Abstract
The effects of different amino acid (AA) supplementations of milk protein-based milk replacers in pre-ruminant calves from 3 days to 7 weeks of age were studied. Animals were divided into 4 groups: Ctrl) Control group fed with milk protein-based milk replacer without supplementation; GP) supplementation with 0.1% glycine and 0.3% proline; FY) supplementation with 0.2% phenylalanine and 0.2% tyrosine; MKT) supplementation with 0.62% lysine, 0.22% methionine and 0.61% threonine. For statistical analysis, t-test was used to compare AA-supplemented animals to the Ctrl group. At week 7, body weight and average daily gain (ADG) were measured and blood samples and skeletal muscle biopsies were taken. Blood biochemistry analytes related to energy metabolism were determined and it was shown that MKT group had higher serum creatinine and higher plasma concentration of three supplemented AAs as well as arginine compared with the Ctrl group. GP group had similar glycine/proline plasma concentration compared with the other groups while in FY group only plasma phenylalanine concentration was higher compared with Control. Although the AA supplementations in the GP and FY groups did not affect average daily gain and metabolic health profile from serum, the metabolome analysis from skeletal muscle biopsy revealed several differences between the GP-FY groups and the Ctrl-MKT groups, suggesting a metabolic adaptation especially in GP and FY groups.
Similar content being viewed by others
Introduction
Traditionally, bovine milk is considered to be the standard for feeding dairy cattle1 and before 1956, whole milk was the main feed for calves2. Due to the increasing demand for dairy products, many dairy cattle farms started feeding calves with commercial milk replacers (MRs)3. MR quality can vary according to protein source. In milk protein-based MR (MPMR), the protein source originates from milk protein derivatives, and they are considered to have the best quality as they are easier to be digested by young calves4. However, due to differences in the levels of certain amino acids (AAs) between MRs and bovine milk, MR AA composition may be inadequate for optimal calf growth, as has been described for infant formulas and human milk5,6.
Among the AAs required for pre-ruminant calf’s growth, methionine (M), lysine (K) and threonine (T) are the most studied due to their low concentration in both milk and milk replacers7,8,9,10. There are indications of improved average daily gain (ADG) and feed efficiency intake in MPMR with MKT supplementation10 but other studies suggested no performance benefits comparing MPMR supplemented with methionine1, or with threonine and isoleucine11.
In humans the comparison of human milk amino acid profile with that of infant formula has been used to propose AA supplementation or ingredient composition in infant formula5,6. The comparison of cow milk AA profile12 with the skimmed-milk based MR (used as a control in the present study) evidenced than some AAs are deficient in MR compared with cow milk. For instance, proline (P), tyrosine (Y) and phenylalanine (F) were 7.6, 36.9, and 17.5% lower in the control MR in comparison with bovine whole milk12. Furthermore, these AAs may be interesting because of their role in the organism, for example proline and glycine (G) are very abundant in collagen13, the main structural protein of connective tissue. Phenylalanine and tyrosine play a major role in the neural system as well as the hormonal regulation of metabolism as they are precursors for the synthesis of catecholamines (epinephrine, norepinephrine and dopamine) and thyroid hormones13. Moreover, a recent study concludes that phenylalanine and tryptophan could be limiting growth in calves fed following an enhanced-growth feeding program14.
Even though many studies described the growth effects of AA supplementation, the effect on tissue composition has rarely been addressed, particularly from the metabolomics perspective. The hypothesis of the present work is that inclusion of these AAs would be able to modify the metabolome of the skeletal muscle since they affect growth, are important components of collagen or are precursors of anabolic or catabolic hormones. The metabolome, the complete set of small-molecules found within a biological sample, is directly related to the metabolic activity, protein activity and/or gene expression15,16. The skeletal muscle is suitable for metabolomic approaches, as several studies have been applied not only in differentiating meat characters of different cattle breeds17, geographic origins18,19, post mortem periods20 and sheep breeds and farming conditions21, but also in evaluating meat conservation and aging through meat exudate22.
The objective of this study is to determine the effects of a MPMR-based diet supplemented with three groups of amino acids (FY, GP, MKT) fed to dairy calves from 3 days up to 7 weeks of age on the ADG and blood biochemical analytes. The second goal is, by using the NMR-metabolomic technology, to monitor the impact of AA supplementations of MPMR on the skeletal muscle metabolome, hence asserting their importance for the dairy industry.
Results
Performance data
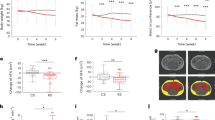
Initial and biopsy time body weight and ages were measured for the four groups in the study, as shown in Table 1. No differences were observed between groups, although a statistical tendency for higher ADG was observed in MKT group compared with Ctrl (p = 0.096) and FY groups (p = 0.097).
Blood profile and multivariate analysis of data
At the end of the treatment (week 7), several serum biochemical analytes related to health and nutritional status as well as plasma free AA profile were determined (Table 2).
