Abstract
Gut microbiota and blood neutrophil-to-lymphocyte ratio (NLR) are associated with systemic inflammation; however, data on the association between gut microbiota and NLR are lacking. We investigated the association between gut microbiota and NLR. A total of 1,309 subjects who had available data on NLR and 16 S rRNA sequencing of gut microbiota were included in this study. They were grouped according to NLR quartile (Q) as follows: lower Q (n = 328, <25% of NLR range), middle 2Q (n = 653, ≥25% to <75%) and upper Q (n = 328, ≥75%). The diversity and composition of the human gut microbiota in the groups were calculated. The phylogenetic diversity of gut microbiota in the lower group was significantly higher than in the middle 2Q group (P = 0.040). The beta-diversity was significantly different among the three groups (P = 0.043), between the lower and middle 2Q groups (P = 0.029), and between the lower and upper groups (P = 0.026). Bacteroides eggerthii showed a positive correlation with NLR (q = 0.015). The diversity and composition of the gut microbiome were different between the NLR groups. Particularly, patients with a lower NLR had a greater diversity of gut microbiota.
Similar content being viewed by others
Introduction
The human intestinal tract contains a varied and complex microbiotic environment that consists of more than 1,000 different species whose DNA encode the proteins for 3 million microbiomes1. Because the composition of gut microbiota is affected by multiple factors, such as host genetics, diet, geographic location, environment, early microbial exposure, and use of medication2,3,4,5, the diversity and abundance of the gut microbiota greatly differ between healthy individuals. However, functional metabolic pathways remain relatively consistent despite variations in the microbiota6. The crucial role of gut microbiota consists of its association with development and maintenance of the human immune system via metabolism7,8. Recently, several studies have reported that the gut microbiota influences the pathogenesis of immune-mediated inflammatory diseases such as inflammatory bowel disease, multiple sclerosis, rheumatoid arthritis, and ankylosing spondylitis7,9.
Neutrophil-to-lymphocyte ratio (NLR), which is calculated by dividing the absolute count of neutrophils by the number of lymphocytes in the complete blood count, is a simply-measured and reproducible biomarker used to evaluate systemic inflammation10,11. NLR is not only highly correlated with the prognosis of cancer patients, but it is also correlated with the prognosis of multiple diseases including inflammatory disease, cardiovascular disease, and tuberculosis12,13,14,15,16,17,18,19. Patients with an elevated NLR show a relatively decreased number of lymphocytes and an increased number of neutrophils, suggesting that cell-mediated immunity is impaired and systemic inflammation is increased in inflammatory processes13,16,20. Although Forget et al. reported that the normal value of NLR ranged from 0.78 to 3.53 in the healthy population21, the range of normal NLR is still controversial due to limited evidence that subjects with a normal NLR range have similar immunity and inflammatory statuses.
We hypothesized that the composition and diversity of human gut microbiota are different depending on the range of NLR since both gut microbiota and NLR are affected by systemic inflammation and immunity in healthy individuals. The purpose of our study is to investigate the association between NLR and gut microbiome in a large-scale population.
Results
Baseline characteristics of the subjects
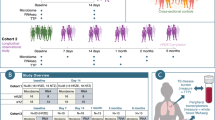
Of the total 1,309 subjects (mean age, 45.7 years; males, 62.3%; mean NLR, 1.67), 328 (25.1%), 653 (49.9%), and 328 (25.1%) were classified as the lower Q, middle 2Q, and upper Q groups, respectively (Table 1 and Fig. 1). The values of the first and third NLR quartiles were 1.25 and 1.98, respectively. No significant differences in demographics were observed between the three groups, except BMI and smoking status. In laboratory findings, the mean NLR was 1.1, 1.6, and 2.6 in the lower Q, middle 2Q, and upper Q groups, respectively. The total white blood cell count (5.3 103/mm3 [lower Q] vs. 5.7 103/mm3 [middle 2Q] vs. 6.5 103/mm3 [upper Q], P < 0.001) and the proportion of neutrophils were significantly higher in the upper Q group than in the lower Q and middle 2Q groups (45.1% vs. 55.2% vs. 65.0%, P < 0.001), whereas the proportions of lymphocytes (44.7% vs. 35.3% vs. 26.3%, P < 0.001), eosinophils (3.0% vs. 2.6% vs. 2.2%, P < 0.001), basophils (0.5% vs. 0.5% vs. 0.4%, P < 0.001), and monocytes (6.6% vs. 6.4% vs. 6.1%, P < 0.001) were higher in the lower Q group compared with upper Q and middle 2Q groups. C-reactive protein (CRP) in the upper group was also significantly higher than in the other groups (0.09 mg/dL vs. 0.09 mg/dL vs. 0.10 mg/dL, P = 0.002). Differences in comorbidities and nutritional intake were not identified between the three groups (Supplement Table S1 and S2).
