Abstract
To clarify the establishment process of coral-algal symbiotic relationships, coral transcriptome changes during increasing algal symbiont densities were examined in juvenile corals following inoculation with the algae Symbiodinium goreaui (clade C) and S. trenchii (clade D), and comparison of their transcriptomes with aposymbiotic corals by RNA-sequencing. Since Symbiodinium clades C and D showed very different rates of density increase, comparisons were made of early onsets of both symbionts, revealing that the host behaved differently for each. RNA-sequencing showed that the number of differentially-expressed genes in corals colonized by clade D increased ca. two-fold from 10 to 20 days, whereas corals with clade C showed unremarkable changes consistent with a slow rate of density increase. The data revealed dynamic metabolic changes in symbiotic corals. In addition, the endocytosis pathway was also upregulated, while lysosomal digestive enzymes and the immune system tended to be downregulated as the density of clade D algae increased. The present dataset provides an enormous number of candidate symbiosis-related molecules that exhibit the detailed process by which coral-algal endosymbiosis is established.
Similar content being viewed by others
Introduction
The association between scleractinian corals and algal symbionts (Symbiodinium spp.), is essential to primary production and reef building in tropical and subtropical oceans. This symbiotic relationship has attracted significant attention due to its sensitivity to environmental changes, often breaking down due to environmental stressors, such as high temperature, high CO2 levels or pollution1,2,3,4. The breakdown of coral-algal endosymbiosis leads to the “coral bleaching” phenomenon, whereby corals appear colorless and damage may ultimately be fatal1,2,5. Corals and the endosymbiont dinoflagellate Symbiodinium live in a mutualistic relationship, corals receiving amino acids, fatty acids and photosynthetic products, such as glucose, from the symbiont, which in turn effectively uses nitrogen and phosphorus from host cells6,7,8,9,10,11,12,13,14,15,16. In addition, Symbiodinium are protected against harmful UV radiation by ultraviolet-absorbing substances, such as mycosporine-like amino acids (MAAs), in the corals17,18,19. Although the details of coral–algal endosymbiotic relationships are becoming better known, the mechanisms of endosymbiont establishment are still remain the questions.
Recently, information on the mechanisms involved in the symbiosis of a cnidarian and Symbiodinium is gradually accumulating20,21. During the initiation of endosymbiosis, coral lectin binds specific glycans on the algal cell wall, the lectin/glycan interaction being a key process for the acquisition of specific algae22,23,24. Acquired algae in corals are surrounded by membrane, a so-called symbiosome, on which Rab family proteins enable the persistence of healthy algal cells and exclusion of dysfunctional algae25,26,27. In addition, previous studies have shown that the TGFβ signaling pathway and Tsr proteins are also involved in the maintenance of algae in cnidarian tissue28,29. Moreover, genes whose expression is up or downregulated between aposymbiotic and symbiotic stages have been identified as candidate symbiosis-related molecules. Genes encoding transporters, carbonic anhydrase, cell membrane proteins, metabolism-associated proteins and peroxidase, plus those related to apoptosis and the inflammatory response, have been identified as endosymbiosis-related genes30,31,32,33,34,35. Although studies involving transcriptomic analyses have become more common over the past decade, the detection of symbiosis-related genes is difficult, some reports showing little difference between symbiotic and non-symbiotic states31,32. Previous studies focused on the very early symbiotic state of planura larvae31 or on the corals colonized with symbiont at high density34,35, but few studies have investigated the process of increasing algal density in the corals. Because the latter process may involves key mechanisms for establishing endosymbiosis being expressed inside corals, it is possible that the molecular process underlying such will be further clarified by studying the genes whose expression changes with increasing Symbiodinium density.
In a previous study, we experimentally prepared corals (Acropora tenuis) associated with monoclonal Symbiodinium [S. goreaui (clade C1) or S. trenchii (clade D1a)] in the laboratory36. Their closely related genotype, clade C1, C2, and D, are naturally associated with A. tenuis in the field, and clade D has been characterized as stress tolerant37,38,39. In this trial, clade C symbionts hardly increased in corals during the first 2 months after inoculation, whereas clade D symbionts increased quickly36, implying that the process of symbiont establishment differs considerably depending on the genetic type (clade) of Symbiodinium. A comparison of these two symbiotic systems therefore provides a clue to understanding the complicated mechanisms underlying endosymbiosis establishment. Next generation sequencing provides larger scale transcriptomic information about both corals and Symbiodinium. In addition to large transcriptomic datasets, whole coral and algal genome sequences have also become available40,41,42,43,44,45,46, which are helpful for distinguishing the transcriptomes of host corals and symbionts from the coral–algae complex. Here, we generated RNA-seq data using the scleractinian coral Acropora tenuis in the early endosymbiosis stage (approximately 10 days and 20 days after the start of co-incubation following inoculation) with clade C and clade D Symbiodinium. We investigated the algal symbiont-dependent changes in gene expression by comparing transcriptomes between the aposymbiotic and symbiotic to clarify the establishment of symbiosis.
Results and Discussion
Algal cells were broken during the early symbiosis stage
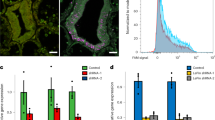
To provide corals in the early symbiosis stage for RNA-seq analyses, A. tenuis juvenile corals (corals a few weeks after settlement) were maintained in artificial seawater, with subsequent introduction of S. goreaui (clade C) and S. trenchii (clade D) [hereafter, corals inoculated with S. goreaui (clade C) are described as “C-corals”; those inoculated with S. trenchi (clade D) are described as “D-corals”]. The establishment process of the coral–algal endosymbiosis was observed using a stereoscopic microscope for the first 20 days after Symbiodinium inoculation (Fig. 1a). Additionally a coral homogenate was observed under a microscope and the number of endosymbiotic algae counted (Fig. 1b). C-corals had almost no algae after 10 days (n = 10), with the average number of clade C cells per polyp at 20 days being 63.3 ± 15.4 (mean ± SE, n = 7). However, all of the clade C endosymbionts were abnormally shaped (swollen or fragmented) (Fig. 1b). Clade D cells in the polyps numbered 292.4 ± 61.6 (n = 5) at 10 days, increasing to 578.3 ± 17.4 (n = 6) by 20 days. Abnormal shapes were observed in 16.4% of the clade D symbionts at 10 days and 11.4% of symbionts at 20 days (Fig. 1b). In past studies, abnormally broken algal cells have been observed in histological sections of primary polyps47 and adult corals exposed to higher temperature48, indicating that symbiotic algae could be digested in host cells, especially at the onset of endosymbiosis and under bleaching conditions.
