Abstract
Cyrtorhinus lividipennis Reuter (Hemiptera: Miridae) is an important egg predator of planthoppers which are destructive rice pests. The chemosensory genes in the mirid antennae play important roles in mating and prey-seeking behaviors. To gain a better understanding of the olfaction of C. lividipennis, we sequenced the antennal transcriptomes of the predator to identify the key olfaction genes. We identified 18 odorant binding proteins (OBPs), 12 chemosensory proteins (CSPs), 1 Niemann-Pick C2 protein (NPC2), 15 odorant receptors (ORs), 6 ionotropic receptors (IRs), 3 gustatory receptors (GRs) and 3 sensory neuron membrane proteins (SNMPs). Quantitative real-time PCR results showed that the relative transcript levels of three ClivORs (ClivOR6, 7 and 14) in the female antennae were 3 to 6 folds higher than that in the male antennae, indicating these genes were more related to oviposition site selection. The relative transcript levels of ClivCSP8 and ClivOR11 were 2.6 and 2.7 times higher in the male antennae than that of the female, respectively, indicating that these genes might be involved in mate searching. Moreover, the responses of dsorco treated predators to volatiles emitted from infested rice were significantly reduced, indicating these volatiles might serve as crucial cues in the host searching of C. lividipennis.
Similar content being viewed by others
Introduction
Natural enemies of herbivorous insects often depend on volatile chemical cues to locate their concealed prey in the complex environment1. For example, some parasitoid species are attracted by herbivore-induced plant volatiles during the foraging process2. Cyrtorhinus lividipennis Reuter (Hemiptera: Miridae) is an important egg predator of planthoppers and leafhoppers which are destructive rice pests in Asia3,4,5. Some studies have reported the role of rice volatiles in regulating the behavior of natural enemies6. C. lividipennis were found to be attracted by volatiles emitted from herbivore-infested plants, suggesting that olfaction played an essential role in their prey search6,7. The antenna, covered with different types of chemosensory sensilla, is the specialized organ for olfaction in insects8. Olfactory perceptions of Hemipteran species, such as Tropidothorax elegans9 and Apolygus lucorum8,10, rely largely on chemosensory genes. Identification of chemosensory genes in C. lividipennis can provide better understanding of how the predator utilizes chemical cues in their search behavior in agricultural systems11.
Chemical cues are transformed into electrical signals by olfactory receptor neurons (ORNs) housed within the sensilla and then these signals are transmitted to the brain to finally elicit distinct behaviors8,12,13,14. The key olfactory proteins involved in the perception of odorants in insects are odorant-binding proteins (OBPs), chemosensory proteins (CSPs), Niemann-Pick C2 protein (NPC2), odorant /ionotropic receptors (ORs and IRs), gustatory receptors (GRs) and sensory neuron membrane proteins (SNMPs)5,15,16,17,18,19.
OBPs and CSPs are small soluble proteins that are highly abundant in the chemosensilla lymph of insects20. The two soluble proteins can transport hydrophobic odorants through the sensillar lymph to activate membrane-bound ORs15,21. A typical OBP (generally 135–220 amino acids) contains six conserved cysteine residues paired into three disulfide bridges. The OBPs undergo ligand-induced conformational shifts that trigger the firing of ORNs22,23. Some studies have showed that OBPs play different roles by binding with various odorants24. For instance, CquiOBP1 was reported to bind with a oviposition pheromone in Culex pipiens quinquefasciatus25. CsupOBP8 was found to be associated with the recognition of plant volatiles in Chilo suppressalis26 and OBP99a in Bactrocera dorsalis was found to be involved in the selection of ovipostion hosts24. The CSP (normally 100–120 residues) has four conserved cysteines that form two disulfide bridges and bear no sequence similarity to OBPs20,27,28. The CSPs have been reported to perform different functions, such as leg regeneration and development5,29,30,31. Liu et al. also found that CSP4 acted as surfactant in the proboscis of two Helicoverpa species32. CSPs were also reported to act as carriers for visual pigments in insects33.
NPC2 are highly divergent between species in arthropods19,28. These proteins share some structrual and functional characteristics with OBPs and CSPs19. Some studies have shown that NPC2 proteins act as carriers for semiochemicals and other hydrophobic compounds19,34.