We made additional comparisons between the lower Q and higher 3Q groups (Supplement Table S3); no significant differences in demographics were identified. In the laboratory findings, the trends were similar to those of the three groups, except there was no statistical difference in the level of CRP (0.09 [lower Q] vs. 0.11 [higher 3Q], P = 0.222)
Comparison of alpha diversity within and between the NLR groups
The alpha diversity of gut microbial taxa between the lower Q, middle 2Q, and upper Q groups did not show statistically significant differences in observed OTU (93.9 ± 45.3 [lower Q] vs. 88.3 ± 45.5 [middle 2Q] vs. 90.4 ± 44.0 [Upper Q], P = 0.216), PD (14.8 ± 3.7 vs. 14.3 ± 3.5 vs. 14.5 ± 3.5, P = 0.166), evenness (0.81 ± 0.07 vs. 0.81 ± 0.07 vs. 0.81 ± 0.0, P = 0.986), and Shannon’s index (5.14 ± 0.76 vs. 5.06 ± 0.75 vs. 5.08 ± 0.75, P = 0.313) (Fig. 2). However, PD was significantly higher in the lower Q group than in the middle 2Q group (P = 0.040) in the post hoc test. These results indicate that richness, including phylogenetic diversity, was different among the NLR groups, but evenness was not.
Box plots of alpha-diversity indices comparing neutrophil-to-lymphocyte-ratio groups. (A) Observed OTU, (B) phylogenetic diversity, (C) Pielou evenness, and (D) Shannon’s index. OTU, operational taxonomic unit; Lower Q, < 25% of NLR range; Middle 2Q, ≥25% to <75% of NLR range; Upper Q ≥ 75% of NLR range. *P < 0.05; #P < 0.25.
In the comparison between the lower Q and higher 3Q groups, the lower Q group showed marginal trends of significance of alpha diversity in PD (14.8 ± 3.7 [lower Q] vs. 14.3 ± 3.4 [higher 3Q], P = 0.055) and observed OTU (93.9 ± 45.3 vs. 89.0 ± 43.0, P = 0.090) compared with the higher 3Q group (Supplement Figure S1).
Comparison of beta diversity within and between the NLR groups
Unweighted UniFrac-based diversity was significantly different among the lower Q, middle 2Q, and upper Q groups (P = 0.043); between the lower Q and upper Q groups (P = 0.029); and between the lower Q and middle 2Q groups (P = 0.026; Fig. 3A). However, the unweighted UniFrac-based diversity was not significantly different between the middle 2Q and upper Q groups (P = 0.939). In weighted UniFrac-based beta diversity, no significant differences were revealed among the three groups (P = 0.455) or within each of the two groups (Fig. 3B).
Comparison distance of beta diversity between neutrophil-to-lymphocyte-ratio groups. (A) Distance from the lower group using unweighted UniFrac and (B) Distance from the lower group using weighted UniFrac. Lower Q, <25% of NLR range; Middle 2Q, ≥25% to <75% of NLR range; Upper Q ≥ 75% of NLR range. *P < 0.05.
There was a significant difference in unweighted UniFrac-based diversity between the lower Q and higher 3Q groups, but the differences were very small (P = 0.013; Supplement Figure S2). Results of weighted UniFrac-based beta diversity were not significantly different between the lower Q and higher 3Q groups (P = 0.175; Supplement Figure S2B).
Associations between gut microbiota composition and NLR and total WBC count using quantitative analysis
In the analysis of correlation between gut microbial taxa abundance and NLR after adjusting for age, sex, smoking status, and BMI, one species (Bacteroides eggerthii which belongs to the family Bacteroidaceae) was positively correlated with NLR (CE, 0.00605; P = 0.001; q = 0.015; Table 2 and Fig. 4A) and showed a positive association trend with total WBC count (CE, 0.00154; P = 0.049; q = 0.232; Fig. 4B). The genera Dialister and Prevotella stercorea had trends of a positive (CE, 0.01059; P = 0.024; q = 0.132) and negative correlation (CE, −0.00864; P = 0.037; q = 0.190) with NLR, respectively (Table 2).