For serum biochemistry analytes, there were no differences between Ctrl and the GP or FY group, whereas MKT group presented higher values of serum creatinine compared to the Ctrl group (p = 0.023) as well as a tendency for higher total protein (p = 0.059). As for the plasma AA concentration, the three supplemented AAs were higher in the MKT group than in the Ctrl group, as well as the concentration of plasma arginine. In the case of FY group, the concentration is significantly higher for phenylalanine supplemented compared with Ctrl group. In the case of GP, there were no significant differences compared to the Ctrl group.
Multivariate analysis of the combined data from serum biochemistry and plasma AA using PCA agreed with the univariate results since the different treatment groups did not show major differences (Fig. 1). Pairwise PLS-DA could only discriminate between the Ctrl and the MKT treatment (NC = 2, R2Y = 0.91, Q2 = 0.60). The VIP variables with values higher than 1.5 and therefore most responsible for the difference were, in decreasing VIP order, methionine, threonine, arginine, creatinine and lysine.
PCA of combined serum biochemical analytes and plasma AAs. Data from dairy calves fed with control milk replacer (Ctrl); milk replacer supplemented with glycine and proline (GP); milk replacer supplemented with phenylalanine and tyrosine (FY) and milk replacer supplemented with lysine, methionine and threonine (MKT) (n = 8 per group). Ellipse Hotelling’s T2 (95%).
Muscle tissue metabolomics profile
NMR spectra analysis and compound identification
Prior to the metabolomics analysis of the muscle tissue extracts, the assignment of the features of the NMR spectrum to specific compounds was made. Selected spectra were assigned using database assisted spectral deconvolution with the Chenomx software platform using its internal database as well as the human metabolite database data (http://www.hmdb.ca/). 2D TOCSY spectra and literature reports21,22 were also used to verify the identity of other metabolites. A total of 70 compounds could be successfully identified (Supplementary Table S1). From these, a subset of 36 metabolites was selected for quantification using Chenomx. The selection of this group was made based on the ability to reliably quantify them, their relevance from chemometric multivariate analysis (see chemometric section) and their relevance to the biochemical analytes of this study. The assignment of the selected metabolites on a representative spectrum is shown in Fig. 2.
Representative 800 MHz 1H-NMR spectrum of calf muscle aqueous fraction. 1X zoom from 0.0–4.8 ppm and 8X zoom from 4.8–9.4 ppm. Assignments of the 36 metabolites that were quantified: 1: 3-Hydroxybutyrate; 2: Acetate: 3: ADP/AMP/ATP; 4: Alanine; 5: Anserine/Carnosine; 6:Betaine; 7: Cadaverine/Lysine; 8: Carnitine; 9: Creatine/Creatine Phosphate; 10: Creatinine; 11: Fumarate; 12: Glutamine; 13: Glutamate; 14: Glycine; 15: Glucose/Glucose-6-Phosphate; 16: Residual H2O (not quantified); 17: Isoleucine/Leucine/Valine; 18: Lactate; 19: Mannose; 20: Myo-Inositol; 21: NAD+/NADP+; 22: NADH/NADPH; 23: Nicotinurate; 24: O-Acetylcarnitine; 25: Phenylalanine; 26: Taurine; 27: Tyrosine.
Univariate and multivariate analyses
Upon inspection, two samples (one from GP and one from FY) were found to have very low concentrations. This implies a putative extraction problem and were discarded from the analysis. A total of 30 samples remained and were subsequently used. The mean concentrations of the 36 quantified metabolites corrected for tissue mass are shown in Table 3. According to the p values from the t-test, no differences between Ctrl and the MKT group could be identified, whereas the GP group had higher carnosine and nicotinurate levels than the Ctrl group. In the FY group, a lower concentration of cadaverine and higher concentration of nicotinurate were found.
Multivariate analyses of the tissue metabolite concentrations, using PCA (Fig. 3a) were consistent with the univariate results. As in the case of the blood profile, the different treatment groups did not show any global differences. Pairwise PLS-DA could only discriminate between the Ctrl-FY groups (NC = 3, R2Y = 0.92, Q2 = 0.58) and FY-MKT groups (NC = 3, R2Y = 0.86, Q2 = 0.45). The VIP variables with values higher than 1.4 reveal that the most responsible for the differentiation were, in decreasing VIP order, cadaverine and nicotinurate in Ctrl - FY comparison and cadaverine and NAD+ in FY-MKT comparison.
PCAs of a Quantified muscle metabolites and b Chemometrics data from muscle biopsies. Data from dairy calves fed with control milk replacer (Ctrl, n = 8); milk replacer supplemented with glycine and proline (GP, n = 7); milk replacer supplemented with phenylalanine and tyrosine (FY, n = 7) and milk replacer supplemented with lysine, methionine and threonine (MKT, n = 8). Ellipse Hotelling’s T2 (95%).
Chemometric analysis
To assess if additional regions of the NMR spectra contained information that could be used to discriminate between the experimental groups, multivariate analysis of the full spectra (chemometric) was also employed. As shown in Fig. 3b, no clear separation or clustering could be observed. This is consistent with the multivariate analysis based on the metabolite concentrations. However, PLS-DA could produce a model with acceptable statistical parameter for the comparison of the FY and Ctrl groups (NC = 3, R2Y = 1.00, Q2 = 0.44). A close look at the VIP data reveals that most compounds with VIP scores over 2.5 are energy-related metabolites, AA, AA derivatives as well as some unassigned compounds (Supplementary Table S2). After a visual inspection of binned spectra, an increase in the total area between 1.422–1.467 ppm and 1.632–1.760 ppm was identified in MKT group (Supplementary Fig. S1). According to the assignment, it was found that these regions mainly contain lysine/cadaverine/arginine/leucine/ornithine as well as some other unassigned metabolites.