Insect ORs belong to the seven-transmembrane domain (TMD) protein family with a reversed topology of having intracellular N-terminus35,36. The conventional insect ORs show great diversity in the DNA sequence levels, which reflect their rapid evolution37. The odorant receptor coreceptor (Orco), highly conserved among insect species, forms ligand-gated ion channels with other ORs to enhance odorant responsiveness15,38,39,40. In fact, Orco can form heterodimeric complexes with conventional ORs that are responsible for binding to diverse odorants41. Disruption of the Orco function can dramatically impair olfactory behavior responses in various insect species, such as A. lucorum, Harpegnathos saltator and Locust amigratoria10,42,43. IRs are relatives of ionotropic glutamate receptors (iGluR) which represent elements for sensing both external and internal chemical cues44,45. IRs are supposed to form two or three subunits coexpressed in the same neuron35,45. They are divided into two major groups, the conserved “antennal IRs” and “divergent IRs”22,46,47. Some GRs are coexpressed in chemosensory neurons which are involved in carbon dioxide detection48. However, GRs are mainly expressed in gustatory receptor neurons in taste organs, which can detect bitter compounds, sugars and contact pheromones22.
Finally, the SNMPs are proteins of the CD36 family that are crucial for pheromone perception18. Insects generally have two SNMP subfamilies (SNMP1 and SNMP2). The SNMP1 subfamily was found to be associated with pheromone detection in Drosophila melanogaster and several lepidopteran species18,49. However, the function of SNMP2 remains poorly understood21.
In this study, we performed Illumina sequencing to identify putative chemosensory genes in the adult C. lividipennis antennae. We identified 18 OBPs, 12 CSPs, 1 NPC2, 15 ORs, 6 IRs, 3 GRs and 3 SNMPs in the transcriptome dataset. The expression patterns of these genes in different tissues were examined by quantitative real-time PCR (qRT-PCR). We further explored the foraging behavior of the predator C. lividipennis by silencing orco in a laboratory experiment.
Results
Illumina sequencing and sequence assembly
A total of 60,658,602 and 53,853,286 clean reads were obtained from the C. lividipennis male and female antennal transcriptome, respectively. The clean reads are available in the NCBI Sequence Read Archive (SRA accession: SRP128761). The combined assembly of all clean reads generated 62,637 unigenes with a mean length of 1,401 bp, and N50 of 2,338 bp and N90 of 588 bp (Supplementary Table S1).
Functional annotation
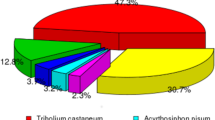
28,147 (44.93%) unigenes were annotated in at least one of the databases. The numbers of unigenes annotated to different databases are shown in Table 1. The largest numbers of unigene annotations are deposited in NR database (23,113, 36.89%). BLASTX homology searched against the NCBI-Nr database showed that the C. lividipennis antennal unigenes were best matched to sequences from Zootermopsis nevadensis (20.6%), followed by Tribolium castaneum (8.0%), and Acyrthosiphon pisum (7.9%) (Fig. 1). Gene Ontology (GO) assignments were used to classify the C. lividipennis antennal transcriptome unigenes into three main functional groups: biological processes, cellular components and molecular functions (Fig. 2). Among the 62,637 unigenes, approximately 34.66% (21,712) of the unigenes could be assigned to GO terms (Table 1). Cellular process (12,485, 20.49%) was the most prevalent terms in the category of biological processes. The cellular components were equally dominated by cell part (6,625, 18.53%) and cell (6,625, 18.53%). Binding (12,700, 48.55%) represented the most abundant GO terms in the molecular function category.
Identification of candidate OBPs
In total, we identified 18 putative OBPs from the C. lividipennis antennal transcriptome. Of these genes, 10 were previously reported in the whole body transcriptome of adult C. lividipennis (Genbank No. KY462016-KY462025). All of the other 8 newly identified OBP sequences (named ClivOBP11-18) had full-length ORFs with predicted signal peptide. The results of BLASTX are shown in Supplementary Table S2. All the ClivOBPs were best matched to known Miridae OBPs. The identities of three pairs of OBPs were higher than 70%: ClivOBP15 and LlinOBP3 (98%), ClivOBP17 and LlinOBP11 (75%), ClivOBP18 and LlinOBP18 (72%). The remaining pairs showed identities ranging from 44 to 67%. Multiple sequence alignments of the newly identified C. lividipennis OBPs indicated that the eight ClivOBPs belongs to classic OBPs (carried six conserved cysteine residues) (Supplementary Fig. S1A)20,50.
