. A total of 31 and 10 differentially accumulating metabolites were identified in ‘Florina’ and ‘Vista Bella’ cell cultures, respectively, as shown in Table 1. Statistically significant differentially accumulating metabolites detected from ‘Florina’ were further analyzed by employing Bonferroni correction (p ≤ 0.05) and their adjusted p-values were shown in Supplementary Table S2. Detected metabolites were further classified into specific metabolite classes such as amino acids, sugar alcohols, sugars, organic acids, vitamins, phenolics and secondary metabolites, on the basis of their chemical nature (Table 2). Metabolite feature areas were normalized using the area of internal standard before performing comparative metabolomics.
As shown in Fig. 1 and Fig. 2, all of the differentially accumulating metabolites did not show distinct accumulation patterns in both SR and SS cell cultures during the time course studied. Interestingly, metabolites belonging to the same biosynthetic pathway often showed distinct accumulation patterns during different post elicitation time points. After VIE-treatment, most of the differential metabolites started enhanced accumulation. At 12 or 36 hpe most of the differential metabolites showed maximum accumulation in SR cultures, including the amino acids, organic acids and phenolics. Following the progression of elicitation, metabolite level first dramatically increased to attain a peak and then decreased thereafter at 72 hpe. Notably, a large number of differentially accumulating metabolites in ‘Florina’ cultures that showed elevated levels, have been identified in more than one time point following elicitation, suggesting activation of the metabolic pathways leading to the biosynthesis of these particular metabolites.
Differentially accumulating primary metabolites in the VIE-treated cell cultures of apple cultivar ‘Florina’ [blue; resistant cultivar (R)] and ‘Vista Bella’ [red, susceptible cultivar (S)]. Data are presented as mean values and error bars represent the standard deviation. The x-axis represents post-elicitation time points (h). The y-axis gives the normalized relative metabolite abundance in terms of the area of internal standard phenylphenol. The ANOVA was performed to assess the statistical significance of differences between samples at different time points (p < 0.05). *Denotes significant difference (p < 0.05); #Denotes non-significant difference (p > 0.05).
Differentially accumulating secondary metabolites detected from the VIE-treated cell cultures of ‘Florina’ (blue) and ‘Vista Bella’ (red). The x-axis represents post-elicitation time points (h). The y-axis gives the normalized relative metabolite abundance in terms of the area of internal standard phenylphenol. The ANOVA was performed to assess the statistical significance of differences between samples at different time points (p < 0.05). *Denotes significant difference (p < 0.05); #Denotes non-significant difference (p > 0.05).
VIE-treatment changes the primary and secondary metabolite signature in ‘Florina’ and ‘Vista Bella’ cultures
VIE-treatment appears to trigger massive metabolic re-programming in ‘Florina’ cell cultures both at the level of primary and secondary metabolites (Fig. 1 and Fig. 2). In comparison, the numbers of metabolites showing a significant differential accumulation in VIE-treated SR compared to SR0 cultures were much lower in VIE-treated SS versus SS0 cultures. Among the 31 differentially accumulating metabolites detected from ‘Florina’, 30 metabolites were up-regulated and one metabolite was down-regulated as shown in Fig. 1. On the contrary only nine metabolites were significantly up- and one metabolite was down-regulated in ‘Vista Bella’ cultures after VIE-treatment (Fig. 1 and Fig. 2). Among the amino acids detected in ‘Florina’, the levels of serine, tryptophan, phenylalanine, proline and aspartic acid were significantly up-regulated after VIE-treatment. Similarly, VIE-treatment triggers up-regulation of glucose, fructose and mannose contents; whereas sucrose content was down-regulated. Among the sugar alcohols, mannitol and sorbitol contents were significantly up-regulated. Pyruvic-, 3-phosphoglyceric-, citric-, succinic-, fumaric-, malic-, and malonic acid levels were up-regulated. Among the vitamins, only ascorbic acid was detected from ‘Florina’ cell cultures, whose level was significantly up-regulated after elicitor treatment. Phenolics and specialized biphenyl and dibenzofuran phytoalexins were the most abundant secondary metabolites detected in the VIE-treated ‘Florina’ cell cultures. Plant phenolics are known to play crucial role in pathogen defense42. Nine phenolics and three inducible phytoalexins (two biphenyl phytoalexins: noraucuparin and aucuparin; one dibenzofuran phytoalexin: eriobofuran) were detected. All these phenolic metabolites showed differential accumulation after the VIE-treatment. The content of these phenolics were first up-regulated after the VIE-treatment and thereafter declined to the basal level (Fig. 2). Interestingly, in our study, biphenyl and dibenzofuran phytoalexins were not detected from the untreated control cells and formed in ‘Florina’ cultures only after the VIE-treatment. On the contrary, in scab susceptible ‘Vista Bella’ cell cultures, only differential accumulation of caffeic-, chlorogenic- and ferulic acids were observed (Fig. 2). Catechin, salicylic acid, noraucuparin, aucuparin and eriobofuran were not detected from ‘Vista Bella’ cultures.