Comparative analyses of metabolites between AA-treatments
While the focus of this work is on the differences between the treatments and the Ctrl group, the identification of differences among treatments has the potential to provide insights into the processes affected by the modifications in feed composition. Therefore, a one-way ANOVA with Tukey post-hoc test was used to identify the most significant changes among AA groups. The results of this analysis are summarized in the scheme shown in Fig. 4, where the upper portion shows the results for blood and the lower those for muscle tissue (complete information in Supplementary Tables S3 and S4).
Schematic overview of the differential metabolite profiles in blood (right-up corner) and muscle (down-left corner) between groups. Data from dairy calves fed with control milk replacer (Ctrl); milk replacer supplemented with glycine and proline (GP); milk replacer supplemented with phenylalanine and tyrosine (FY) and milk replacer supplemented with lysine, methionine and threonine (MKT). Blood analytes are shown in the upper-right corner, and muscle metabolites are shown in the down-left corner. Parameters at higher concentration in the row-group vs the column-group are presented in underlined font, whereas parameters at higher concentration in the column-group vs the row-group are presented in italic font. (*P < 0.05, **P < 0.01, no asterisk: statistical tendency 0.05 ≤ P ≤ 0.1).
Finally, when performing a PCA analysis based on the metabolites cadaverine, carnitine, carnosine, NAD+ and nicotinurate, Ctrl and MKT groups tend to appear on one side while GP and FY groups on the other side (Fig. 5).
PCA based on 5 quantified muscle metabolites (cadaverine, carnitine, carnosine, NAD+ and nicotinurate). Data from dairy calves fed with control milk replacer (Ctrl, n = 8); milk replacer supplemented with glycine and proline (GP, n = 7); milk replacer supplemented with phenylalanine and tyrosine (FY, n = 7) and milk replacer supplemented with lysine, methionine and threonine (MKT, n = 8). Ellipse Hotelling’s T2 (95%).
Discussion
The supplementation of MR with AAs is an important topic in pre-ruminant nutrition. In the present work, the impacts of supplementing MR with three different AA combinations (GP, FY and MKT) on weight gain, blood biochemistry profile and muscle metabolome were analysed after feeding the calves with control or supplemented MR for 7 weeks. The effectiveness of the supplementation has been evaluated by quantifying the plasma free AA profiles. Methionine/lysine/threonine, as well as phenylalanine, increased in the plasma of the supplemented groups as expected (MKT and FY, respectively). On the contrary, there were no significant differences in glycine/proline, and in tyrosine, probably due to the non-essential character of these AA or a difference in doses or intestinal absorption. An increase in plasma arginine has also been observed in the MKT group. A plausible explanation is that arginine and lysine share the same transport system for entering into the cell23. Thus, an increase in plasma lysine could compete with arginine for transport into the muscle leading to a concomitant increase in plasma arginine.
The profile of serum biochemical analytes considered as markers of the nutritional status of the animal (glucose, urea, creatinine, cholesterol, TGs, NEFAs and total protein) did not show major differences between groups. The only significant difference was observed in calves with MKT treatment, which showed higher serum values for creatinine versus the Ctrl group in the t-test and versus the FY group in the Tukey HSD test. These results suggest an increase in protein anabolism since blood creatinine concentration is positively related to muscular mass24. This conclusion may be reinforced by the observed tendency towards an increase in total protein in the MKT group versus the Ctrl group (p = 0.059). The enzymes ALT, AST and GGT were analysed for their role in the hepatic metabolism of AA and as markers of liver function, and no differences were observed between groups. Haptoglobin, marker of inflammation, and insulin and IGF-I, markers of metabolism and growth, were not altered either by AA supplementation.
The semitendinosus muscle NMR-metabolome was then characterized. From a glimpse of the results of muscle profile (Fig. 5), a remarkable finding is that the most distinguishable groups from Ctrl were FY and GP, in contrast with the ADG and serum analysis where MKT was the most divergent. Comparing Ctrl and MKT groups, there were no differences from the t-test and Tukey HSD pairwise comparisons of quantified muscle metabolites. Furthermore, the multivariate PLS-DAs of quantified metabolites and chemometrics results did not show an acceptable discrimination between Ctrl and MKT groups.