Comparison of gut microbiota composition between the NLR groups using qualitative analysis
The relative abundance of gut microbiota was compared between groups using MaAsLine. The upper Q group had a significantly higher proportion of Bacteroides eggerthii compared with the lower Q (CE, 0.0119; P < 0.001; q = 0.001) and middle 2Q groups (CE, 0.00978; P < 0.001; q = 0.003; Fig. 5A). This significant trend of Bacteroides eggerthii held in the quantitative analysis. The relative abundance of the genus Bilophilia was significantly lower in the lower Q group than in the upper Q group (CE, 0.0045; P = 0.004; q = 0.043) and showed a marginal trend toward decrease compared with the middle 2Q group (CE, 0.0034; P = 0.014; q = 0.098; Fig. 5B). The abundance of Dialister organisms at the genus level in the lower Q group also showed a decreasing trend compared with the upper Q (CE, 0.0148; P = 0.045; q = 0.220) and middle 2Q groups (CE, 0.0129; P = 0.043; q = 0.205; Fig. 5C).
Comparison of relative abundance between neutrophil-lymphocyte-ratio groups. (A) Bacteroides eggerthii, (B) genus Bilophila, and (C) genus Dialister. Bars and error bars show the mean ± 95% confidence interval. Lower Q, < 25% of NLR range; Middle 2Q, ≥25% to <75% of NLR range; Upper Q ≥ 75% of NLR range. *q < 0.05; #q < 0.25.
In the comparison between the lower Q and higher 3Q groups, the relative abundance of the genus Bilophila in the lower Q group was significantly lower than that in the higher 3Q group (CE, 0.0038; P = 0.004; q = 0.036; Supplement Figure S3A). The relative abundance of the genera Dialister (CE, 0.0135; P = 0.024; q = 0.143; Supplement Figure S3C) and Bacteroides eggerthii (CE, 0.0054; P = 0.027; q = 0.145; Supplement Figure S3D) had decreasing trends in the lower Q group than in the higher 3Q group, while abundance of Prevotella stercorea (CE, −0.0124; P = 0.019; q = 0.119; Supplement Figure S3B), the genus Phascolarctobacterium (CE, −0.0111; P = 0.035; q = 0.178; Supplement Figure S3E), and the genus Lachnospira (CE, −0.0081; P = 0.048; q = 0.229; Supplement Figure S3F) in the lower Q group showed trends toward increase compared with those in the higher 3Q group.
Discussion
This is the first population-based study that identified correlations between NLR and gut microbiota in a population with a relatively normal NLR range. The gut microbiome in the lower Q NLR group showed higher richness in alpha diversity analysis. We also found differences in beta diversity among the NLR groups, particularly among the lower Q and other upper groups. The relative abundance of Bacteroides eggerthii was positively correlated with the value of NLR and was significantly elevated in the upper Q group compared with the lower Q and middle 2Q groups. When comparing the lower Q and higher 3Q groups, the genus Bilophila was less abundant in the lower Q group than in the higher 3Q group.
NLR can be used as a prognostic marker in multiple diseases12,13,14,17,18,19,22,23. The value of NLR ranges from 0.78 to 3.53 in the healthy adult population21. Although there is controversy on the optimal NLR cut-off values, numerous studies have reported that the NLR cut-off value for mortality in specific diseases is between 3 and 517,23,24,25. The cut-off value of NLR for disease severity and activity in inflammatory bowel disease (IBD) is slightly lower compared with other medical conditions, ranging from 2 to 326,27,28. Our study demonstrated a subtle difference in beta diversity between the lower and upper 3Q groups. Considering that we divided the NLR groups based on a cut-off of 1.25, which was considered a normal value in previous studies, the composition of the microbiota might be different depending on the NLR, even within the normal range. In addition, these subtle differences might be due to the healthy population.
NLR and CRP can be used as markers of systemic inflammation in various medical settings. CRP is an acute-phase protein produced in the hepatocytes, exposed to the inflammatory cytokines and tumor necrosis factor. CRP is rapidly increasing in response to inflammation and used as a marker reflecting prognosis and disease severity in various inflammatory situations including infection, trauma, and tumor29; however, it is difficult to interpret because the cause of inflammation is not well discriminated. On the other hand, because NLR represents the function of lymphocyte, it can more accurately reflect both the inflammatory response and the immune state30,31. Therefore, the NLR can most sensitively reflect the microbiome changes caused by external factors, such as immune systems or inflammatory responses.