The early symbiotic stage of juvenile corals, and the number of differentially expressed genes. (a) Clade D Symbiodinium increased in Acropora tenuis juveniles (D-Symbiosis), whereas clade C colonization (C-Symbiosis) was very slow within 20 days. (b) Abnormally expanded Symbiodinium cells were observed in coral homogenates. Red arrows indicate expanded Symbiodinium cells and black arrow indicates normal Symbiodinium cell. Scale bars = 0.01 mm. (c) Bar graph showing the number of differentially expressed coral genes detected at days 10 and 20 after inoculation with Symbiodinium based on comparisons with aposymbiotic corals (apo).
In contrast to clade C, clade D densities showed a relatively steady increase, which may be attributable to their ability to tolerate or avoid the host’s immune system49. Transcriptome analysis of these corals is one of the most effective ways of determining the nature of changes that occur with the success and failure of endosymbiosis.
Increasing numbers of differentially-expressed coral genes with increasing endosymbiont density
Corals were fixed 10 and 20 days following inoculation with the two Symbiodinium clades (Fig. 1a), for the RNA-seq analyses, performed using an Illumina Hiseq 2000 (Illumina Inc., San Diego, CA, USA). De novo transcriptome assembly from the Illumina sequencing data and Basic Local Alignment Search Tool nucleotide database (BLASTn) searches resulted in 108,247 contigs obtained from A. tenuis, and 22,773 and 22,426 from clade C and clade D Symbiodinium, respectively.
The GC content distribution of the assembled coral and Symbiodinium transcriptomes showed two clear peaks at approximately 43% and 55%, apparently having originated from the coral and symbiont, respectively, since the reference transcriptome data from the coral and algae showed similar GC contents (Supplementary Fig. S2). Gene expression was detected in the coral by mapping the data on a fasta file derived from corals, and the differentially expressed genes (DEGs) between symbiotic and aposymbiotic corals identified using a R package, edgeR (Fig. 1c). Reproducibility among biological replicates was relatively high (correlation coefficient >0.89), although it is necessary to pay attention to the fact that two biological replicates in each condition have insufficient statistical power to detect DEGs.
We identified 991 DEGs in C-corals at 10 days, 592 DEGs in C-corals at 20 days, 3,116 DEGs in D-corals at 10 days, and 7,667 DEGs in D-corals at 20 days (Supplementary Fig. S3). Venn diagrams (Supplementary Fig. S4) revealed overlaps among the four different biological conditions, but identified only three downregulated DEGs [three homologs of green fluorescent protein - (GFP)-like chromoproteins] that were common to all four (Supplementary Table S1), suggesting that coral GFP is more sensitive to contact with symbiotic algae at the onset of endosymbiosis. In order to investigate coral gene expression change correlated with algal density, correlation coefficients (R) between algal density (apo, 10 days, 20 days) and gene expression was calculated for D-corals. The results indicated 919 genes positively correlated (R > 0.90) and 405 genes negatively correlated (R < −0.90) with algal density. Tables S2 and S3 show the 100 genes most highly correlated (R > 0.90, or R < −0.90) with algal density (cells/polyp) and gene expression. Genes for peroxidasin, glutamine synthetase and solute carrier family 26 member 6 (SLC26A6) had a positive slope, indicating they were strongly up-regulated by the increase in algal cell density (Supplementary Table S2). In contrast, genes for Alpha-glucosidase, low-affinity immunoglobulin epsilon Fc receptor and programmed cell death protein, for example, had a steeply negative slope, indicating their strong down-regulation with increasing symbiont density (Supplementary Table S3).
The RNA-seq method was validated using quantitative polymerase chain reaction (qPCR) and the gene expression of 13 randomly selected DEGs compared (Supplementary Fig. S4). The expression patterns observed in the qPCR results were generally consistent with the patterns obtained by RNA-seq. In addition, algal gene expression was also detected from the endosymbionts. Due to a lack of control conditions for Symbiodinium, the detection of DEGs using the Symbiodinium transcriptome could not be statistically analyzed. However, all Symbiodinium genes expressed in each sample were isolated and utilized for the discussion of Symbiodinium-coral interactions. Among the 22,773 clade C contigs and 22,426 clade D contigs, 48.3% and 71.1%, respectively, showed significant similarities (Blastx e-value < 1e–4) to the Swiss-Prot database.
Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed using DAVID (https://david.ncifcrf.gov/)50 and the KEGG mapper (http://www.genome.jp/kegg/mapper.html)51 to identify enriched biological processes from the coral DEGs and algal transcriptome. Fig. 2 shows the GO annotation results for up and downregulated genes. The results showed that the upregulated genes in D-corals included a large number involved in metabolism, indicating that the presence of symbiotic algae has a great influence on coral metabolism. The C-corals, on the other hand, had a considerably smaller number of DEGs, with only a few enriched GO and KEGG pathways being detected. The results of the D-coral analysis provide the basis of the following discussion on the establishment of endosymbiosis, and are compared with the clade C results representing a case in which algae do not increase in density.