On the other hand, when comparing quantified metabolites in GP and FY groups versus Ctrl, the t-test identified nicotinurate being higher in FY and GP, carnosine being higher in GP and cadaverine being lower in FY. Differences between all groups are shown in Fig. 4, which summarizes the results from the ANOVA analysis with Tukey HSD test over the whole dataset. As explained in the figure, it is evident that GP and FY are different from MKT. Thus, cadaverine was lower in FY versus MKT, carnosine was higher in FY versus MKT. Nicotinurate was higher in GP versus Ctrl and MKT, and NAD+ was higher in GP and FY versus MKT. Furthermore, PLS-DAs of quantified metabolites and chemometrics shows an acceptable discrimination test between Ctrl-FY and FY-MKT comparison and PCA analysis based on the metabolites cadaverine, carnitine, carnosine, NAD+ and nicotinurate, grouped Ctrl and MKT group on one side and GP and FY groups on the other side (Fig. 5). Finally, when the surroundings of the two chemical shifts of cadaverine were analysed (between 1.422–1.467 ppm and 1.632–1.760 ppm), an increase in the total area in MKT group was observed versus both GP and FY groups. The increased total areas in MKT group may due to higher concentration of lysine and/or any of these other metabolites.
In summary, the main differential muscle metabolites were cadaverine, carnosine, carnitine, nicotinurate and NAD+. Considering the physiological interpretation of these results, cadaverine is the product of decarboxylation of lysine. Given the fact that MKT group was supplemented with lysine, this may be the reason for higher cadaverine levels.
Carnosine is a dipeptide abundant in skeletal muscle of mammals25,26,27. Amongst other biological functions, it is a powerful antioxidant28,29. It is interesting to notice that the conversion of proline into hydroxyproline during collagen synthesis requires the action of proline hydroxylase, an enzyme that needs molecular O2 as substrate. Likewise, the conversion of phenylalanine into tyrosine requires oxygen as a substrate for phenylalanine hydroxylase. This situation may indicate that the muscle cell in the groups supplemented with GP or FY could be under a situation of oxidative damage. The increase in carnosine concentration could alleviate this condition.
Carnitine is involved in the transport of acyl groups to the mitochondria, and thus in oxidative metabolism. This metabolite was also increased in GP. Similarly, nicotinurate, one of the metabolites of NAD+, is increased in the GP versus Ctrl and MKT groups. NAD+ is the final product of the oxidative respiratory chain and it has increased concentrations in both GP and FY groups.
Altogether, these results suggest a higher importance of aerobic metabolism in GP and FY groups.
Despite the fact that this study reveals the metabolomic changes due to the AA supplementation adaptation, two main limitations need however to be addressed. Firstly, this study is only focused on muscle metabolome, thus the general metabolomic profile is not presented. These results may be extended by analyzing the urine and/or serum metabolome. Secondly, only the aqueous fraction was analysed in the NMR. As such, organic phase remained unstudied. As a result, the lipid metabolism is not included and may omit some important information. Addressing these two points are important follow-up studies.
Conclusions
To the best of our knowledge, this work is the first to report a NMR metabolomics study of skeletal muscle of calves with diets differing in AA supplementation. Herein we conclude that the AA supplementation added to the MR in the conditions given in the present work does not have a marked influence in the ADG in calves from 3 days to 7 weeks of age. Nevertheless, calves supplemented with MKT had higher serum creatinine concentration, considered an indicator of increased muscular mass. These differences between MKT and the Ctrl group are however not reflected in the metabolomics analysis of skeletal muscle, as all profiles are very similar. Nevertheless, the metabolomic analysis of skeletal muscle revealed several differences between the GP/FY groups and the Ctrl/MKT groups, suggesting a metabolic adaptation especially in the GP ad FY groups. In conclusion, our results support the performance improvement after MKT supplementation in MR compared with Ctrl and GP and FY combinations, and reveal that different AA profile of calf MR can modify protein and muscle metabolism. This study may help the dairy industry to take metabolic factors into account when supplementing relevant AAs in MR.
Methods
Animals and housing
Animals were managed according to the recommendations of the Animal Care Committee of Institut de Recerca i Tecnologia Agroalimentàries (IRTA) under the approval research protocol FUE-2017-00587321, authorization code 9733. Thirty-two Holstein male calves (weighing 44.6 ± 1.07 kg of BW at the age of 2.7 ± 0.27 days) were obtained from Granja Murucuc (Gurb, Barcelona, Spain) and housed individually at IRTA Torre Marimon (Caldes de Montbui, Barcelona, Spain). Animals used in the component here described were a part of a larger study with a higher number of animals that aimed to evaluate calf performance. The sample size was chosen based on the previous similar studies18,19,21,30,31,32.
Diet treatments and performance recordings
MR was offered in 3-L nipple bottles twice a day, and all animals followed the same MR feeding program. Calves were fed on 4 L/d commercial milk replacer at 12.5% Dry Matter (DM). Milk replacer (Nukamel, Weert, The Netherlands) contained 25.4% CP (based on skimmed milk protein, and whey protein concentrate) and 20.3% fat from experimental days 1 to 4. Subsequently, the volume of MR was increased to 5 L/d from day 5 to 7, followed by 6 L/d from days 8 to 14, and 6 L/d at 15% DM concentration from day 15 to 49. At day 50, MR was reduced to one feeding per day of 3 L at 15% DM until day 56 when animals were fully weaned.