The phylogenetic tree was constructed to reveal the relationships of ClivOBPs to those of other hemipteran species, including three Miridae (A. lucorum, Lygus lineolaris and Adelphocoris lineolatus) and three Delphacidae species (Nilaparvata lugens, Sogatella furcifera and Laodelphax striatella). The tree revealed that ClivOBPs spread across several branches. Several ClivOBPs (ClivOBP12, 13, 14, 16 and 18) were clustered with AlucOBPs in one subbranch (Supplementary Fig. S2).
Identification of candidate CSPs and NPC2
Twelve putative CSPs and one NCP2 were identified in the C. lividipennis antennal transcriptome. Among them, five CSP sequences were also reported in the whole body transcriptome of adult C. lividipennis (Genbank No. KY462026-KY462030). The remaining seven ClivCSPs were named from ClivCSP6 to ClivCSP12. The results of BLASTX are shown in Supplementary Table S2. All the deduced ClivCSPs sequences had full-length ORFs with the conserved four cystine residues (Supplementary Fig. S1B). ClivCSP7 and ClivCSP12 showed the highest identities to AsutCSP4 (81%) and AlinCSP12 (80%) respectively, while ClivCSP9 and ClivCSP10 showed identities <57% to other known CSPs.
A phylogenetic analysis was performed to show the relationships among ClivCSPs and CSPs from other hemipteran species, including two Miridae (A. lucorum and A. lineolatus) and two Delphacidae species (N. lugens and L. striatella). The ClivCSPs phylogenetic tree showed that ClivCSPs spread across several branches. Some ClivCSPs (ClivCSP9, ClivCSP12) were closely related to NlugCSPs (Supplementary Fig. S3).
Identification of chemoreceptor genes
A total of 15 candidate ORs, 6 candidate IRs and 3 putative GRs were identified in the C. lividipennis antennal transcriptome. The results of BLASTX are shown in Supplementary Table S3. All of the ORs contained full-length ORFs ranged from 314 to 490 amino acid residues with 4–8 transmembrane domains. Seventeen ClivORs shared 28–77% sequence identities with the ORs in A. lucorum. The C. lividipennis Orco sequence showed the highest identity (89%) to the A. lineolatus Orco. Of the 6 candidate IRs, four unigenes were predicted to have full-length ORFs with a least one TMD. The ClivIRs shared 22–57% sequence identities with other insect IRs (Supplementary Table S3). We performed the phylogenetic tree to better understand the relationships of the ClivOR proteins with ORs in other hemipteran species, including one Miridae (A. lucorum), two Aphididae (Myzus persicae and A. pisum) and one Delphacidae species (N. lugens).The phylogenetic tree revealed that the C. lividipennis Orco was clustered with Orcos in other insect species (Fig. 3).
Identification of candidate SNMPs
We found two subfamilies of SNMPs in C. lividipennis (1 ClivSNMP1 and 2 ClivSNMP2, Supplementary Table S3). The ClivSNMPs showed 35–51% identities with other insect species. In addition, the three ClivSNMPs had two transmembran domains.
Sex-specific expression of C. lividipennis chemoreception genes
Results of the qRT-PCR assays indicated that three ClivOBPs (15, 17, 18), four ClivCSPs (6, 9, 11, 12), twelve ClivORs (1, 2, 4, 6–10, 12, 13, 14 and orco), ClivIRs (1–6), three ClivGRs (1–3) and two ClivSNMPs (ClivSNMP2-1 and 2-2) were more highly expressed in the female antennae than in the male. In particular, the relative transcript levels of three ClivORs (6, 7 and 14) in the female antennae were 3 to 6 folds higher than in the male (Fig. 4A). Besides, two ClivOBPs (14, 16), two ClivCSPs (7, 8), ClivOR11 and ClivSNMP1 were highly expressed in the male antennae. Among these genes, the relative expression levels of ClivCSP8 and ClivOR11 were 2.6 and 2.7 times higher in the male antennae than that of the female, respectively (Fig. 4B). In addition, three ClivOBPs (11, 12, 13), ClivCSP10, ClivNPC2 and two ClivORs (3, 5) were expressed in both the male and female antennae with similar transcript accumulations (Supplementary Fig. S4).