Principal component analysis (PCA) reveals metabolic alterations in VIE-treated ‘Florina’ cell cultures
To assess the data reproducibility in the different biological replicates of the metabolites measurements, GC-MS data were subjected to principal component analysis (PCA). For PCA, only those 21 metabolites were included (a marked in the Table 1) which showed differential accumulation only in the scab resistant cultivar ‘Florina’ but not in the scab susceptible cultivar ‘Vista Bella’. PCA helps to reduce the dimensionality of complex data sets, which facilitate better visualization of the inherent patterns in the data. PCA analyses uses, linear orthogonal transformation of the original data variables to generate a new set of uncorrelated variables known as principal components (PCs)43. Results indicated that the biological replicates for each group [control (0 h) and treated (6 h, 12 h, 24 h, 36 h, 48 h and 72 h)] were always clustered together (Fig. 3A), indicating on one hand the high data reproducibility at the different treatment time and on the other hand the significance of their effects on the metabolite level alteration. As shown in Fig. 3A, the first and second PCs of the analyzed PCA score plot represented 48.0% (PC1) and 35.1% (PC 2) of the total variance of the samples. Furthermore, a PCA loading plot (Fig. 3B) was constructed to show the abundant variable (metabolites) contributing to the PCA results. Our results suggest that these differentially accumulating metabolites are the probable reasons contributing to the resistance responses of ‘Florina’ against the VIE-treatment.
Hierarchical clustering analysis of the metabolite profiles
In order to search for any probable discrepancies in the metabolite profiles of all the seven (0–72 h) sample groups, the 21 differentially accumulating metabolites used for the PCA analyses were organized and visualized by heatmap analysis tool of MetaboAnalyst 4.0 (Fig. 4)44. The Heatmap offered excellent separation of the metabolite trend between the non-treated (0 h) and VIE-treated (6–72 h) samples.
Changes in metabolic pathways: metabolic pathway network
A simplified metabolic pathway network was reconstructed using key metabolic pathways, such as the shikimic acid pathway, the phenylpropanoid pathway, the glycolytic pathway, the pentose phosphate pathway, the biphenyl biosynthetic pathway, the flavonoids biosynthesis pathway, the tricarboxylic acid (TCA) pathway, and the amino acid biosynthetic pathway to show the regulated pattern diversity of each detected differentially accumulating metabolites from ‘Florina’ with respect to their proportional incorporation into key metabolic pathways. As shown in Fig. 5, sucrose as the precursor for glucose and fructose was higher in non-elicited cells than that of VIE-treated cells. However, sucrose hydrolysis product, the glucose and the fructose were higher in the VIE-treated cells, suggesting that more active sucrose catabolism occurred in the VIE-treated cells. The level of mannose, sorbitol and mannitol were higher in the VIE-treated cells, suggesting that fructose is metabolized more towards these metabolites rather than re-entering into glycolytic pathway. The level of identified TCA cycle metabolites such as citric, succinic, fumaric and malic acids were up-accumulated in the VIE-treated cells suggesting higher turn-over number of TCA cycle. The metabolites derived from shikimate pathway showed an absolutely distinct accumulation pattern. The level of all the detected shikimate-derived metabolites such as caffeic acid, protocatechuic acid, catechin, chlorogenic acid, benzoic acid, ferulic acid, p-coumaric acid, trans-cinnamic acid and salicylic acid were higher in the VIE-treated cells. Aucuparin, noraucuparin and eriobofuran were synthesized only after the VIE-treatment.
Gene expression analyses
The expression levels of selected genes coding for enzymes involved in phenylpropanoid, flavonoids and biphenyl biosynthesis were studied further as the contents of phenolics, flavonoids and biphenyls were found to be significantly elevated in the VIE-treated ‘Florina’ cell cultures. The expression levels of seven genes after VIE-treatment were investigated by quantitative real-time PCR, and the results are shown in Fig. 6. The expression of phenylalanine ammonia lyase (MdPAL) gene was markedly increased after the VIE-treatment. The expression level of cinnamate-4-hydroxylase (MdC4H), 4-coumarate:CoA ligase (Md4CL), chalcone flavanone isomerase (MdCHI), flavanone 3-hydroxylase (MdF3H), biphenyl synthase 3 (MdBIS3), and alternative oxidase (MdAOX) were also significantly up-regulated after VIE-treatment demonstrating that phenylpropanoid, flavonoid and biphenyl biosynthesis were triggered by the VIE-treatment. These gene expression data were well correlated with the enhanced accumulation of phenylpropanoids such as cinnamic acid, flavonoids and biphenyls in the elicited cell cultures of ‘Florina’.
Inhibition of conidial germination of V. inaequalis with apple cell culture metabolites
The incubation of V. inaequalis conidial suspension with combinations of aucuparin, noraucuparin and eriobofuran, three induced metabolites detected from the VIE-treated ‘Florina’ cell culture resulted into 74% inhibition of conidial germination in contrast to the untreated control (Supplementary Fig. S2). Treatment of conidial suspension with noraucuparin (5 µM) alone resulted into 28% inhibition of conidial germination. Individual treatment with aucuparin (5 µM) and eriobofuran (5 µM) showed 46% and 34% inhibition in conidial germination, respectively. Combinations of aucuparin (5 µM) and noraucuparin (5 µM) resulted into relatively higher inhibition of conidial germination. For conidia germination assay, pH range of 5 to 5.6 was found to be optimum. These results clearly showed synergistic effect aucuparin, noraucuparin and eriobofuran on inhibition of V. inaequalis conidial germination. However the underlying mechanisms of actions for this inhibitory effect remain elusive.