Transition of the coral immune system to accept algal endosymbionts
A number of immune system-related genes were detected as symbiosis-associated genes. GO terms associated with regulation of the immune effector process, defense response, immune system processes, and positive regulation of lymphocyte-mediated immunity were downregulated in D-corals (Fig. 2 and Supplementary Fig. S4). These GO terms include tumor necrosis factor receptor-associated factor 6, matrix metalloproteinase 9, low-affinity immunoglobulin epsilon Fc receptor, lipopolysaccharide binding protein, and toll-like receptor 2. The decrease in expression of gene encoding low-affinity immunoglobulin epsilon Fc receptor showed a strongly negative correlation with increasing clade D algal cell density, suggesting that a decrease in the gene expression may be caused by the link to algal cell density (Supplementary Table S3). In this context, genes related to the TGFβ signaling pathway were also upregulated in D-corals (TGFβ acting as a key regulator of immune tolerance and inflammatory responses) (Supplementary Fig. S5c). In Aiptasia, adding anti-TGFβ to block the putative TGFβ pathway results in immune stimulation and a failure of the symbiont to colonize the host28. Such changes in the host immune system could be important for maintaining symbionts in the host cells, as well as during viral and bacterial infections52. In the case of C-corals, such changes relating to immune system reduction were not detected. The different expression patterns of the host immune system depended on the symbiont type, revealing that a decline in the immune system is necessary for the acceptance of many symbionts into host tissue.
Activation of endocytosis and deactivation of lysosomal acid hydrolase
The results of the KEGG pathway analysis revealed dynamic changes in the endocytosis pathway in D-corals (Supplementary Fig. S5abc). Clathrin-dependent endocytosis, as well as recycling of endosome- and late endosome-associated proteins, were upregulated with an increase in the number of algae. Since the clathrin-mediated pathway of endocytosis has an upper size limit for internalization of approx. 200 nm, it could not be directly associated in the incorporation of endosymbiotic algae, but be associated in a recycling of symbiosome membrane53,54. Our results showed that the expression levels of Rab GTPase family proteins (Rab proteins) 7, 8 were changed significantly in D-corals (Fig. 3, Supplementary Fig. S6a). Studies of Aiptasia have demonstrated that several types of Rab proteins (Rab 3, 5, 7, and 11) are localized in the symbiosome, each playing an important role in the maintenance, degradation and recycling of endosymbionts25,26,27. These studies show that Rab 7 is absent in the majority of phagosomes containing live and healthy zooxanthellae in Aiptasia, whereas phagosomes containing heat-killed or -damaged zooxanthellae stain positively for Rab 725. In corals, Rab 7 was reported to be expressed at significantly higher levels under high-light/high-temperature conditions associated with oxidative stress and coral bleaching55. Accordingly, Rab 7 may be involved in the elimination of damaged Symbiodinium. Another upregulated Rab protein, Rab 8, is a membrane trafficking protein with functions in the membrane recycling process56. The high expression of these Rab proteins in D-corals may be associated with eliminating broken algal cells found during the early stages of endosymbiosis (Fig. 1b).
Heatmap of differentially expressed genes (DEGs) in aposymbiotic corals (aposymbiotic), corals associated with clade C (clade C-Symbiosis) and those associated with clade D (clade D-Symbiosis). There are selected notable treatment-specific genes (FDR P < 0.05) form each functional categories. The color shows expression levels normalized to z-score in all biological replicates; blue and yellow colors denote low and high intensities, respectively. The original heatmaps of each functional category are shown in Supplementary.
A large number of Rab proteins were also identified in the algal transcriptome, including Rab 1, 2, 3, 4, 5, 6, 7, 11, 23 and 28 (Supplementary Fig. S6b). Among them, Rab 5b is a candidate symbiosis-related algal gene, having been identified from the Plasmodium parasite, where it was localized in the parasitophorous vacuole membrane surrounding intracellular Plasmodium57. In the case of the symbiosome membrane of a cnidarian host, the symbiosome is partially derived from Symbiodinium itself58. The identification of Rab 5b from Symbiodinium suggests the possibility that Symbiodinium-derived Rab proteins may also become localized in the symbiosome membrane, being involved in the establishment of endosymbiosis.
Although some endocytosis-related genes were expressed at greater levels in D-corals, lysosomal acid hydrases, such as proteases, lipases, sulfatases, glucosidases, sphingomyelinases and ceramidases, tended to decrease (Fig. 3, Supplementary Fig. S7). In particular, downregulation of several digestive enzymes targeting glycan is possibly related to the establishment of endosymbiosis. Because glycan is a cell wall component of Symbiodinium59, such declines in digestive enzymes would seem essential to avoid the digestion of endosymbiotic Symbodinium cells. Our observations indicated that symbiotic algal cells were broken 10 days after inoculation, the proportion of broken cells thereafter having decreased at 20 days. Such a decrease was probably associated with a decrease in the expression of digestive enzymes in the corals.
Nonsense transcripts expressed in the symbiont
As mentioned above, a decline in the coral immune system and digestive enzyme activities occurs at the beginning of endosymbiosis, although the trigger for such transcriptome changes remains unclear. However, examining characteristic categories from the Symbiodinium transcriptome provides an insight, for example, of genes involved in microRNA expression in Symbiodinium. A gene cluster associated with RNA interference, including a regulator of the nonsense transcript 1 homolog and a probable ATP-dependent RNA helicase spindle-E, was enriched in symbiotic clade D Symbiodinium on day 20 (Supplementary Fig. S8). This suggests that nonsense-mediated RNA decay occurred in the symbiont as density of the latter increased and that symbiont nonsense RNA modulates the cnidarian host transcriptome, including immune system-related and digestive enzyme-related genes. Some studies have suggested that cnidarian protein-coding genes are predicted targets of symbiont miRNA-mediated post-transcriptional regulation42,60. In plant root endosymbioses (e.g., the legume Rhizobium with arbuscular mycorrhizal fungi) and human bacterial infections, symbiont-derived miRNA sequences significantly regulate the expression of targeted host genes61. Thus, although further verification of the function of miRNA in endosymbiosis is needed, RNAi via miRNA may well be important in the intracellular symbiotic system.