The alpha diversity indices were also higher in the lower Q group than in the upper 3Q group. These findings suggest that, even within the normal rage of NRL, alpha diversity (particularly, the richness) of gut microbiota decreases as NLR increases. We also found that the abundance of several gut microorganisms had a linear correlation with NLR in the healthy population. This is consistent with previous reports that the activity scores of several diseases have a linear correlation with NLR32,33. In conclusion, even within the normal range, the lower is the NLR, the lower is the risk for inflammatory disease, which is similar to the findings that the lower is the blood glucose or blood pressure, the lower is the risk of diabetes mellitus or cardiovascular-related mortality, respectively34,35.
B. eggerthii is a less commonly isolated species of Bacteroide, which is a genus of Gram-negative, anaerobic, non-spore-forming bacteria that is part of the normal human gastrointestinal flora36,37,38. Using in vivo mouse models, Dziarski et al. indicated that an abundance of B. eggerthii is associated with dextran sulfate sodium-induced colitis39. In addition, several reports have suggested that changes in abundance of Bacteroide are observed in subjects with IBD, including patients with Crohn’s disease and ulcerative colitis40,41,42. On the other hand, NLR is a biomarker of disease activity and prognosis in patients with IBD14,26,27. Considering previous reports indicating the relationships between colitis and NLR and between colitis and B. eggerthii, our study showed a significant association between NLR and B. eggerthii. Therefore, there is likely a link among NLR, B. eggerthii, and intestinal inflammation.
Biophilia spp. are Gram-negative, anaerobic, urease-positive, bile-resistant bacteria associated with an animal-based protein and high-saturated fat diet, and they promote T helper type 1 (Th1)-mediated immunity43. The most frequently isolated species of Biophilia is B. wadsworthia, and although it is found in < 0.1% of the normal human gastrointestinal flora, it is found in abundance in infectious conditions such as colitis, perforation, and abscesses43,44. Devkota et al. demonstrated that B. wadsworthia induced colitis in IBD-prone interleukin (IL)-10 mice by activating Th1-mediated colonic inflammation45. We identified a lower abundance of Biophilia in the lower Q NLR group than in the higher 3Q group. Considering that NLR is a marker of prognosis and severity of IBD, an elevated abundance of B. eggerthii and genus Biophilia might be associated with the status of bowel inflammation, and NLR could reflect these microbiota changes, even though our subjects were relatively healthy.
Our results suggest that several organisms of the gut microbiota differ in abundance between the lower Q and higher Q3 groups (q-value < 0.2). The genus Dialister, which showed an increasing trend of abundance in the higher Q3 NLR group, has been associated with irritable bowel syndrome46,47. Organisms of the genus Prevotella, considered to have a beneficial role due to its association with a plant-rich diet48, showed a decreased quantity in the higher Q3 NLR group. These results suggest that NLR decreases as the gut microbiota changes within a beneficial environment. Furthermore, since NLR is related to the prognosis of various medical conditions12,13,14,17,18,19,22,23, it is suggested that small changes can reflect the early progression of disease.
Our study has several limitations. First, this was a single-center, retrospective, cross-sectional study, and it is unable to provide a causal relationship between gut microbiota and NLR. Second, we only included a relatively young and healthy population, and data on subjects with disease are limited. Third, the clinical parameters such as symptoms or diagnosis were not acquired because we used health check-up materials, not medical records. Instead, data on medical, past, family, and medication history from the self-reported questionnaire were used. Some subjects tend to misreport or forget to include their history. However, this is the largest population-based study that demonstrated the association between the gut microbiome and NLR, showing differences in the diversity and abundance of bacteria between NLR groups.
This study investigated the association between the gut microbiota and NLR in a large healthy population. The diversity of the gut microbiota increased in subjects with a lower NLR. Furthermore, a higher NLR was associated with increased abundance of IBD-related gut microbiota and decreased abundance of beneficial bacteria. The findings of the present study highlight the role of NLR as a marker of intestinal inflammation because it reflects changes in the gut microbiota. Further long-term chronological studies are needed to determine the mechanism of the gut microbiota’s influence on NLR.
Method
Study population and design
A total of 1,463 Korean men and women between the ages of 25 and 78 years with a comprehensive annual or biennial physical examination between June 2014 and September 2014 at Kangbuk Samsung Hospital Healthcare Screening Center in the Republic of Korea were initially screened (Fig. 1)49. Among them, subjects who met the following criteria were excluded: 1) Subjects who received antibiotics within six weeks of enrollment or lipid-lowering drugs or probiotics within four weeks of enrollment due to the medication’s influence on gut microbiota (n = 141); 2) Subjects who did not have NLR data obtained during the physical examination (n = 5); and 3) Subjects whose fecal samples contained < 1,000 sequences per sample (n = 8). Finally, 1,309 subjects were included for analysis.