Discussion
The present GC-MS-based metabolomics analysis was performed to identify signature metabolites associated with apple resistance to scab fungus V. inaequalis. This study is the first report on the systemic identification of metabolic changes in response to VIE-treatment in the cell cultures of scab resistant cultivar ‘Florina’ and scab susceptible cultivar ‘Vista Bella’, before and after treatment. In this study we have used apple cell culture as model system because cell cultures offer an uniform plant material for studying metabolic changes and typically plant cell cultures exhibit relatively less metabolic complexity due to lack of pigments and lignified tissues. Earlier plant cell cultures have been successfully used for analyzing plant-microbe interactions in apple and other plant systems29,45,46. The metabolomics data were then correlated with the expression patterns of genes linked to selected secondary metabolite biosynthesis to obtain a comprehensive insight of the changes occurring after VIE treatment.
From the metabolic point of view, VIE-treatment appears to cause massive metabolic re-programming in ‘Florina’ both at the level of primary and secondary metabolisms. In this study we have detected only 60 metabolites from the VIE-treated cell cultures of ‘Florina’ which is relatively less as compared to the previously reported metabolites from apple38. This is possibly explained by the fact that cell culture system used in this study is devoid of complex metabolites such as chlorophyll, pigments and lignified tissues, and thereby represents a metabolically simple system as compared to intact apple plants and fruits.
The changes in the metabolites concentration suggested that six biochemical pathways were mainly altered by VIE-treatment. Considering the limited number of metabolites reported in this study, the most altered pathways were amino acid, sugar, sugar alcohols, organic acids, vitamin, phenolics and biphenyl-dibenzofuran phytoalexin biosynthesis. Earlier studies have shown that up-regulation of primary metabolites, mainly in the level of amino acids, sugar and sugar alcohols occur during plant-pathogen interactions47. Notably, VIE-treated ‘Florina’ cultures showed differential accumulation of selected organic acids. The elevated level of organic acids might be associated with the higher turnover number of glycolysis and TCA cycles, which generates more abundant biosynthetic intermediates. High organic acid concentrations are known to facilitate better ion absorption and thereby known to be associated with enhanced disease resistance48. Likewise, ascorbic acid is known to be associated with the superior defense responses in many plant species49.
Among secondary metabolites, phenolic and flavonoid contents were significantly up-regulated in the VIE-treated ‘Florina’ cell cultures as compared to ‘Vista Bella’ cultures. Similar array of polyphenols were previously detected in ‘Florina’ leaves50. However, quercitin and avicularin as detected earlier from ‘Florina’ leaves were not detected from VIE-treated ‘Florina’ cell cultures. Phenolic acids are known to be deposited along the cell wall to provide first line of defense against infection51. Previously it was reported that differential accumulations of flavonols (catechins and proanthocyanidins) were key factors in providing scab resistance in the apple cultivars27,52,53. In our study, high basal level of catechin content was also observed in the ‘Florina’ cell culture. Earlier finding showed that scab-resistant apple cultivars are particularly rich in the content of caffeic-, chlorogenic-, and ferulic acids and their concentration rapidly increases after the scab-infection as compared to susceptible cultivars54. It is known that benzoic acid serves as the precursor for biphenyl and dibenzofuran phytoalexins biosynthesis in apple7. Therefore, enhanced biosynthesis of benzoic acid in the VIE-treated ‘Florina’ cultures could be linked with bipehnyl and dibenzofuran phytoalexins formation in the ‘Florina’. It has already been reported that trans-cinnamic acid serves as the precursor of many defense metabolites, such as phenylpropanoids, benzoates, lignans, and flavonoids55. It was worth speculating that after the VIE-treatment; first trans-cinnamic acid level is up-regulated, which in turn get metabolized to biphenyl and dibenzofuran phytoalexins through intermediate formation of benzoic acid. Moreover, considerable up-regulation of salicylic acid (SA) biosynthesis was also observed in the VIE-treated ‘Florina’ cell culture. The salicylic acid can induce local resistance in the form of hypersensitive reaction or long distance systemic acquired resistance (SAR) by triggering the production of pathogenesis-related proteins56. The crucial role of SA as signaling molecule in plant defense response has already been reported earlier57. VIE-treatment to ‘Florina’ cell culture led to the production of biphenyl and dibenzofuran phytoalexins which are known to inhibit the growth of V. inaequalis7. Interestingly, in our study, biphenyl and dibenzofuran phytoalexins were absent from the untreated controls cells and from the scab susceptible cultivar ‘Vista Bella’. Notably, malusfuran, a glucosylated dibenzofuran phytoalexin detected earlier from the elicited cell culture of resistant apple cultivar ‘Liberty’, has not been detected in this study. On the contrary, eriobofuran (free dibenzofuran) has been detected from the VIE-treated ‘Florina’ cell cultures. The glucosylated dibenzofurans are possibly broken-down to provide free dibenzofuran such as eriobofuran upon pathogen infection. An identical breakdown of phloridzin into phloretin was observed in several scab resistant apple cultivars58.
During the present study, PCA was performed to evaluate the dynamics of the metabolic re-programming following VIE treatment (Fig. 3A). The metabolite profiles at 6, 12, 24, 36, 48 and 72 h reflected VIE-induced enhanced accumulation of metabolites as compared to non-treated 0 h control. Only, three metabolites, noraucuparin, aucuparin and eriobofuran were newly formed after VIE-treatment. The 21 detected metabolites that showed significant differential accumulation in SR cultivar ‘Florina’ indicates a strong defense response of SR cultivar to VIE-treatment. In addition, heatmap analyses (Fig. 4) demonstrated that the VIE-treated cells were the major sources of variance in the metabolic pool, which indicated significant metabolic re-programming by the VIE-treatment.