Sex-specific expression of C. lividipennis chemoreception genes. (A) The female-dominantly expressed olfactory genes. (B) The male-dominantly expressed olfactory genes. Gene expression patterns in antennae were normalized relative to legs (male and female mixture). Data were presented as the mean of three replicates (n = 3) ± SE. Different lower cases indicate significant differences (p < 0.05). FA: female antennae, MA: male antennae, L: legs.
Responses of C. lividipennis to different odors after silencing orco gene
We used RNAi to investigate the functions of orco in the foraging behavior of C. lividipennis. dsorco treated predators showed an approximately 80% decrease in the transcripts of orco compared to dsGFP treatments (Fig. 5A). These dsorco and dsGFP treated mirids were further used for olfactory response study.There were significant differences in the foraging behavior between the dsorco and the dsGFP treated insects. The responses of the dsorco treated predators to volatiles emitted by infested plants were significantly lower than those of the dsGFP treated individuals (t = 5.9285, df = 7.189, p < 0.01). The number of mirids showing no response was higher under the dsorco treatment than the dsGFP treatment (t = 2.6848, df = 4.451, p < 0.05). No obvious differences of dsorco and dsGFP treated predators in response to volatiles released by healthy plants were observed, indicating that some other genes might be involved in olfactory responses (t = 0.8801, df = 4.587, p = 0.4225) (Fig. 5B).
Responses to different odor sources by C. lividipennis after dsRNA silencing treatment. (A) Relative transcript accumulation of orco after RNAi were quantified by qRT-PCR. (B) Responses of C. lividipennis to different odor sources after dsGFP and dsorco treatment. Infested plants, healthy plants denote volatiles emitted by gravid female-damaged rice seedlings and healthy rice seedlings respectively. *, **, and n.s. refereed to the difference between two treatments (dsGFP and dsorco) is significant (p < 0.05), highly significant (p < 0.01), and not significant (p > 0.05) (t-test), respectively.
Discussion
C. lividipennis is one of the most important natural enemies of planthoppers in Asian rice fields3. However, studies on the insect predator’s olfactory system are scarce36. It has been shown that C. lividipennis rely on herbivore-induced rice volatiles to identify eggs of N. lugens6 and the antennae are the main olfactory organs for this insect. The C. lividipennis antennal transcriptome dataset would be able to provide a better molecular understanding of its olfactory systems that can improve the effectiveness of the predator in biological control.
In this study, we identified 58 putative olfactory genes (18 OBPs, 12 CSPs, 1 NPC2, 15 ORs, 6 IRs, 3 GRs and 3 SNMPs) based on the transcriptome analysis of male and female antennae of C. lividipennis. The number of OBPs identified in C. lividipennis was more than those in Sitobion avenae (13 OBPs), N. lugens (11 OBPs) and A. pisum (15 OBPs)50,51,52, but less than those in Tessaratoma papillosa (33 OBPs) and A. lucorum (38 OBPs)8,46. The number of the CSPs in C. lividipennis was close to the previous findings in other hemipteran insects such as A. pisum (13CSPs), S. furcifera (9 CSPs) and N. lugens (17CSPs)50,52,53. In addition, we identified 24 chemosensory receptors (15 ORs, 6 IRs and 3 GRs) in C. lividipennis, which was fewer than that in other insect species such as Anoplophora chinensis (44 ORs, 23 IRs and 19 GRs)13 and Cylas formicarius (54 ORs, 15 IRs and 11 GRs)54. The number of identified olfactory genes varied in different species, which might be the limitation of the Illumina sequencing methods and depth55. The transcriptome data may only represent part of expressed chemosensory genes in the cell but not those genes with low transcript abundance or no expression56,57.
The chemosensory genes of C. lividipennis showed various similarities with the genes from other hemipteran species, which might be caused by their different host preferences53. In the phylogenetic trees, the OBPs and CSPs in C. lividipennis clustered with olfactory genes in other species (like A. lucorum, L. lineolaris), which suggested that these genes might have similar functions in general odorant perception21. In addition, most of the ClivORs were clustered with ORs in A. lucorum, which might be involved in the detection of host plant volatiles8.