Calves were divided in four different treatments according to AA supplementation in the MR formula (which replaced whey protein concentrate in the control MR). All AAs were provided by Livzon Group (Fuxing Pharmaceutical, Ningxia (China)). Treatments were previously defined in a list, which indicated the MR treatment in each individual calf box. Then, as calves were arriving to the facilities, they were allocated correlatively to the individual boxes with a predetermined treatment. In this way, the only identification of the calves was the eartag number, which allowed the blinding of researchers and care-takers to treatment allocation. The four treatments were:
A (Ctrl): Control group with no AA supplementation
B (GP): supplementation with 0.1% glycine plus 0.3% proline in the MR formula
C (FY): supplementation with 0.2% phenylalanine plus 0.2% tyrosine in the MR formula
D (MKT): supplementation with 0.62% lysine plus 0.22% methionine plus 0.61% threonine in the MR formula
Ingredients and chemical composition are shown in Table 4. All animals had free access to water, and a pelleted concentrate starter feed (Pinallet, Cardona, Spain) was limited to minimize the impact of concentrate feeding on amino acid absorption: 100 g/day from day 1 to day 17 of study, 200 g/d from days 18 to 21, 300 g/d from day 22 to day 24. Subsequently, 400 g/d were used from day 25 to day 28, 500 g/d, from day 29 to day 31, 600 g/d from day 32 to day 33 d, 700 g/d at day 34, 800 g/d at day 35, 1,000 g/d from day 36 to day 42, 1,200 g/d from day 43 to day 49, and ad libitum afterwards until 63 d of study, when the study finished. Information about composition and AA concentration in the starter is provided in Table 5. Chopped barley straw was offered ad libitum during this study (Table 6). Calves were weighed weekly with an electronic scale (Mobba SC-01, Badalona, Spain) until the end of the study.
Blood and muscle sample collection
At the 7th week of the study, blood samples were obtained from jugular vein 2 h after the morning MR feeding, and were kept in 10 ml Vacutainer tubes without anticoagulant to obtain the serum for biochemistry profiles or with lithium heparin for plasma AA analyses. Serum and plasma were obtained by centrifugation at 1,500 g for 10 min and stored in aliquots at −20 °C until further analysis.
At 47.9 ± 0.46 days of age, a biopsy was obtained from semitendinosus muscle. Briefly, animals were injected with xylazine intravenously at the dose of 0.005 mL/kg BW, once the animals were sedated, the hind leg was shaved and disinfected with iodine povidone. Then, 2 mL of 0.02 g/mL lidocaine were subcutaneously injected as a local anaesthetic. A 3-cm cut in the skin and subcutaneous layer was done with a scalpel, and a 2-cm zone of the muscle was delimited with suture, and it was dissected with curve scissors. Finally, the muscular, subcutaneous and cutaneous layers were sutured. Muscle samples were immediately frozen into liquid nitrogen and subsequently kept at −80 °C until further analysis.
Quantification of plasma AAs
Plasma AAs were measured by HPLC following a modified protocol33. At the day of assay, 100 µl of thawed plasma were transferred to a 2.0 mL micro-centrifuge tube and 10 µl of 10 mM DTT in PBS (pH 7.4) and 5,79 µl 2.5 mM GABA as internal standard were added. The solution was vortexed for 30 s and an equivalent volume of 10% sulfosalicylic acid was added to precipitate the proteins, followed by mixing for 2 min. The samples were centrifuged at 14,000 g for 10 min at 4 °C and the supernatant collected34.
For derivatization, the AccQ Fluor (Waters, Milford, MA, USA) method was used following the instructions provided by the manufacturer. A total of 10 μL standard solution or supernatant of deproteinized plasma sample was taken into a 1.5 mL micro-centrifuge tube, buffered with 70 µL Waters AccQFluor Borate Buffer and derivatized by addition of 20 µL of AccQ-Fluor reagent, vortexed and heated for 10 min at 55 °C.
HPLC was performed on an Elite LaCHrom (Hitachi, Tokyo, Japan) equipped with an UV detector (Hitachi L-24200, Tokyo, Japan) with a Novapak C18 column (300 mm × 3.9 mm) from Waters. The flow rate was 1.0 ml/min and the column temperature was kept at 28 °C. The injection volume was 10 μL and the detection wavelength was set at 254 nm. The solvent system consisted of two eluents: (A) AccQ > Tag eluent A concentrate (10%, v/v) and water (90%, v/v) and (B) 60% acetonitrile/40% water. The following gradient elution was used: 0 to 0.50 min, 100% A; 0.5 min, 98% A to 2% B; 20 min, 93% A to 7% B; 32 min, 91.5% A to 8.5% B; 40 min, 82% A to 18% B; 47.5 min: 79% A to 21% B; 55 min: 60% A to 40% B; 60 min: 100% B. Pure AAs were used as standards and GABA was used as internal control (Sigma, St. Louis, MO, USA). Standards were prepared at 2.5 mM stock solutions in 0.1 M HCl and was serially diluted from 500 µM to 7.81 µM to perform the standard curve in order to quantify AA of plasma samples. EZChrom Elite system V3.1.7 software (Agilent, Santa Clara, CA, USA) was used for system control and data acquisition. Validation of the technique showed a good precision (Coefficients of Variation from 1% to 5% for all AA), good linearity (R2 > 0.998) except for serine (R2 = 0.990) and cysteine (R2 = 0.989) and a limit of quantification lower than 10 μM (except for serine (11 μM) and histidine/glutamine (21 μM).