Changes in metabolic processes
The transcriptome data revealed that coral metabolism increased dramatically as a result of the increase in the number of Symbiodinium in corals (Figs 2,3). The pathways related to carbon metabolism and biosynthesis of amino acids were enriched as the genes encoding their components were upregulated in corals colonized with a high density of clade D Symbiodinium (Fig. 3, Supplementary Figs S9, S10). Upregulation of coral carbon metabolic processes is closely related to the photosynthetic pathway of Symbiodinium. Symbiodinium possesses the crassulacean acid metabolism (CAM) photosynthetic pathway62, which largely influences coral metabolism because genes related to the metabolism of pyruvate, malate, and oxaloacetate, the CAM photosynthetic metabolites, were upregulated in symbiotic corals (Supplementary Fig. S11). The expression of genes related to fructose metabolism, such as the fructose-1,6-bisphosphatase gene, increased remarkably (Supplementary Fig. S11), indicating that corals obtain a large amount of fructose from algae. One of the glucose metabolic pathways, the pentose phosphate pathway, was also upregulated in D-corals (Supplementary Fig. S12). The pentose phosphate pathway and its intermediate products are involved in the production of many bioactive substances, including nucleotides and MAAs, which are UV-absorbing molecules63. These results revealed that coral could receive source of carbon hydrate and nucleic acids from symbiont.
The existence of glucose metabolism regulating systems was also suggested by the transcriptome data obtained here. A series of insulin signaling pathways linked to lipogenesis, glycogenesis, anti-lipolytic functions, insulin-like growth factor and insulin-like growth factor receptor homologs were up- or down- regulated in D-corals (Figs 2,3 and Supplementary Fig. S13). Although such signaling pathways have not yet been elucidated in corals, insulin plays an important role in regulating developmental processes in Hydra64,65. Additionally, Symbiodinium may also be indirectly affected by insulin-like growth factor, since genes encoding insulin-induced or insulin-degrading enzymes were detected among the Symbiodinium transcripts. Although further confirmation of the presence of insulin-like substances is necessary, our data suggested that such substances could regulate coral glucose metabolism, since a series of genes related to insulin were detected in both coral and zooxanthella. Considering that Symbiodinium provides glucose to corals11, the coral insulin signaling pathway may have a role in restricting the use of sugars obtained from the algae.
Symbiodinium also possesses a pathway for ammonia assimilation into glutamate, such metabolic processes facilitating the effective use of amino acids in corals (Figs 2,3, Supplementary Fig. S14). Glutamine synthesis and related metabolic processes, such as proline metabolism, were upregulated in D-corals at 10 days, and it appears that corals promote the metabolism of glutamine (Supplementary Fig. S15). The results were similar to past studies involving RNA-seq analysis of algal-symbiotic and non-symbiotic anemones, which reported that transcripts for glutamine synthetase and an NADPH-dependent glutamate synthase are upregulated in the symbiotic state50. Proline synthetic pathways of corals were also enhanced in D-corals and their symbiont (Supplementary Fig. S15). Proline mignt be transported to endosymbiotic algae because osmoregulated proline transporters are expressed in endosymbiotic algae. As the transporter name implies, proline is a common osmolyte in plants and bacteria66. Some osmoregulated proline transporters were expressed in clade D symbionts, but not in those of clade C, suggesting that proline is necessary for algae to survive in host cells under higher-osmolality conditions. The results further showed that glutathione peroxidase, which catalyzes the reduction of hydroxyperoxides by glutathione, is expressed at higher levels in D-corals than in C-corals (Supplementary Fig. S16). Glutathione is synthesized from glutamate, the metabolism of which is activated by algal symbiosis. Hence it is possible that glutathione peroxidase utilizing glutamate may be activated in symbiotic corals. Glutathione peroxidase acts as a stress marker gene in corals, its expression increasing under stressful conditions67. Because symbiotic Symbiodinium itself produces reactive oxygen species, especially under high temperature and light stress, corals are generally at risk of oxidative stress by acquiring algae. The ability to modulate antioxidation by enzymes, such as glutathione peroxidase, is indispensable for accepting endosymbiotic algae, the synthesis of such enzymes being activated by the algae.
In addition, sulfur assimilation was also upregulated by the clade D symbiont (Fig. 3, Supplementary Fig. S17a). As past studies have demonstrated, sulfate utilization by corals may be influenced by algal endosymbiosis, the up-regulation of the sulfate assimilation process possibly being a consequence of algal-coral endosymbiosis68. The sulfate assimilation related enzyme bifunctional 3′-phosphoadenosine 5′phosphosulfate synthase underwent increased expression in D-corals, but the homologous gene could not be detected in Symbiodinium, which instead had a gene encoding the adenosine 3′-phospho 5′-phosphosulfate transporter (Supplementary Fig. S17b). It is therefore conceivable that corals with algal cells actively uptake sulfate ions and synthesize Adenosine 3′-phospho 5′-phosphosulfate, which is transported to Symbiodinium.
Other metabolic processes enhanced by algal symbiosis include biosynthesis of the neurotoxins69, γ-glutamyl-β-cyanoalanine and γ-glutamyl-β-aminopropionitrile, biosynthesis of plasmanylcholine and metabolism of thiamine (vitamin B1). Plasmanylcholine is involved in acclimation to a temperature-fluctuating environment in Screlactinian corals70 (Fig. 3, Supplementary Figs S18–S20). Although most corals cannot alone synthesize these substances, they may be synthesized using metabolites of the symbiont, and accelerated synthesis of these substances could affect in coral health and defense mechanism.
Taking our findings together, we detected the processes required to establish endosymbiosis by focusing on transcriptomic changes during the early stage of symbiosis (Fig. 4). The following processes were considered to occur during the establishment of symbiosis: 1) Upregulation of the endocytosis pathway and a decrease in the expression of digestive enzymes for uptake and maintenance of algal symbionts. 2) The coral immune response system was partially inactivated. 3) miRNA of the endosymbiotic algae was overexpressed during the early stage of endosymbiosis. Additionally, the following coral metabolic processes changed dramatically: metabolism of sugar, pyruvate, malate, oxaloacetate, glutamate, thiamine and proline, and biosyntheses of γ-glutamyl-β-cyanoalanine, γ-glutamyl-β-aminopropionitrile, plasmanylcholine. The transcriptome data also suggested that corals regulate sugar metabolic processes via an insulin-like signaling pathway.
Schematic summary of events presumably occurring in the early symbiosis of reef-building corals and alga, based on the transcriptome data of D-corals. Red arrows indicate up-regulated genes, and blue arrow indicate down-regulated genes with with increasing algal symbiont. The metabolic pathway and signaling pathway influenced by the presence of algal symbionts are represented by thin black arrows and dashed arrows, respectively.