We additionally examined the qRT PCR-based selected gene expressions of ‘Florina’ cultures before and after the VIE-treatment. The expression levels of all the seven genes studied were up-regulated after the VIE-treatment, thereby indicating up-regulation of phenylpropanoid, flavonoids and biphenyl biosyntheses after VIE-treatment. Enhanced expressions of PAL, C4H and 4CL genes are known to be associated with enhanced phenylpropanoid biosynthesis59,60. Likewise, high CHI and F3H expressions are linked to higher accumulation of catechin. Similar to PAL, a massive up-regulation of BIS3 expression was also observed. BIS3 is known to be the main BIS isoenzyme responsible for phytoalexin biosynthesis in apple shoots7. The expression of BIS3 preceded the accumulation of noraucuparin, aucuparin and eriobofuran, suggesting involvement of BIS3 in biphenyl-dibenzofuran biosynthesis. High AOX expression level in the VIE-treated ‘Florina’ cultures is probably associated with the enhanced biosynthesis of phenylpropanoids, as shown earlier in other plant species61. The elevated AOX expression is further known to be associated with the plant’s ability to facilitate metabolic re-programming to cope-up with stress conditions62.
The inhibitory effect of noraucuparin, aucuparin and eriobofuran on conidial germination of V. inaequalis provides further support for our assumption that biphenyl-dibenzofuran phytoalexins play crucial role in scab resistance in SR cultivar ‘Florina’. Here we observed that conidial germination is reduced by addition of these phytoalexins together (Supplementary fig. S2), suggesting a synergistic mode of actions of these phytoalexins. Together with the observation that these biphenyl and dibenzofuran phytoalexins were induced by VIE-treatment in ‘Florina’, this result supports the hypothesis that these specialized phytoalexin accumulation in planta have a role in prohibiting the scab infection and disease progression. However, the exact mode-of-actions of these phytoalexin need to be further investigated for better understanding of resistance mechanism in apple.
Conclusions
This is the first report to use non targeted GC-MS metabolomics analyses of apple cell cultures treated with elicitor prepared from the scab fungus V. inaequalis. Our results showed that the developed metabolomics method has excellent sensitivity and specificity to analyze metabolites from apple cell cultures. Further, our results indicated that apple cell cultures could be used as a model system to understand apple-Venturia interactions in terms of metabolic re-programming. We were able to identify significant changes in the secondary metabolites in ‘Florina’ cell cultures, indicating metabolic re-programming of specific biosynthetic pathways after VIE-treatment. Based on these results we can conclude that biphenyl and dibenzofuran biosynthesis pathway plays a crucial role in providing scab resistance in the ‘Florina’ cell cultures. In the present study, we analyzed metabolic re-programming in the V. inaequalis elicitor treated apple cell culture system. However, future metabolomics analysis using scab-infected apple plants is required to get a complete picture of metabolic basis of scab resistance in apple. Our results would provide a strong basis for the future metabolomics analysis of apple plants to decipher marker metabolites associated with the scab resistance in apples.
Methods
Chemicals
Solvents used for extraction were of high-performance liquid chromatography grade. 2-phenylphenol (internal standard for gas chromatography) was purchased from SRL chemicals (India). N-methyl-N-(trimethylsilyl)-trifluoroacetamide (MSTFA), pyridine and all metabolite standards were obtained from Sigma.
Plant material and elicitor-treatment
One scab resistant (SR) apple (Malus domestica) cultivar ‘Florina’ and one scab-susceptible (SS) cultivar ‘Vista Bella’ was used in this study to develop cell suspension culture. Cell suspension cultures were developed as described before2. Cell cultures were grown in liquid LS medium containing 2 µM 2,4-D and 1 µM NAA at 26 °C in the dark on an orbital shaker at 120 rpm. An elicitor prepared from apple scab fungus, V. inaequalis was used in this study. V. inaequalis strain (MTCC No.: 1109) was purchased from microbial type culture collection and gene bank (MTCC), Chandigarh, India. V. inaequalis elicitor (VIE) was prepared by homogenizing the fungal hyphae as described before63. Briefly, 10 g of ground fungal mycelium was added to 1 L of acidified water (pH 2.0). Water extract was then boiled for 1 h on a hot plate, cool down to room temperature and filter sterilized. After filtration, the pH of the fungal extract solution was adjusted to 5.0 and the final volume was adjusted to 1 L by adding distilled water. This solution was used as the VIE. Elicitor-treatment was performed by adding 2.5 mL of the VIE (~70 mg of fungal polysaccharide) to the 50 mL of seven-day-old cell apple suspension cultures of both ‘Florina’ and ‘Vista Bella’. Upon elicitor-treatment, cells were harvested at defined time points: 0, 6, 12, 24, 36, 48, and 72 hpe. In control treatment, similar volume of sterile distilled water was added in lieu of the VIE. All experiments were performed with at least three biological repeats.