Demographics, laboratory data, past medical history, and other clinical data were obtained from the medical record, and questionnaires at the time of physical examination. This study protocol was approved by the Institutional Review Board of Kangbuk Samsung Hospital (KBSMC 2013-01-245-12). After explaining the nature and possible consequences of the study, we obtained written informed consent from all study participants. All applicable institutional and governmental regulations concerning the ethical use of human volunteers were followed during this research.
DNA extraction from fecal samples and bacterial 16S rRNA gene sequencing
Fecal samples were frozen at −20 °C immediately after collection and stored at −70 °C within 24 hours. Within one month of receiving the samples, the MO Bio PowerSoil® DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA, USA) was used for DNA extraction from the fecal samples according to the manufacturer’s instructions. For amplification and sequencing of the DNA to analyze the bacterial communities, the methods described in a previous study were used49. Amplified genomic DNA was obtained using fusion primers targeting the variable V3 and V4 regions of the 16 S rRNA genes with indexing barcodes. The Illumina Miseq platform (Illumina, San Diego, CA, USA) was pooled for sequencing all samples according to the manufacturer’s specifications50,51.
16S rRNA gene compositional analysis
The DADA2 pipeline of the QIIME2 package (https://qiime2.org)52 was performed to generate unique sequence variants by filtering low quality samples and chimera. Since unique grouping sequences produce the “operational taxonomic units (OTUs)” from DADA2, they are regarded as 100% OTU and are typically referred to as sequence variants. The Feature Table was generated from the QIIME2 software, and it was the equivalent of the biom table and the representational sequence file. Almost identical (99%) sequence which was homology to an optimized version of the gene from the GreenGenes database (version 13.8) containing the V3-V4 region to detect taxonomies, mapped the sequences.
Statistical analysis
The values of NLR were transformed using the natural logarithm (ln) to construct a normal distribution. After transformation, we divided the subjects into three groups according to NLR quartile (Q): lower Q (the lowest 25% of the NLR range), middle 2Q (from the next lowest 25% of the NLR range to the second highest 25% of the NLR range), and upper Q (the highest 25% of the NLR range). We also compared the lower Q to the higher 3Q, which included the middle 2Q and upper Q groups (the highest 75% of the NLR range) to investigate various aspects of the microbiota. All basic statistical analyses were performed with SPSS version 24.0 (SPSS Inc., Chicago, IL, USA). QIIME2 (version 2018.04) was utilized for exploratory and differential microbial composition analyses52. Alpha diversity measures of richness, community diversity, evenness, and phylogenetic diversity of gut microbial taxa were presented as observed OTU, Shannon index53, Pielou’s evenness54, and Faith’s phylogenetic diversity (PD)55, respectively. For measuring beta diversity, unweighted and weighted UniFrac56 values were calculated to determine the dissimilarity between groups. Diversity between the NLR groups was compared using pairwise PERMANOVA57. A P-value < 0.05 was considered statistically significant.
Correlation and comparison between the abundance of taxa and NLR were calculated using the Multivariate Association with Linear Models (MaAsLin) software package (https://huttenhower.sph.harvard.edu/maaslin)58 of RStudio (version 0.98.983). Confounding variables (age, sex, smoking status, and body mass index [BMI])-adjusted coefficients (CE) were estimated using MaAslin. All analyses of MaAslin were conducted using the default options. The FDR (Benjamini-Hochberg) method was used to adjust multiple comparisons on each phylogenetic level. A q-value < 0.05 was considered statistically significant.
Data Availability statement
All the supporting data is provided as supplementary files.
References
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65, https://doi.org/10.1038/nature08821 (2010).
Erwin, G. Zoetendal, A. D. L. A. W. M. A.-v. V. J. A. G. M. d. V. W. M. d. V. The Host Genotype Affects the Bacterial Community in the Human Gastronintestinal Tract. Microbial Ecology in Health and Disease 13, 129–134, https://doi.org/10.1080/089106001750462669 (2001).
Turnbaugh, P. J. et al. A core gut microbiome in obese and lean twins. Nature 457, 480, https://doi.org/10.1038/nature07540, https://www.nature.com/articles/nature07540#supplementary-information (2008).