In this study, we clarified the endosymbiosis process by comparing two algal symbiotic systems with different rates of increase in the number of symbionts. The study investigated endosymbiosis establishment based on corals colonized by Symbiodinium trenchi (clade D). Accordingly, it is necessary to verify whether or not the symbiosis-related changes described here would occur generally in response to other Symbiodinium clades. Notwithstanding, the changes in expression of host genes with an increased density of symbiotic algae is expected to further clarify our understanding of endosymbiosis-related pathways and metabolism.
Materials and Methods
Animals and algae
Acropora tenuis were collected around Akajima Island, Okinawa by Akajima Marine Science Laboratory (Okinawa, Japan). Collection of A. tenuis larvae was performed as previously described in Iwao et al.71. Several days after spawning, metamorphosis was induced by exposure of larvae to 2 μM Hym 248 in containers (55-mm diameter)34. The Symbiodinium strains CCMP2466 (clade C1) and CCMP2556 (clade D) were obtained from the Bigelow Laboratory for Ocean Sciences (West Boothbay Harbor, ME, USA; https://ccmp.bigelow.org/) and cultured in f/2 medium (Wako Chemicals, Osaka, Japan) with antibiotics (kanamycin 20 μg/mL and ampicillin 50 μg/mL) at 24 °C under a 12-h light (20 µE/m2/s), 12-h dark cycle. Juvenile polyps were cultured in Petri dishes containing artificial seawater (KUMARINE SeaSalt α, Marine route one, Kanagawa, Japan) at 24 °C under a 12-h light (70 µE/m2/s), 12-h dark cycle. Cells of each strain of Symbiodinium algae (approximately 1,000 cells per polyp) were introduced to A. tenuis primary polyps 10 days after settlement. Each Symbiodinium culture was subsequently introduced into Petri dishes containing polyps every day. Approximately 40 larvae settled in each dish. Some of the juvenile polyps were maintained in an aposymbiotic state and used for experiments. Five dishes were used for each treatment (inoculation with clade C1, inoculation with clade D, aposymbiotic), and seawater was changed daily.
Microscopic observations
Juvenile polyps were observed with a stereomicroscope during the incubation period. Color micrographs of several polyps were taken with a digital scanning microscope (model VHX-1000; Keyence, Tokyo, Japan) using a digital camera (Digital Slight DA-L1; Nikon) to evaluate the density of symbionts. Micrographs of polyps were taken 10 and 20 days after inoculation with Symbiodinium algal cultures. Approximately five polyps were photographed in each treatment.
Symbiodinium cell counts
Polyps were fixed in 3% formaldehyde 10 and 20 days after inoculation with Symbiodinium. Subsequently, samples were decalcified using decalcification solution containing 0.5 M ethylenediaminetetraacetic acid (EDTA) for 2 days36. Each polyp was placed in a 1.5-mL tube containing 0.01% TritonX, and homogenized. Algal cells in the homogenate were counted using a hemocytometer (Thomas Scientific, Swedesboro, NJ). Three or four polyps were used for counting of Symbiodinium algal cells in 10 days and 20 days after inoculation with Symbiodinium. Polyps associated with clade C1 algae contained a small number of Symbiodinium cells. These polyps were crushed using a cover glass onto a glass slide to count their endosymbiotic Symbiodinium cells, because a very small number of algal cells could be lost in the homogenate and hemocytometer.
RNA extraction
Polyps collected 10 and 20 days after inoculation with clade C or clade D Symbiodinium cultures were fixed in RNAlater (Ambion, Austin, TX, USA). Polyps in four replicates of each treatment were fixed and used for RNA extraction, and one or two replicates were used for each set of biological observations. For RNA-seq analysis, two biological replicate were prepared for each day (one replicate derived from one petri dish). Total RNA was extracted from juvenile polyps using a PureLink RNA Mini kit (Life Technologies corporation, Carlsbad, CA). The total RNA was treated with DNase I (TAKARA, Ohtsu, Japan) to digest genomic DNA, and then mRNA was purified from the samples using the NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB, Ipswich, MA).
RNA-seq
cDNA libraries were generated from mRNA samples using the NEBNext mRNA Library Prep Master Mix Set for Illumina (NEB). Pared-end sequencing of 100 bp was performed by Macrogen Japan using a HiSeq 2000 sequencer (Illumina, San Diego, CA), which resulted in over 35 million reads per sample. Short reads were first pre-processed, trimming bases with a Phred quality score below Qv = 20 from the 5′ end and 3′ end of each read, and retaining reads ≥25 bp, while reads with 30% of bases having Qv ≤ 15 were filtered out. This processing was performed using the DDBJ pipeline (https://p.ddbj.nig.ac.jp/pipeline/Login.do). All sequence data were deposited in the DDBJ/EMBL/GenBank databases under accession number DRA006413.
Identification of differentially expressed genes between coral samples
Trinity (version, 2.1.1) software was used to assemble de novo transcripts from all reads in 12 A. tenuis samples (aposymbiotic corals, clade C symbiotic corals, and clade D symbiotic corals). Then, TransDecoder (http://transdecoder.sourceforge.net/) was used to identify candidate regions from the assembled transcripts or contigs. To detect contig sequences originating from the host coral, we built custom coral and Symbiodinium databases. The coral database included an A. digitifera genome40, non-symbiotic A. hyacinsus and A. tenuis transcriptomes (http://www.bio.utexas.edu/research/matz_lab/matzlab/Data.html), and de novo A. tenuis transcriptomes obtained from aposymbiotic polyps in this study. The Symbiodinium database includes a Symbiodinium genome41, and Symbiodinium clade C and clade D transcriptome data from the PALUMBI Lab72. The nucleotide sequences of the assembled contigs were aligned to each custom database using BLASTn, and contigs that aligned only to the coral database were annotated as A. tenuis contigs. Different e-value cutoffs (1e−1, 1e−2, 1e−3, 1e−4, 1e−5, and 1e−10) were examined and the cutoff value (1e−4) that maximized the number of A. tenuis contigs was adopted with reference to Shinzato et al.73. Reads were aligned to coral contigs using bowtie2 (version 2.2.4) software and mapped reads were counted using eXpress (version, 1.5.1) software. The R package edgeR was used to compare gene expression between inoculated polyps and non-treated polyps after 10 or 20 days, and to identify detected contigs as differentially expressed if the adjusted false-discovery rate was P ≤ 0.05.