Extraction of polar metabolites
The VIE-treated apple cell cultures were harvested by vacuum filtration at defined time points (0–72 h) and kept in hot air oven at 60 °C for 4 h. Dried cell mass (2 g) was crushed in liquid nitrogen and fine powdered samples were used for metabolite extraction. The extraction of polar metabolites for GC-MS analyses was performed following the protocol described before64 with suitable modifications. An extraction mixture was prepared by adding methanol/water/chloroform in the ratio of 2.5:1:1 (v/v/v) and stored at −20 °C. Pre-cooled extraction mixture (1 mL) was then added to 200 mg powdered samples in a 1.5 mL micro centrifuge tube and vortexed vigorously at room temperature for 2 min. In order to identify extraction efficiency, 50 µL of 2-phenylphenol (from 2 mg.mL−1 methanol stock) was spiked in the extraction mixture as the internal standard (IS) and vortexed again for 1 min. The extracts were then centrifuged at 14000 g for 5 min. The resulting supernatant (0.8 mL) was transferred into a new 1.5 mL tube and then 0.4 mL of water was added to the supernatant, whole mixture was vortexed for 10 s and subsequently centrifuged at 14000 × g for 5 min. The polar upper phase (methanol/water) was transferred to a new micro-centrifuge tube and then dried out in a vacuum concentrator (Labconco, Centrivap; USA) at 20 °C for 2 h followed by 12 h freeze drying in a lyophilizer. Finally dried material was subjected to double derivatization for GC-MS analyses65. First, derivatization was performed by adding 40 μL of methoxyamine hydrochloride (stock solution: 20 mg.mL−1 in pyridine) to the dried sample and incubating the solutions at 37 °C for 2 h. Then second derivatization was performed by adding 80 μL of N-methyl-N-(trimethylsilyl)-trifluoroacetamide (MSTFA) at 37 °C for 30 min. Derivatization reaction prepared with empty tube served as the control.
GC-MS analysis
GC-MS analysis was performed on Agilent 7890A gas chromatograph (Agilent technologies, CA, USA) coupled with an Agilent 5975C mass detector (Agilent technologies, CA, USA). Double-derivatized sample (1 μL) was injected into GC-MS by automatic sampler (7683B series, Agilent Technologies) with a split ratio of 1:5. DB-5 MS column (5% phenyl methyl polysiloxane: 30 m × 0.25 mm i.d. × 0.25 µm, Agilent technologies) was used for metabolite separation. The temperature program was as follows: Initial temperature of 80 °C for 1 min, followed by temperature increase to 220 °C at the ramp rate of 10 °C.min−1, followed by temperature increase to 310 °C at the ramp rate of 20 °C.min−1 and finally a 10 min hold at 320 °C. Total run time calculated was 39 min. Helium was used as carrier gas at a flow rate of 1 mL.min−1. The inlet temperature and interface temp was set 280 °C. The MS unit was tuned to its maximum sensitivity and the mass range for total ion current was m/z 80–700, and the detector voltage was set at 1700 V. Each sample was replicated three times. Scan was started after solvent delay of 7 min with scan frequency of 4 S−1 (2.0 HZ).
Metabolite identification
Metabolites were identified by comparing the mass-to-charge ratios and abundance of each metabolite detected against a standard NIST-17 mass spectral library (National Institute of Standards and Technology), and our in-house mass spectral database that include several secondary metabolites, amino acids, organic acids, and sugar standards. Metabolite identity was reported only when the matching value of the mass spectra comparison was more than 70 percent, and an increase in the area of the corresponding peak was observed when spiking the sample with the corresponding pure standard. Each mass spectrum was carefully analyzed for co-elution detection. Co-elution was not detected in any of the identified peaks.
Metabolite data pre-processing and statistical analysis
Raw GC-MS data files obtained from Agilent ChemStationTM software were deconvoluted by Automated Mass Spectral Deconvolution and Identification System (AMDIS) using tools available with WsearchPro (www.wsearch.com.au). Metabolite data obtained were further converted into.csv (comma separated values) format before uploading in MetaboAnalyst 4.0 (http://www.metaboanalyst.ca). TIC values were normalized using internal standard. After that, pareto scaling (mean-centered and divided by the square root of standard deviation of each variable) was performed followed by normalization before statistical analyses. Significant differences in metabolite levels were calculated by one-way analysis of variance (ANOVA) using Statistical Package for the Social Sciences (SPSS) software followed by Tukey’s significant-difference test. Statistical significance level was set at p < 0.05. Principal component analyses (PCA) were performed by using interactive online tool of MetaboAnalyst 4.0. The output for PCA data consisted of score plots for visualizing the contrast between various time points of VIE-treated samples and loading plots to explain the cluster separation. A heatmap was created using interactive heatmap tool of MetaboAnalyst 4.0. A simplified metabolic pathway was manually constructed using information from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database via pathway analysis in MetaboAnalyst 4.0.
Quantitative Real-Time PCR
Total RNA was isolated from the VIE-treated ‘Florina’ cultures at defined time points (0–48 hpe) using RNeasy Plant Mini Kit from Qiagen (www.qiagen.com). Total RNA (1 µg) was reverse transcribed at 42 °C using Oligo (dT) primers and RevertAid H Minus reverse transcriptase (Fermentas; www.thermoscientificbio.com) to form cDNA. Quantitative RT-PCR was performed with the QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific) using Power UpTM SYBR Green Master Mix (Thermo Fisher Scientific) following the manufacturer’s instruction. PCR program: 40 cycles at 90 °C for 15 sec followed by 55 °C for 1 min and final extension at 72° C for 1 min. Melt-curve analysis was performed to evaluate gene-specific amplification. Amplification and correlation efficiencies of each PCR were determined using six serial dilutions of cDNA from all samples. To derive relative quantification, the PCR efficiency was used to transform the cycle threshold values into raw data. Expression levels of M. domestica phenylalanine ammonia-lyase (MdPAL), cinnamate-4-hydroxylase (MdC4H), 4-coumarate CoA ligase (Md4CL), chalcone flavanone isomerase (MdCHI), flavanone 3-hydroxylase (MdF3H), biphenyl synthase 3 (MdBIS3), and alternative oxidase (MdAOX) were evaluated using the gene-specific primers (Supplemental Table S3). All samples were normalized using apple actin gene. Scaling of expression level was performed in relation to the respective mRNA expression levels in the control (0 h) cells, which were set to 1. Three technical repeats were performed. Published mathematical model was used for the estimations of efficiency and gene expression levels66.