The Human Microbiome Project, C. Structure, function and diversity of the healthy human microbiome. Nature 486, 207, https://doi.org/10.1038/nature11234, https://www.nature.com/articles/nature11234#supplementary-information (2012).
Khachatryan, Z. A. et al. Predominant Role of Host Genetics in Controlling the Composition of Gut Microbiota. PLoS ONE 3, e3064, https://doi.org/10.1371/journal.pone.0003064 (2008).
Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214, https://doi.org/10.1038/nature11234 (2012).
Tlaskalová-Hogenová, H. et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cellular And Molecular Immunology 8, 110, https://doi.org/10.1038/cmi.2010.67 (2011).
Rowland, I. et al. Gut microbiota functions: metabolism of nutrients and other food components. European Journal of Nutrition 57, 1–24, https://doi.org/10.1007/s00394-017-1445-8 (2018).
Forbes, J. D., Van Domselaar, G. & Bernstein, C. N. The Gut Microbiota in Immune-Mediated InflammatoryDiseases. Frontiers in Microbiology 7, 1081, https://doi.org/10.3389/fmicb.2016.01081 (2016).
Zahorec, R. Ratio of neutrophil to lymphocyte counts–rapid and simple parameter of systemic inflammation and stress in critically ill. Bratislavske lekarske listy 102, 5–14 (2001).
Li, W., Ai, X., Ni, Y., Ye, Z. & Liang, Z. The Association Between the Neutrophil-to-Lymphocyte Ratio and Mortality in Patients with Acute Respiratory Distress Syndrome: A Retrospective Cohort Study. Shock (Augusta, Ga.), https://doi.org/10.1097/shk.0000000000001136 (2018).
Donskov, F. Immunomonitoring and prognostic relevance of neutrophils in clinical trials. Seminars in cancer biology 23, 200–207, https://doi.org/10.1016/j.semcancer.2013.02.001 (2013).
Faria, S. S. et al. The neutrophil-to-lymphocyte ratio: a narrative review. ecancermedicalscience 10, 702, https://doi.org/10.3332/ecancer.2016.702 (2016).
Acarturk, G. et al. Neutrophil-to-lymphocyte ratio in inflammatory bowel disease - as a new predictor of disease severity. Bratislavske lekarske listy 116, 213–217 (2015).
Rifaioglu, E. N., Bulbul Sen, B., Ekiz, O. & Cigdem Dogramaci, A. Neutrophil to lymphocyte ratio in Behcet’s disease as a marker of disease activity. Acta dermatovenerologica Alpina, Pannonica, et Adriatica 23, 65–67 (2014).
Abakay, O., Abakay, A., Sen, H. S. & Tanrikulu, A. C. The relationship between inflammatory marker levels and pulmonary tuberculosis severity. Inflammation 38, 691–696, https://doi.org/10.1007/s10753-014-9978-y (2015).
Han, Y. et al. High blood neutrophil-lymphocyte ratio associated with poor outcomes in miliary tuberculosis. Journal of Thoracic Disease 10, 339–346, https://doi.org/10.21037/jtd.2017.12.65 (2018).
Isaac, V. et al. Elevated neutrophil to lymphocyte ratio predicts mortality in medical inpatients with multiple chronic conditions. Medicine 95, e3832, https://doi.org/10.1097/md.0000000000003832 (2016).
Bhat, T. et al. Neutrophil to lymphocyte ratio and cardiovascular diseases: a review. Expert review of cardiovascular therapy 11, 55–59, https://doi.org/10.1586/erc.12.159 (2013).
Sato, Y. et al. Increased neutrophil-to-lymphocyte ratio is a novel marker for nutrition, inflammation and chemotherapy outcome in patients with locally advanced and metastatic esophageal squamous cell carcinoma. Biomedical Reports 7, 79–84, https://doi.org/10.3892/br.2017.924 (2017).
Forget, P. et al. What is the normal value of the neutrophil-to-lymphocyte ratio? BMC Research Notes 10, 12, https://doi.org/10.1186/s13104-016-2335-5 (2017).
Walsh, S. R., Cook, E. J., Goulder, F., Justin, T. A. & Keeling, N. J. Neutrophil-lymphocyte ratio as a prognostic factor in colorectal cancer. Journal of surgical oncology 91, 181–184, https://doi.org/10.1002/jso.20329 (2005).
Gu, X.-B., Tian, T., Tian, X.-J. & Zhang, X.-J. Prognostic significance of neutrophil-to-lymphocyte ratio in non-small cell lung cancer: a meta-analysis. Scientific Reports 5, 12493, https://doi.org/10.1038/srep12493 (2015).