Analysis of the Symbiodinium tnranscriptome
To detect contig sequences originating from clade C or clade D Symbiodinium, a de novo transcript was assembled using Trinity (version, 2.1.1) from 522 million reads from 8 A. tenuis samples associated with clade C Symbiodinium and 493 million reads from 8 A. tenuis samples associated with clade D Symbiodinium. Then, TransDecoder was used to identify candidate regions, and contigs that aligned only to the Symbiodinium database were annotated as Symbiodinium clade C or clade D transcripts by the same method used for identification of coral genes. For analysis of Symbiodinium transcriptomes, reads from samples collected 10 and 20 days after inoculation were aligned to Symbiodinium contigs and counted using bowtie2 (version, 2.2.4) and eXpress (version, 1.5.1). Contigs that were aligned ≥1 time were annotated against the SwissProt protein database and NCBI non-redundant protein database (nr) using the blastx program (BLAST 2.2.30) to determine their functions.
Blastx search, GO enrichment analysis, and KEGG pathway analysis of DEGs, and quantitative PCR can be found in SI Materials and Methods.
References
Glynn, P. W., Maté, J. L., Baker, A. C. & Calderón, M. O. Coral bleaching and mortality in panama and ecuador during the 1997–1998 El niño–southern oscillation event: spatial/temporal patterns and comparisons with the 1982–1983 event. Bull. Mar. Sci. 69, 79–109 (2001).
Bruno, J., Siddon, C., Witman, J., Colin, P. & Toscano, M. El Niño related coral bleaching in Palau, Western Caroline Islands. Coral reefs 20, 127–136 (2001).
Anthony, K. R. N., Kline, D. I., Diaz-Pulido, G., Dove, S. & Hoegh-Guldberg, O. Ocean acidification causes bleaching and productivity loss in coral reef builders. Proc. Natl. Acad. Sci. USA 105, 17442–17446 (2008).
Wooldridge, S. A. Water quality and coral bleaching thresholds: Formalising the linkage for the inshore reefs of the Great Barrier Reef, Australia. Mar. Pollut. Bull. 58, 745–751 (2009).
Gates, R. D., Baghdasarian, G. & Muscatine, L. Temperature stress causes host cell detachment in symbiotic cnidarians: implications for coral bleaching. Biol Bull. 182, 324–332 (1992).
Muscatine, L. The role of symbiotic algae in carbon and energy flux in reef corals. Coral Reefs 25, 75–87 (1990).
Muscatine, L. & Ceiwichmfu, E. Assimilation of photosynthetic products of zooxanthellae by a reef coral. Biol. Bull. 137, 506–523 (1969).
Trench, R. K. The physiology and biochemistry of zooxanthellae symbiotic with marine coelenterates. I, The assimilation of photosynthetic products of zooxanthellae by two marine coelenterates. Proc. R. Soc. Lond. B 177, 225–235 (1971a).
Trench, R. K. The physiology and biochemistry of zooxanthellae symbiotic with marine coelenterates II. Liberation of fixed 14C by zooxanthellae in vitro. Proc. R. Soc. Lond. B 177, 237–250 (1971b).
Battey, J. F. & Patton, J. S. A reevaluation of the role of glycerol in carbon translocation in zooxanthellae-coelenterate symbiosis. Mar. Biol. 79, 27–38 (1984).
Whitehead, L. F. & Douglas, A. E. Metabolite comparisons and the identity of nutrients translocated from symbiotic algae to an animal host. J. Exp. Biol. 206, 3149–3157 (2003).
Wang, J. T. & Douglas, A. E. Essential amino acid synthesis and nitrogen recycling. in an alga-invertebrate symbiosis. Mar. Biol. 135, 219–222 (1999).
Papina., M., Meziane, T. & Van Woesik, R. Acclimation effect on fatty acids of the coral Montipora digitata and its symbiotic algae. Comp. Biochem. Physiol. Biochem. Mol. Biol. 147, 583–589 (2007).
Piniak, G. A., Lipschultz, F. & McClelland, J. Assimilation and partitioning of prey nitrogen within two anthozoans and their endosymbiotic zooxanthellae. Mar. Ecol. Prog. Ser. 262, 125–136 (2003).
Tanaka, Y., Grottoli, A. G., Matsui, Y., Suzuki, A. & Sakai, K. Partitioning of nitrogen sources to algal endosymbionts of corals with long-term 15N-labelling and a mixing model. Ecol. Modell. 309, 163–169 (2015).
Tanaka, Y., Miyajima, T., Koike, I., Hayashibara, T. & Ogawa, H. Translocation and conservation of organic nitrogen within the coral-zooxanthella symbiotic system of Acropora pulchra, as demonstrated by dual isotope-labeling techniques. J. Exp. Mar. Bio. Ecol. 336, 110–119 (2006).
Rosic, N. N. & Dove, S. Mycosporine-like amino acids from coral dinoflagellates. Appl. Environ. Microbiol. 77, 8478–8486 (2011).
Jokiel, P. L., Lesser, M. P. & Ondrusek, M. E. UV-absorbing compounds in the coral Pocillopora damicornis: Interactive effects of UV radiation, photosynthetically active radiation, and water flow. Linmol. Oceanorg. 42, 1468–1473 (1997).
Dunlap, W. C., Chalker, B. E., Bandaranayake, W. M. & Wu Won, J. J. Nature’s sunscreen from the Great Barrier Reef, Australia. Int. J. Cosmet. Sci. 20, 41–51 (1998).
Fransolet, D., Stéphane, R. & Plumier, J. C. Establishment of endosymbiosis: the case of cnidarians and Symbiodinium. J. Exp. Mar. Biol. Ecol. 420-421, 1–7 (2012).