Determination of serum biochemical analytes
Serum biochemistry analytes were analysed with the Olympus AU400 analyser with following techniques and OSR reagents (Olympus System Reagent)35,36: glucose (HK method), urea (GLDH method), creatinine (Jaffé method), cholesterol (CHOP-PAP-method), triglyceride (GPO-PAP method), total protein (Biuret method), ALT (International Federation of Clinical Chemistry (IFCC) method), AST (IFCC method) and GGT (IFCC method). NEFAs were determined with NEFA-C reagent (Wako Chemicals GmbH, Neuss, Germany). Quality control protocols were based on the daily quantification of two control sera of low and high concentration (Control serum I and Control serum II, Beckman Coulter) Haptoglobin was determined by a colorimetric method (Tridelta, Ireland). Insulin and IGF-I were determined by ELISA (Bovine Insulin ELISA from Mercodia, Uppsala, Sweden and Human IGF-I ELISA E20 from Mediagnost, Reutlingen, Germany). This reagent is suitable for bovine samples, as declared by the manufacturer, but the cross-reactivity was not assessed by the authors.
Intra-assay Coefficients of Variation (CV) were: glucose (1.7%), urea (2.1%), creatinine (1.4%), cholesterol (1.1%), triglycerides (3.0%), total protein (0.9), ALT (1.7%), AST (1.0%), GGT (0.6%), NEFAs (2.7%), Haptoglobin (4.1%), insulin (4.1%) and IGF-1 (4.4%).
Muscle extraction and NMR data acquisition
Muscle tissues were transferred from −80 °C freezer to dry ice. All muscle tissues were ground into fine powder while mixed with liquid nitrogen in a mortar and pestle. The extraction of the aqueous metabolites was performed using a modified chloroform/methanol method previously described21. In brief: approximately 0.5 g of muscle tissue was mixed with 1.2 mL of chloroform/methanol (1:2 v/v) in a glass tube and was vortexed for 1 min. Then an extra 0.4 mL of chloroform was added to compensate the evaporated chloroform and the tube was vortexed for an additional 1 minute. Afterwards 0.4 mL of double distilled water was added and the tube vortexed again for 1 min. The homogenate was centrifuged at 1935 g for 20 min at 4 °C. 1000 μL of the top (methanol/water) fraction was transferred into a 2 mL polypropylene tube and dried in a centrifugal vacuum concentrator. To resuspend the dried sample for NMR analysis, 600 µL of phosphate buffer in D2O (150 mM; pH 6.6 (pD 7.0); with 100 μM of TSP-D4 (deuterated Trimethylsilylpropanoic acid)) were added, followed by 1 min vortexing and 10 min centrifugation at 14,100 g at room temperature. The supernatants were transferred into NMR borosilicate tubes rated for 800 MHz (New Era, Vineland, NJ, USA).
1D 1H NOESY spectra were collected on an 800 MHz NMR spectrometer equipped with a room temperature HCN inverse Z-gradient probe (Bruker Biospin, Billerica, MA, USA). The pulse sequence used for 1D gradient NOESY with water pre-saturation was “noesygppr1D” with the Bruker standard acquisition parameters for profiling (spectral width: 16393.443 Hz, recycling delay: 4 s, acquisition time: 1.9988480 s, mixing time 10 ms). 96 scans and 8 dummy scans were used for each spectrum resulting in a 10-min experiment. For select samples, additional homonuclear and heteronuclear spectra (1H J-resolved, 1H-1H COSY, and 1H-13C HSQC) spectra were also collected to assist with compound identification.
NMR data processing and analysis
Spectra were processed with 1 Hz exponential appodization, no additional zero filling and polynomial baseline correction with the TopSpin 3.2 software (Bruker Biospin, Billerica, MA, USA). Compound quantification in reference to the TSP internal standard was performed with the Chenomx 8.1 NMR Suit (Edmonton, Alberta, Canada). The limit used for compound quantification in Chenomx was set at 0.002 mM.
Metabolomics data have been deposited to the EMBL-EBI MetaboLights database (DOI: 10.1093/nar/gks1004. PubMed PMID: 23109552) with the identifier MTBLS720. The complete dataset can be accessed here https://www.ebi.ac.uk/metabolights/MTBLS720.
Statistical analysis
Performance and blood data
The univariate analysis was carried out with the SPSS 24 (IBM, Armonk, NY, USA). The significance level was declared at P < 0.05 and a tendency was considered at 0.05 ≤ P ≤ 0.1. Descriptive data are presented with the means and the standard error (mean ± SE). Shapiro-Wilks test was used to assess normality of data. Means were compared to the Ctrl group by a t-test and for pairwise comparisons, ANOVA with Tukey HSD post hoc was applied. The multivariate analyses were performed by SIMCA 13.0.3 (Umetrics AB, Umeå, Sweden) after applying auto-transform to reduce data skewness and UV scaling to reduce relative importance of large values.