Conidia germination inhibition assay
V. inaequalis mycelium was grown on potato dextrose agar plates covered by cellophane paper and conidia were harvested as described before67. Conidia were suspended in autoclaved water in a concentration of 1 × 105 conidia.mL−1. Conidia germination inhibition assay was performed as described earlier68. An aliquot (40 µL in 50% methanol) comprised of aucuparin, noraucuparin and eriobofuran, 5 µM each, either individually or in combinations, was mixed with 10 µL of conidial suspension (1 × 105 conidia.mL−1). After mixing well, 10 µL was immediately plated on 2% water-agar slides. In control slides conidia were incubated in presence of 50% methanol. After 24 h incubation at 20 °C the number of germinated conidia of both treated and control slides were counted under microscope.
The inhibition percentage of conidial germination was calculated using following the formula:
C1 = Number of germinated conidia in treated slides.
C2 = Number of germinated conidia in control slides.
Data Availability Statement
All data generated and analyzed in this study are included in this manuscript either as main data or as supplementary information files.
References
Luby, J. J. Taxonomic classification and brief history in apples: botany, production and uses. CABI, 1–14 (2003).
Sarkate, A., Banerjee, S., Mir, J. I., Roy, P. & Sircar, D. Antioxidant and cytotoxic activity of bioactive phenolic metabolites isolated from the yeast-extract treated cell culture of apple. Plant Cell, Tissue Organ Cult. 130, 641–649 (2017).
Boyer, J. & Liu, R. H. Apple phytochemicals and their health benefits. Nutr. J. 3, 5 (2004).
Lee, K. W., Kim, Y. J., Kim, D. O., Lee, H. J. & Lee, C. Y. Major phenolics in apple and their contribution to the total antioxidant capacity. J. Agric. Food Chem. 51, 6516–20 (2003).
Tsao, R., Yang, R., Young, J. C. & Zhu, H. Polyphenolic profiles in eight apple cultivars using high-performance liquid chromatography (HPLC). J. Agric. Food Chem. 51, 6347–6353 (2003).
Wolfe, K., Wu, X. & Liu, R. H. Antioxidant activity of apple peels. J. Agric. Food Chem. 51, 609–614 (2003).
Chizzali, C. et al. Formation of biphenyl and dibenzofuran phytoalexins in the transition zones of fire blight-infected stems of Malus domestica cv. ‘Holsteiner Cox’ and Pyrus communis cv. ‘Conference’. Phytochemistry 77, 179–185 (2012).
He, X. & Liu, R. H. Triterpenoids isolated from apple peels have potent antiproliferative activity and may be partially responsible for apple’s anticancer activity. J. Agric. Food Chem. 55, 4366–70 (2007).
Gerhauser, C. Cancer chemopreventive potential of apples, apple juice, and apple components. Planta Med. 74, 1608–1624 (2008).
Hyson, D. A. A comprehensive review of apples and apple components and their relationship to human health. Adv. Nutr. 2, 408–420 (2011).
Jha, G., Thakur, K. & Thakur, P. The Venturia apple pathosystem: pathogenicity mechanisms and plant defense responses. J. Biomed. Biotechnol. 1–10 (2009).
Bus, V. G. M., Rikkerink, E. H. A., Caffier, V., Durel, C. E. & Plummer, K. M. Revision of the nomenclature of the differential host-pathogen interactions of Venturia inaequalis and Malus. Annu. Rev. Phytopathol. 49, 391–413 (2011).
MacHardy, W. E., Gadoury, D. M. & Gessler, C. Parasitic and biological fitness of Venturia inaequalis: relationship to disease management strategies. Plant Dis. 85, 1036–1052 (2001).
Cusin, R., Revers, L. F., Maraschin, F. & dos, S. New biotechnological tools to accelerate scab-resistance trait transfer to apple. Genetics and Molecular Biology 40, 305–311 (2017).
Bonaterra, A., Badosa, E., Cabrefiga, J., Francés, J. & Montesinos, E. Prospects and limitations of microbial pesticides for control of bacterial and fungal pomefruit tree diseases. Trees 26, 215–226 (2012).
Laurens, F. et al. Review on apple genetics and breeding programmes and presentation of a new European initiative to increase fruit breeding efficiency. J Fruit Sci. 27, 102–107 (2011).
Schouten, H. J. et al. Cloning and functional characterization of the Rvi15 (Vr2) gene for apple scab resistance. Tree Genet. & Genomes 10, 251–260 (2014).
Galli, P., Broggini, G. A. L., Gessler, C. & Patocchi, A. Phenotypic characterzation of the Rvil5 (Vr2) apple scab resistance. J. Plant Pathol. 92, 219–226 (2010).
Würdig, J., Flachowsky, H., Saß, A., Peil, A. & Hanke, M. V. Improving resistance of different apple cultivars using the Rvi6 scab resistance gene in a cisgenic approach based on the Flp/FRT recombinase system. Mol. Breed. 35, 95 (2015).
Flor, H. H. The complementary genic system in flax and flax rust. Adv. Genet. 8, 29–54 (1956).
Komjanc, M. et al. A leucine-rich repeat receptor-like protein kinase (LRPKm1) gene is induced in Malus × domestica by Venturia inaequalis infection and salicylic acid treatment. Plant Mol. Biol. 40, 945–957 (1999).