Meyer, E. & Weis, V. M. Study of Cnidarian-algal symbiosis in the “omics” age. Biol. Bull. 223, 44–65 (2012).
Jimbo, M. et al. The D- galactose-binding lectin of the octocoral Sinularia lochmodes: characterization and possible relationship to the symbiotic dinoflagellates. Comp. Biochem. Physiol. B 125, 227–236 (2000).
Wood-Charlson, E. M., Hollingsworth, L. L., Krupp, D. A. & Weis, V. M. Lectin/glycan interactions play a role in recognition in a coral/dinoflagellate symbiosis. Cell. Microbiol. 8, 1985–1993 (2006).
Koike, K. et al. Octocoral chemical signaling selects and controls dinoflagellate symbionts. Biol. Bull. 207, 80–86 (2004).
Chen, M. C., Cheng, Y. M., Sung, P. J., Kuo, C. E. & Fang, L. S. Molecular identification of Rab7 (ApRab7) In Aiptasia pulchella and its exclusion from phagosomes harboring zooxanthellae. Biochem. Biophys. Res. Commun. 308, 586–595 (2003).
Chen, M. C. et al. ApRab11, a cnidarian homologue of the recycling regulatory protein Rab11, is involved in the establishment and maintenance of the Aiptasia-Symbiodinium endosymbiosis. Biochem. Biophys. Res. Commun. 338, 1607–1616 (2005).
Chen, M. C., Cheng, Y. M., Hong, M. C. & Fang, L. S. Molecular cloning of Rab5 (ApRab5) In Aiptasia pulchella and its retention in phagosomes harboring live zooxanthellae. Biochem. Biophys. Res. Commun. 324, 1024–1033 (2005).
Detournay, O., Schnitzler, C. E., Poole, A. & Weis, V. M. Regulation of cnidarian-dinoflagellate mutualisms: Evidence that activation of a host TGF innate immune pathway promotes tolerance of the symbiont. Dev. Comp. Immunol. 38, 523–537 (2012).
Neubauer, E. F. et al. A diverse host thrombospondin-type-1 repeat protein repertoire promotes symbiont colonization during establishment of cnidarian-dinoflagellate symbiosis. eLife 6, e24494 (2017).
DeBoer, M. L., Krupp, D. A. & Weis, V. M. Proteomic and transcriptional analyses of coral larvae newly engaged in symbiosis with dinoflagellates. Comp. Biochem. Physiol. D. 2, 63–73 (2012).
Voolstra, C. R. et al. The host transcriptome remains unaltered during the establishment of coral–algal symbioses. Mol. Ecol. 18, 1823–1833 (2009).
Schnitzler, C. E. & Weis, N. M. Coral larvae exhibit few measurable transcriptional changes during the onset of coral-dinoflagellate endosymbiosis. Mar. Genom. 3, 107–116 (2010).
Lehnert, E. M. et al. Extensive differences in gene expression between symbiotic and aposymbiotic cnidarians. G3 4, 277-295 (2014).
Yuyama, I. et al. Identification of symbiotically expressed coral mRNAs using a model infection system. Biochem Biophys. Res. Commun. 36, 793–798 (2005).
Yuyama, I., Watanabe, T. & Takei, I. Profiling differential gene expression of symbiotic and aposymbiotic coral using a high coverage gene expression profiling (HiCEP) analysis. Mar. Biotech. 13, 1436–2236 (2011).
Yuyama, I. & Higuchi, T. Comparing the effects of symbiotic algae (Symbiodinium) clades C1 and D on early growth stage of Acropora tenuis. Plos One 9, e98999 (2014).
van Oppen, M. J., Palstra, F. P., Piquet, A. M. & Miller, D. J. Patterns of coral-dinoflagellate associations in Acropora: significance of local availability and physiology of Symbiodinium strains and host-symbiont selectivity. Proc. Biol. Sci. 268, 1759–1767 (2001).
Abrego, D., van Oppen, M. J. & Willis, B. L. Onset of algal endosymbiont specificity varies among closely related species of Acropora corals during early ontogeny. Mol. Ecol. 18, 3532–3543 (2009).
Yuyama, I., Nakamura, T., Higuchi, T. & Michio, H. Different stress tolerances of juvenile polyps of the coral Acropora tenuis associated with clades C1 and D Symbiodinium. Zool. Stud. 55, 2016–4 (2016).
Shinzato, C. et al. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323 (2011).
Shoguchi, E. et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 23, 1399–1408 (2013).
Lin, S. et al. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 350, 691–694 (2015).
Voolstra, C. R. et al. Comparative analysis of the genomes of Stylophora pistillata and Acropora digitifera provides evidence for extensive differences between species of corals. Sci. Rep. 7, 17583 (2017).
Celis, J. S. et al. Binning enables efficient host genome reconstruction in cnidarian holobionts. Gigascience 7, 7 (2018).
Shoguchi, E. et al. Two divergent Symbiodinium genomes reveal conservation of a gene cluster for sunscreen biosynthesis and recently lost genes. BMC Genomics 19, 458 (2018).
Aranda, M. et al. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 6, 39734 (2016).
Hirose, M., Yamamoto, H. & Nonaka, M. Metamorphosis and acquisition of symbiotic algae in planula larvae and primary polyps of Acropora spp. Coral Reefs 27, 247–254 (2008).
Sammarco, P. W. & Strychar, K. B. Responses to High Seawater Temperatures in Zooxanthellate Octocorals. PLoS One 8, e54989 (2013).
Ratzka, C., Gross, R. & Feldhaar, H. Endosymbiont Tolerance and Control within Insect Hosts. Insects 3, 553–572 (2012).
Dennis, G. Jr. et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome. Biol. 4, P3 (2003).
Kanehisaa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28, 27–30 (2000).
Maizels, R. M. & McSorley, H. J. Regulation of the host immune system by helminth parasites. J Allergy Clin Immunol 138, 666–675 (2012).
McMahon, H. T. & Boucrot, E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 12, 517–533 (2011).
Sorkin, A. & von Zastrow, M. Endocytosis and signaling: intertwining molecular networks. Nat Rev Mol Cell Biol 10, 609–622 (2009).