Muscle data
For Chemometric statistical analysis, processed spectra were loaded into “R” where peak alignment was performed with SPEAQ. The data were then exported into a coma-separated value (CSV) file and loaded into SIMCA 13.0.3. All spectra were treated with a log transform and by Pareto scaling. For the whole groups, unsupervised Principal Components Analysis (PCA) was performed to detect outliers, patterns and trends. For pairwise Partial Least Square Discriminant Analyses (PLS-DAs), Q2 > 0.4 were selected as acceptable models and Variable Importance in Projection (VIP) scores were used to identify metabolites responsible for the separation. These metabolites together with others that are relevant for the project were then quantified using database assisted spectral deconvolution that was performed using Chenomx. The resulting concentration were normalized based on the mass tissue used for extraction and then further performed univariate analysis by SPSS 24 and multivariate analysis by SIMCA 13.0.3 after applying auto-transform and UV scaling.
Availability of Data and Materials
All data generated or analysed during this study are included in this published article and its Supplementary files.
References
Castro, J., Hwang, G., Saito, A., Vermeire, D. & Drackley, J. Assessment of the effect of methionine supplementation and inclusion of hydrolyzed wheat protein in milk protein-based milk replacers on the performance of intensively fed Holstein calves. J. Dairy Sci. 99, 6324–6333 (2016).
Otterby, D. E. & Linn, J. G. Advances in Nutrition and Management of Calves and Heifers. J. Dairy Sci. 64, 1365–1377 (1981).
Davis, C. L. & Drackley, J. K. The development, nutrition, and management of the young calf. (Iowa State University Press, 1998).
Montagne, L., Toullec, R. & Lallès, J. P. Intestinal digestion of dietary and endogenous proteins along the small intestine of calves fed soybean or potato. J. Anim. Sci. 79, 2719–2730 (2001).
Heine, W. E. Qualitative Aspects of Protein in Human Milk and Formula: Amino Acid Pattern. Protein Metabolism During Infancy, edited by Niels C. R. Raiha. Nestle Nutrition Workshop Series, Vol. 33. (Nestec Ltd. Vevey/Raven Press, Ltd., New York, 1994).
Agostoni, C., Carratù, B., Boniglia, C., Riva, E. & Sanzini, E. Free Amino Acid Content in Standard Infant Formulas: Comparison with Human Milk. J. Am. Coll. Nutr. 19, 434–438 (2000).
Jenkins, K. J. & Emmons, D. B. Fortification of calf milk replacers with amino acids in free form or plastein-bound. Can. J. Anim. Sci. 63, 893–903 (1983).
Kanjanapruthipong, J. Supplementation of milk replacers containing soy protein with threonine, methionine, and lysine in the diets of calves. J. Dairy Sci. 81, 2912–2915 (1998).
Pelaez, R. & Walker, D. Milk replacers for preruminant lambs: limiting amino acids in two soybean protein isolates determined with a change-over design. Aust. J. Agric. Res. 30, 125–134 (1979).
Hill, T. M., Bateman, H. G., Aldrich, J. M., Schlotterbeck, R. L. & Tanan, K. G. Optimal concentrations of lysine, methionine, and threonine in milk replacers for calves less than five weeks of age. J. Dairy Sci. 91, 2433–2442 (2008).
Morrison, S., Campbell, J. & Drackley, J. Amino acid supplementation of calf milk replacers containing plasma protein. J. Dairy Sci 100, 4637–4649 (2017).
Ceballos, L. S. et al. Composition of goat and cow milk produced under similar conditions and analyzed by identical methodology. J. Food Compos. Anal. 22, 322–329 (2009).
Wu, G. Amino acids: Metabolism, functions, and nutrition. Amino Acids 37, 1–17 (2009).
Terré, M., Bach, A. & Devant, M. Performance and behaviour of calves reared in groups or individually following an enhanced-growth feeding programme. J. Dairy Res. 73, 480–486 (2006).
Nicholson, J. K. & Wilson, I. D. Understanding ‘Global’ Systems Biology: Metabonomics and the Continuum of Metabolism. Nat Rev Drug Discov 2, 668–676 (2003).
Bernini, P. et al. Standard operating procedures for pre-analytical handling of blood and urine for metabolomic studies and biobanks. J. Biomol. NMR 49, 231–243 (2011).
Ritota, M., Casciani, L., Failla, S. & Valentini, M. HRMAS-NMR spectroscopy and multivariate analysis meat characterisation. Meat Sci. 92, 754–761 (2012).
Shintu, L., Caldarelli, S. & Franke, B. M. Pre-selection of potential molecular markers for the geographic origin of dried beef by HR-MAS NMR spectroscopy. Meat Sci. 76, 700–707 (2007).
Jung, Y. et al. Discrimination of the geographical origin of beef by 1H NMR-based metabolomics. J. Agric. Food Chem. 58, 10458–10466 (2010).
Graham, S. F. et al. The application of NMR to study changes in polar metabolite concentrations in beef longissimus dorsi stored for different periods post mortem. Metabolomics 6, 395–404 (2010).
Palma, M. et al. The hepatic and skeletal muscle ovine metabolomes as affected by weight loss: a study in three sheep breeds using NMR-metabolomics. Sci. Rep. 6, 39120 (2016).