Palin, R. P., Devine, A. T., Hicks, G. & Burke, D. Association of pretreatment neutrophil-lymphocyte ratio and outcome in emergency colorectal cancer care. Annals of the Royal College of Surgeons of England 100, 308–315, https://doi.org/10.1308/rcsann.2017.0232 (2018).
Vano, Y.-A. et al. Optimal cut-off for neutrophil-to-lymphocyte ratio: Fact or Fantasy? A prospective cohort study in metastatic cancer patients. PLOS ONE 13, e0195042, https://doi.org/10.1371/journal.pone.0195042 (2018).
Gao, S. Q. et al. Neutrophil-lymphocyte ratio: a controversial marker in predicting Crohn’s disease severity. International journal of clinical and experimental pathology 8, 14779–14785 (2015).
Demir, A. K. et al. The relationship between the neutrophil-lymphocyte ratio and disease activity in patients with ulcerative colitis. The Kaohsiung journal of medical sciences 31, 585–590, https://doi.org/10.1016/j.kjms.2015.10.001 (2015).
Torun, S. et al. Assessment of neutrophil-lymphocyte ratio in ulcerative colitis: a promising marker in predicting disease severity. Clinics and research in hepatology and gastroenterology 36, 491–497, https://doi.org/10.1016/j.clinre.2012.06.004 (2012).
Du Clos, T. W. Function of C-reactive protein. Annals of medicine 32, 274–278 (2000).
Kitayama, J., Yasuda, K., Kawai, K., Sunami, E. & Nagawa, H. Circulating lymphocyte number has a positive association with tumor response in neoadjuvant chemoradiotherapy for advanced rectal cancer. Radiation oncology 5, 47 (2010).
Scarsi, M. et al. Reduction of peripheral blood T cells producing IFN-gamma and IL-17 after therapy with abatacept for rheumatoid arthritis. Clinical and experimental rheumatology 32, 204–210 (2014).
Buyukkaya, E. et al. Correlation of neutrophil to lymphocyte ratio with the presence and severity of metabolic syndrome. Clinical and applied thrombosis/hemostasis: official journal of the International Academy of Clinical and Applied Thrombosis/Hemostasis 20, 159–163, https://doi.org/10.1177/1076029612459675 (2014).
Velissaris, D., Pantzaris, N. D., Bountouris, P. & Gogos, C. Correlation between neutrophil-to-lymphocyte ratio and severity scores in septic patients upon hospital admission. A series of 50 patients. Romanian journal of internal medicine = Revue roumaine de medecine interne. https://doi.org/10.2478/rjim-2018-0005 (2018).
Hansson, L. et al. Effects of intensive blood-pressure lowering and low-dose aspirin in patients with hypertension: principal results of the Hypertension Optimal Treatment (HOT) randomised trial. HOT Study Group. Lancet (London, England) 351, 1755–1762 (1998).
Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). UK Prospective Diabetes Study (UKPDS) Group. Lancet (London, England) 352, 837–853 (1998).
Holdeman, L. V., Good, I. J. & Moore, W. E. Human fecal flora: variation in bacterial composition within individuals and a possible effect of emotional stress. Applied and environmental microbiology 31, 359–375 (1976).
Madigan, M. (SciELO Espana, 2005).
Liu, C. et al. Rapid identification of the species of the Bacteroides fragilis group by multiplex PCR assays using group-and species-specific primers. FEMS microbiology letters 222, 9–16 (2003).
Dziarski, R., Park, S. Y., Kashyap, D. R., Dowd, S. E. & Gupta, D. Pglyrp-Regulated Gut Microflora Prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii Enhance and Alistipes finegoldii Attenuates Colitis in Mice. PLOS ONE 11, e0146162, https://doi.org/10.1371/journal.pone.0146162 (2016).
De Cruz, P. et al. Association between specific mucosa-associated microbiota in Crohn’s disease at the time of resection and subsequent disease recurrence: a pilot study. Journal of gastroenterology and hepatology 30, 268–278, https://doi.org/10.1111/jgh.12694 (2015).
Noor, S. O. et al. Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. BMC gastroenterology 10, 134, https://doi.org/10.1186/1471-230x-10-134 (2010).
Lucke, K., Miehlke, S., Jacobs, E. & Schuppler, M. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. Journal of medical microbiology 55, 617–624, https://doi.org/10.1099/jmm.0.46198-0 (2006).