Downs, C. A. et al. xSymbiophagy as a cellular mechanism for coral bleaching. Autophagy 5, 211–216 (2005).
Peränen, J., Auvinen, P., Virta, H., Wepf, R. & Simons, K. Rab8 promotes polarized membrane transport through reorganization of actin and microtubules in fibroblasts. J. Cell. Biol. 135, 153–167 (1996).
Ebine, K. et al. Plasmodium Rab5b is secreted to the cytoplasmic face of the tubovesicular network in infected red blood cells together with N-acylated adenylate kinase 2. Malar. J. 15, 323 (2016).
Wakefiel, T. S. & Kempf, S. C. Development of host- and symbiont-specific monoclonal antibodies and confirmation of the origin of the symbiosome membrane in a cnidarian-dinoflagellate symbiosis. Biol. Bull. 200, 127–143 (2001).
Lin, K. L., Wang, J. T. & Fang, L. S. Participation of glycoproteins on zooxanthellal cell walls in the establishment of a symbiotic relationship with the sea anemone. Aiptasia pulchella. Zool. Stud. 39, 172–178 (2000).
Liew, Y. J. et al. Identification of MicroRNAs in the Coral Stylophora pistillata. PLoS One 9, e91101 (2014).
Lelandais-Brière, C., Moreau, J., Hartmann, C. & Crespi, M. Noncoding RNAs, Emerging Regulators in Root Endosymbioses. Mol. Plant. Microbe. Interact. 29, 170–180 (2016).
Streamer, M., McNeil, Y. R. & Yellowlees, D. Photosynthetic carbon dioxide fixation in zooxanthellae. Mar Biol 115(2), 195–198 (1993).
Balskus, E. P. & Walsh, C. T. The genetic and molecular basis for sunscreen biosynthesis in cyanobacteria. Science 329, 1653–1656 (2010).
Tarrant, A. M. Endocrine-like Signaling in Cnidarians: Current Understanding and Implications for Ecophysiology. Integr. Comp. Biol. 45, 201–214 (2005).
Steele, R. E., Lieu, P., Mai, N. H., Shenk, M. A. & Sarras, M. P. Jr. Response to insulin and the expression pattern of a gene encoding an insulin receptor homologue suggest a role for an insulin-like molecule in regulating growth and patterning in Hydra. Dev. Genes. Evol. 206, 247–259 (1996).
Burg, M. B. & Ferraris, J. D. Intracellular organic osmolytes: function and regulation. J. Biol. Chem. 283, 7309–7313 (2008).
Anithajothi, R., Duraikannu, K., Umagowsalya, G. & Ramakritinan, C. M. The presence of biomarker enzymes of selected Scleractinian corals of Palk Bay, southeast coast of India. Biomed. Res. Int. 2014, 684874 (2014).
Yuyama, I., Higuchi, T. & Takei, Y. Sulfur utilization of corals is enhanced by endosymbiotic algae. Biology Open 5, 1299–1304 (2016).
Strong, F. M. Naturally occurring toxic factors in plants and animals used as food. Can. Med. Assoc. J. 94, 568–573 (1966).
Tang, C. H. et al. Cellular membrane accommodation to thermal oscillations in the coral Seriatopora caliendrum. PLoS One 9, e105345 (2014).
Iwao, K., Fujisawa, T. & Hatta, M. A cnidarian neuropeptide of the GLW amide family induces metamorphosis of reef-building corals in the genus Acropora. Coral Reefs 21, 127–129 (2002).
Ladner, J. T., Barshis, D. J. & Palumbi, S. R. Protein evolution in two co-occurring types of Symbiodinium: an exploration into the genetic basis of thermal tolerance in Symbiodinium clade D. BMC Evol. Biol. 2, 217 (2014).
Shinzato, C., Inoue, M. & Kusakabe, M. A Snapshot of a Coral “Holobiont”: A Transcriptome Assembly of the Scleractinian Coral, Porites, Captures a Wide Variety of Genes from Both the Host and Symbiotic Zooxanthellae. PLoS One 9, e85182 (2014).
Acknowledgements
We are grateful to the members of the Akajima Marine Science Laboratory, especially Mr. Kenji Iwao, for provision of A. tenuis larvae and Dr. Tomihiko Higuchi for supporting statistical analysis about correlation analysis. We also thank the members of the Laboratory for DNA data analysis in NIG for valuable discussion and supporting our experiments, especially Hisako Tashiro, Chie Iwamoto. RNA-seq data analysis was partially performed on the NIG supercomputer at ROIS National Institute of Genetics. This work was supported by a research grant by the Japan Society for the Promotion of Science (IY, #14J40135 and #15K18744), and the Environment Research and Technology Development Fund (No. 4–1806) from the Ministry of the Environment in Japan.
Author information
Authors and Affiliations
Contributions
I.Y. performed the experiments, analyses NGS data, and wrote manuscripts, M.I., M.N., M.Y., K.I., helped with analyses NGS data and manuscripts preparation.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yuyama, I., Ishikawa, M., Nozawa, M. et al. Transcriptomic changes with increasing algal symbiont reveal the detailed process underlying establishment of coral-algal symbiosis. Sci Rep 8, 16802 (2018). https://doi.org/10.1038/s41598-018-34575-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-34575-5
Keywords
This article is cited by
-
Algae-coral symbiosis: fragility owing to anthropogenic activities and adaptive response to changing climatic trends
Environment, Development and Sustainability (2024)
-
Genes possibly related to symbiosis in early life stages of Acropora tenuis inoculated with Symbiodinium microadriaticum
Communications Biology (2023)
-
Larval transcriptomic responses of a stony coral, Acropora tenuis, during initial contact with the native symbiont, Symbiodinium microadriaticum
Scientific Reports (2022)
-
Increased incompatibility of heterologous algal symbionts under thermal stress in the cnidarian-dinoflagellate model Aiptasia
Communications Biology (2022)
-
Photosynthesis acclimation under severely fluctuating light conditions allows faster growth of diatoms compared with dinoflagellates
BMC Plant Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.