Castejón, D., García-Segura, J. M., Escudero, R., Herrera, A. & Cambero, M. I. Metabolomics of meat exudate: Its potential to evaluate beef meat conservation and aging. Anal. Chim. Acta 901, 1–11 (2015).
Luiking, Y. C. & Deutz, N. E. P. Biomarkers of arginine and lysine excess. J. Nutr. 137, 1662S–1668S (2007).
Uemura, O. et al. Creatinine-based equation to estimate the glomerular filtration rate in Japanese children and adolescents with chronic kidney disease. Clin. Exp. Nephrol. 18, 626–633 (2014).
Christman, A. A. Factors affecting anserine and carnosine levels in skeletal muscles of various animals. Int. J. Biochem. 7, 519–527 (1976).
Dunnett, M., Harris, R., Soliman, M. & Suwar, A. A. Carnosine, anserine and taurine contents in individual fibres from the middle gluteal muscle of the camel. Res. Vet. Sci. 62, 213–216 (1997).
Imanari, M., Higuchi, M., Shiba, N. & Watanabe, A. Accurate analysis of taurine, anserine, carnosine and free amino acids in a cattle muscle biopsy sample. Anim. Sci. J. 81, 369–376 (2010).
Boldyrev, A. A., Aldini, G. & Derave, W. Physiology and Pathophysiology of Carnosine. Physiol. Rev. 93, 1803–1845 (2013).
Boldyrev, A. A., Dupin, A. M., Bunin, A. Y. A., Babizhaev, M. A. & Severin, S. E. The antioxidative properties of carnosine, a natural histidine containing dipeptide. Biochem. Int. 15, 1105–1113 (1987).
Wan, J. et al. Amniotic fluid metabolomics and biochemistry analysis provides novel insights into the diet-regulated foetal growth in a pig model. Sci. Rep. 7, 1–10 (2017).
Artegoitia, V. M., Foote, A. P., Lewis, R. M. & Freetly, H. C. Rumen fluid metabolomics analysis associated with feed efficiency on crossbred steers. Sci. Rep. 7, 1–14 (2017).
Carrillo, J. A. et al. Integrated metabolomic and transcriptome analyses reveal finishing forage affects metabolic pathways related to beef quality and animal welfare. Sci. Rep. 6, 1–16 (2016).
Castellanos, M., Van Eendenburg, C., Van Gubern, C. & Sanchez, J. M. Ethyl-bridged hybrid column as an efficient alternative for HPLC analysis of plasma amino acids by pre-column derivatization with 6-aminoquinolyl- N -hydroxysuccinimidyl carbamate. J. Chromatogr. B 1029–1030, 137–144 (2016).
Sharma, G. et al. Analysis of 26 amino acids in human plasma by HPLC using AQC as derivatizing agent and its application in metabolic laboratory. Amino Acids 46, 1253–1263 (2014).
Saco, Y. et al. Evaluation of serum cortisol, metabolic parameters, acute phase proteins and faecal corticosterone as indicators of stress in cows. Vet. J. 177, 439–441 (2008).
Marco-Ramell, A. et al. Biochemical and proteomic analyses of the physiological response induced by individual housing in gilts provide new potential stress markers. BMC Vet. Res. 12, 265 (2016).
Acknowledgements
The authors gratefully thank all the staff of the IRTA Farm (Torre Marimon, Caldes de Montbui, Spain) for their assistance in feeding and care of the animals. This research was supported by the Spanish Ministry of Economy and Competitivity (AGL2015-68463-C2-1-P to MT and AGL2015-68463-C2-2-P to AB). Funding from Short Term Scientific Mission between UAB and ITQB from DairyCare (European Cooperation in Science and Technology, COST FA 1308) is acknowledged (COST-STSM-FA1308-36644). Part of the work was also supported by ONEIDA project (LISBOA-01-0145-FEDER-016417) co-funded by FEEI - “Fundos Europeus Estruturais e de Investimento” from “Programa Operacional Regional Lisboa 2020” and by national funds from FCT - “Fundação para a Ciência e a Tecnologia”. The NMR spectrometers at CERMAX are integrated in the National NMR Network (PTNMR) and are partially supported by Infrastructure Project No 022161 (co-financed by FEDER through COMPETE 2020, POCI and PORL and FCT through PIDDAC). The author Kuai Yu is funded by China Scholarship Council (CSC) graduate scholarship program.
Author information
Authors and Affiliations
Contributions
K.Y. performed the experimental metabolomics work and data analysis, sample measurements and participated in the manuscript writing. M.M. directed and participated in the metabolomic analysis. D.V. participated in sample measurements. Y.S. participated in sample measurements. A.A. contributed to the manuscript writing and metabolomics study design. M.T. participated in the experimental design and was in charge of the animal feeding trial and revised the manuscript. A.B. participated in the experimental design, supervised the analytical work and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yu, K., Matzapetakis, M., Valent, D. et al. Skeletal muscle metabolomics and blood biochemistry analysis reveal metabolic changes associated with dietary amino acid supplementation in dairy calves. Sci Rep 8, 13850 (2018). https://doi.org/10.1038/s41598-018-32241-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-32241-4
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.