Clark, M. D., Bus, V. G. M., Luby, J. J. & Bradeen, J. M. Characterization of the defence response to Venturia inaequalis In ‘Honeycrisp’ apple, its ancestors, and progeny. Eur. J. Plant Pathol. 140, 69–81 (2014).
Win, J., Greenwood, D. R. & Plummer, K. M. Characterisation of a protein from Venturia inaequalis that induces necrosis in Malus carrying the V m resistance gene. Physiol. Mol. Plant Pathol. 62, 193–202 (2003).
Nicholson, R. L., Van Scoyoc, S. & Williams, E. B. Response of detached apple leaves to Venturia inaequalis. Phytopathology 63, 649–650 (1973).
Kokubun, T. & Harborne, J. B. A survey of phytoalexin induction in leaves of the rosaceae by copper ions. Sect. C J. Biosci. 49, 628–634 (1994).
Mikulic Petkovsek, M., Slatnar, A., Stampar, F. & Veberic, R. Phenolic compounds in apple leaves after infection with apple scab. Biol. Plant. 55, 725 (2011).
Mayr, U., Michalek, S., Treutter, D. & Feucht, W. Phenolic compounds of apple and their relationship to scab resistance. Journal of Phytopathology 145, 69–75 (1997).
Hrazdina, G., Borejsza-Wysocki, W. & Lester, C. Phytoalexin production in an apple cultivar resistant to Venturia inaequalis. Phytopathology 87, 868–876 (1997).
Sarkate, A., Saini, S. S., Pranav, K., Sharma, A. K. & Sircar, D. Salicylaldehyde synthase activity from Venturia inaequalis elicitor-treated cell culture of apple. J. Plant Physiol. 221, 66–73 (2018).
Holzapfel, C., Meisel, B., Thümmler, F., Leser, C. & Treutter, D. Differential gene expression in leaves of a scab susceptible and a resistant apple cultivar upon Venturia inaequalis inoculation. Trees 26, 121–129 (2012).
Bowen, J. K. et al. Venturia inaequalis: the causal agent of apple scab. Mol Plant Pathol 12, 105–122 (2011).
Dunemann, F. & Egerer, J. A major resistance gene from Russian apple ‘Antonovka’ conferring field immunity against apple scab is closely linked to the Vf locus. Tree Genet. Genome 6, 627–633 (2010).
Gau, A. E., Koutb, M., Piotrowski, M. & Kloppstech, K. Accumulation of pathogenesis-related proteins in the apoplast of a susceptible cultivar of apple (Malus domestica cv. Elstar) after infection by Venturia inaequalis and constitutive expression of PR genes in the resistant cultivar Remo. European Journal of Plant Pathology. 110, 703–711 (2004).
Bolar, J. P., Norelli, J. L., Harman, G. E., Brown, S. K. & Aldwinckle, H. S. Synergistic activity of endochitinase and exochitinase from Trichoderma atroviride (T. harzianum) against the pathogenic fungus (Venturia inaequalis) in transgenic apple plants. Transgenic Research 10, 533–543 (2001).
Wu, Q., Chen, M., Zhou, H., Zhou, X. & Wang, Y. Metabolite profiles of populus in response to pathogen stress. Biochem. Biophys. Res. Commun. 465, 421–6 (2015).
Shuman, J. L. et al. Plant metabolomics by GC-MS and differential analysis. Methods Mol. Biol. 678, 229–46 (2011).
Schauer, N. & Fernie, A. R. Plant metabolomics: towards biological function and mechanism. Trends Plant Sci. 11, 508–16 (2006).
Cuthbertson, D., Andrews, P. K., Reganold, J. P., Davies, N. M. & Lange, B. M. Utility of metabolomics toward assessing the metabolic basis of quality traits in apple fruit with an emphasis on antioxidants. J. Agric. Food Chem. 60, 8552–8560 (2012).
Aprea, E. et al. Metabolite profiling on apple volatile content based on solid phase microextraction and gas-chromatography time of flight mass spectrometry. J. Chromatogr. A 1218, 4517–4524 (2011).
Vanzo, A., Jenko, M., Vrhovsek, U. & Stopar, M. Metabolomic profiling and sensorial quality of ‘golden delicious’, ‘liberty’, ‘santana’, and ‘topaz’ apples grown using organic and integrated production systems. J. Agric. Food Chem. 61, 6580–6587 (2013).
Sciubba, F. et al. Nuclear Magnetic Resonance-based metabolic comparative analysis of two apple varieties with different resistances to apple scab attacks. J. Agric. Food Chem. 63, 8339–8347 (2015).
Fotirić Akšić, M. M. et al. Polyphenolic profile of pear leaves with different resistance to pear psylla (Cacopsylla pyri). J. Agric. Food Chem. 63, 7476–7486 (2015).
Park, Y. J. et al. Triterpene and flavonoid biosynthesis and metabolic profiling of hairy roots, adventitious roots, and seedling roots of Astragalus membranaceus. J. Agric. Food Chem. 63, 8862–9 (2015).
Shen, J. et al. Metabolite profiling of tea (Camellia sinensis L.) leaves in winter. Sci. Hortic. 192, 1–9 (2015).
Sarkate, A. et al. Molecular cloning and functional analysis of a biphenyl phytoalexin-specific O-methyltransferase from apple cell suspension cultures. Planta, https://doi.org/10.1007/s00425-018-3031-6 (2018)
Saini, S. S. et al. Benzaldehyde dehydrogenase-driven phytoalexin biosynthesis in elicitor treated Pyrus pyrifolia cell cultures. J. Plant Physiol. 215, 154–62 (2017).