Singh, R. K. et al. Influence of diet on the gut microbiome and implications for human health. Journal of Translational Medicine 15, 73, https://doi.org/10.1186/s12967-017-1175-y (2017).
Baron, E. J. Bilophila wadsworthia: a unique Gram-negative anaerobic rod. Anaerobe 3, 83–86, https://doi.org/10.1006/anae.1997.0075 (1997).
Devkota, S. et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature 487, 104–108, https://doi.org/10.1038/nature11225 (2012).
Fourie, N. H. et al. The microbiome of the oral mucosa in irritable bowel syndrome. Gut microbes 7, 286–301, https://doi.org/10.1080/19490976.2016.1162363 (2016).
Lopetuso, L. R. et al. Gut Microbiota in Health, Diverticular Disease, Irritable Bowel Syndrome, and Inflammatory Bowel Diseases: Time for Microbial Marker of Gastrointestinal Disorders. Digestive diseases (Basel, Switzerland) 36, 56–65, https://doi.org/10.1159/000477205 (2018).
Ley, R. E. Gut microbiota in 2015: Prevotella in the gut: choose carefully. Nature reviews. Gastroenterology & hepatology 13, 69–70, https://doi.org/10.1038/nrgastro.2016.4 (2016).
Kim, H. N. et al. Correlation between gut microbiota and personality in adults: A cross-sectional study. Brain, behavior, and immunity 69, 374–385, https://doi.org/10.1016/j.bbi.2017.12.012 (2018).
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K. & Schloss, P. D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Applied and environmental microbiology 79, 5112–5120, https://doi.org/10.1128/aem.01043-13 (2013).
Fadrosh, D. W. et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2, 6, https://doi.org/10.1186/2049-2618-2-6 (2014).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods 7, 335–336, https://doi.org/10.1038/nmeth.f.303 (2010).
Neisser, U. Cognitive psychology: Classic edition, (Psychology Press, 2014).
Pielou, E. C. The measurement of diversity in different types of biological collections. Journal of theoretical biology 13, 131–144 (1966).
Faith, D. P. Conservation evaluation and phylogenetic diversity. Biological conservation 61, 1–10 (1992).
Lozupone, C., Lladser, M. E., Knights, D., Stombaugh, J. & Knight, R. UniFrac: an effective distance metric for microbial community comparison. The ISME journal 5, 169 (2011).
Navas-Molina, J. A. et al. Advancing our understanding of the human microbiome using QIIME. Methods in enzymology 531, 371–444, https://doi.org/10.1016/b978-0-12-407863-5.00019-8 (2013).
Lee, S.-W., Kuan, C.-S., Wu, L. S.-H. & Weng, J. T.-Y. Metagenome and Metatranscriptome Profiling of Moderate and Severe COPD Sputum in Taiwanese Han Males. PLOS ONE 11, e0159066, https://doi.org/10.1371/journal.pone.0159066 (2016).
Acknowledgements
We are thankful for the computing resources provided by the Global Science experimental Data hub Center (GSDC) Project and the Korea Research Environment Open NETwork (KREONET) at the Korea Institute of Science and Technology Information (KISTI). This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (NRF-2016R1A6A3A11932719, NRF-2018R1D1A1B07050067), and Intramural Research Support Program for 2016/2017 of Ewha Womans University School of Medicine. The study was also supported by the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare (HI14C0072) and by an NRF grant funded by the Korean government (MSIT) (2010-0027945).
Author information
Authors and Affiliations
Contributions
J.H.L. and H.L.K. designed the study, and H.Y.Y. and H.N.K. performed the clinical research and the statistical and microbiota analyses. H.Y.Y., H.N.K., H.L.K. and J.H.L. wrote the initial manuscript. S.H.L., S.J.K., Y.C., S.R. and H.S. supervised the study. All authors read, commented on, and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yoon, HY., Kim, HN., Lee, S.H. et al. Association between Neutrophil-to-Lymphocyte Ratio and Gut Microbiota in a Large Population: a Retrospective Cross-Sectional Study. Sci Rep 8, 16031 (2018). https://doi.org/10.1038/s41598-018-34398-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-34398-4
Keywords
This article is cited by
-
Gut OncoMicrobiome Signatures (GOMS) as next-generation biomarkers for cancer immunotherapy
Nature Reviews Clinical Oncology (2023)
-
High intake of vegetables is linked to lower white blood cell profile and the effect is mediated by the gut microbiome
BMC Medicine (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.