Rojas, C. M., Senthil-Kumar, M., Tzin, V. & Mysore, K. S. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Frontiers in Plant Science 5, 17 (2014).
Hudina, M. & Śtampar, F. Sugars and organic acids contents of European Pyrus comminus L. and Asian Pyrus serotina r Rehd. pear cultivars. Acta Aliment. 29, 217–230 (2000).
Pastori, G. M. et al. Leaf vitamin C contents modulate plant defense transcripts and regulate genes that control development through hormone signaling. Plant Cell 15, 939–51 (2003).
Petkovickek, M. M., Stampar, F. & Veberic, R. Increased phenolic content in apple leaves infected with the apple scab pathogen. J. Plant Pathol. 90, 49–55 (2008).
Schwalb, P. & Feucht, W. Changes in the concentration of phenolic substances in the bark during the annual development of the cherry tree (Prunus avium L.). Adv. Hortic. Sci. 13, 71–75 (1999).
Treutter, D. & Feucht, W. The pattern of flavan-3-ols in relation to scab resistance of apple cultivars. J. Hortic. Sci. 65, 511–517 (1990).
Picinelli, A., Dapena, E. & Mangas, J. J. Polyphenolic pattern in apple tree leaves in relation to scab resistance. A preliminary study. J. Agric. Food Chem. 43, 2273–2278 (1995).
Treutter, D. Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol. 7, 581–91 (2005).
Widhalm, J. R. & Dudareva, N. A familiar ring to it: biosynthesis of plant benzoic acids. Mol. Plant 8, 83–97 (2015).
Vasyukova, N. I. & Ozeretskovskaya, O. L. Induced plant resistance and salicylic acid: a review. Appl. Biochem. Microbiol. 43, 367–373 (2007).
Vlot, A. C., Dempsey, D. A. & Klessig, D. F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206 (2009).
Lattanzio, V. et al. Low temperature metabolism of apple phenolics and quiescence of Phlyctaena vagabunda. J. Agric. Food Chem. 49, 5817–5821 (2001).
Mukherjee, C., Samanta, T. & Mitra, A. Redirection of metabolite biosynthesis from hydroxybenzoates to volatile terpenoids in green hairy roots of Daucus carota. Planta 243, 305–20 (2016).
Cavallini, E. et al. The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiology 167, 1448–1470 (2015).
Sircar, D., Cardoso, H. G., Mukherjee, C., Mitra, A. & Arnholdt-Schmitt, B. Alternative oxidase (AOX) and phenolic metabolism in methyl jasmonate-treated hairy root cultures of Daucus carota L. J. Plant Physiol. 169, 657–663 (2012).
Arnholdt-Schmitt, B., Costa, J. H. & de Melo, D. F. AOX-a functional marker for efficient cell reprogramming under stress? Trends Plant Sci. 11, 281–287 (2006).
Zhang, Y. et al. Salicylic acid confers enhanced resistance to Glomerella leaf spot in apple. Plant Physiol. Biochem. 106, 64–72 (2016).
Kim, J. K. et al. Metabolic differentiation of diamondback moth (Plutella xylostella (L.) Resistance in cabbage (Brassica oleracea L. ssp. capitata). J. Agric. Food Chem. 61, 11222–11230 (2013).
Lisec, J., Schauer, N., Kopka, J., Willmitzer, L. & Fernie, A. R. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 1, 387–396 (2006).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001).
Parker, D. M. et al. Production and transformation of conidia of Venturia inaequalis. Phytopathology 85, 87–91 (1995).
Hossain, M. B., Piotrowski, M., Lensing, J. & Gau, A. E. Inhibition of conidial growth of Venturia inaequalis by the extracellular protein fraction from the antagonistic bacterium Pseudomonas fluorescens Bk3. Biol. Control 48, 133–139 (2009).
Acknowledgements
This work was supported by a research grant (NASF/ABP 6023/2017–18 to D. Sircar) from the National Agricultural Science Fund (ICAR-NASF), India. A.S and S.S.S are grateful to the Indian Institute of Technology Roorkee for providing MHRD-scholarships. DT acknowledge CSIR-India for providing junior and senior research fellowship. This work was supported by a research grant (NASF/ABP 6023/2017–18 to D. Sircar) from the National Agricultural Science Fund (ICAR-NASF), India.
Author information
Authors and Affiliations
Contributions
D.S. conceived and designed the study. A.S. and S.S.S. carried out the experiments and data processing. D.S., S.S.S., P.K.A. and P.R. performed statistical analyses. S.S.S. and M.G. prepared the figures and tables. D.T. performed metabolomics analyses of susceptible cultivar. D.S., J.I.M., P.R. and P.K.A. interpreted the results. D.S., P.R., S.S.S. wrote and revised the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sarkate, A., Saini, S.S., Teotia, D. et al. Comparative metabolomics of scab-resistant and susceptible apple cell cultures in response to scab fungus elicitor treatment. Sci Rep 8, 17844 (2018). https://doi.org/10.1038/s41598-018-36237-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-36237-y
This article is cited by
-
Biphenyls and dibenzofurans of the rosaceous subtribe Malinae and their role as phytoalexins
Planta (2023)
-
Simultaneous removal of arsenic, fluoride, and manganese from synthetic wastewater by Vetiveria zizanioides
Environmental Science and Pollution Research (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.