Several chemosensory genes were reported to have sex biased transcript accumulation in other insects, such as in T. papillosa and A. lucorum8,46. Both mating and feeding behavior strongly rely on chemical cues. Some studies showed that the chemosensory genes were associated with the perception of plant volatiles and sex pheromones26,58. Our study showed that the expression patterns of several chemosensory genes in C. lividipennis had sexual differences. The genes more highly expressed in the female antennae (ClivOR6, 7 and 14) might encode proteins involved in oviposition site selection6,46. Some genes (such as ClivOBP14 and ClivOR11) were male-dominantly expressed, indicating the preferential functions in the detection and discrimination in mate searching21,59. There was no significant difference in the expression of the other genes (such as ClivOBP11 and ClivOR3), which might have more basic functions in binding general volatiles21,51.
Orco is the highly conserved olfactory co-receptor that plays important roles in OR-mediated chemosensation43. Orco has been identified in most insect species, including D. melanogaster, A. pisum and B. dorsalis41,60,61,62. In the study, the ClivOrco grouped with other Orcos, indicating that Orco was highly conserved within these hemipteran species. The orco gene does not function directly in odor recognition but rather encodes the obligate co-receptor of all ORs, which significantly impacts olfaction10. Some studies showed that the disruption of orco resulted in reductions in olfactory sensitivity in Drosophila and other insects42,43,63. It was also reported that orco mutations impaired social behavior plasticity, reproduction and development of ORNs in ants38,43. In this study, the transcripts of Clivorco were much more abundant compared to conventional ORs, which was consistent to the findings from Chrysoperla sinica36. After silencing of orco gene, around half of the treated predators showed no response to the volatiles emitted by healthy plants or infested plants. The formation of the OR-Orco dimers could be disrupted in the dsorco treated mirids, which could lead to the reduction of OR-mediated chemosensation10. Thus, many dsorco treated predators could not respond to volatiles emitted from the healthy or infested plants. In addition, dsorco treated C. lividipennis showed lower responses to the volatiles emitted by BPH-infested rice plants compared with the dsGFP treated insects, indicating the orco gene may play crucial roles in the host searching of C. lividipennis. Our study provides a foundation for further investigations into the functions of the specific chemosensory genes associated with different olfactory cues. EAG (or single sensillum recording) test of dsorco-treated and dsGFP-treated predators might provide solid conclusions about behavioral responses of the predator to plant volatiles64.
Materials and Methods
Insects rearing
The C. lividipennis individuals used in this study were originally collected from rice fields in Zi Jin Gang campus of Zhejiang University in Hangzhou, China. The laboratory colony was reared in a climate room at 26 ± 1 °C and 70% relative humidity under a photoperiod of 16:8 h light: dark for several generations. The fifth instar nymphs were kept in separate cages for eclosion. The C. lividipennis were checked daily for emergence and supplied with sufficient eggs of N. lugens.
RNA isolation and Illumina sequencing
For transcriptome analysis, approximately 300 pairs of adult antennae from each gender were individually dissected and frozen in liquid nitrogen immediately, then stored at −80 °C till to the RNA isolation. Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. RNA degradation and contamination was monitored on 1% agarose gels. The purity and concentration of RNA were measured using NanoPhotometer® spectrophotometer (IMPLEN, CA, USA) and Qubit® 2.0 Flurometer (Life Technologies, CA, USA). RNA integrity was further assessed using the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). cDNA library construction and Illumina sequencing for antennae samples were performed at Novogene (Beijing, China). A total amount of 1.5 µg RNA per sample was used, and sequencing libraries were generated using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer’s instructions. Briefly, poly-T oligo-attached magnetic beads were used to purify mRNA from total RNA. Fragmentation was carried out using divalent cations under elevated temperature in fragmentation buffer. First strand cDNA was synthesized using random hexamer primer, followed by second strand cDNA synthesis using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After end-repair and ligation of adaptors, the products were amplified by PCR and purified with AMPure XP system (Beckman Coulter, Beverly, USA). The library quality was assessed on the Agilent Bioanalyzer 2100 system. Then the two libraries created from the antennae of male and female C. lividipennis were sequenced on an Illumina Hiseq platform and paired-end reads were generated.
Transcriptome assembly and functional annotation
Raw data (raw reads) of fastq format were firstly processed through in-house perl scripts. In this step, clean data (clean reads) were obtained by removing reads containing adapter, reads containing ploy-N and low quality reads from raw data. At the same time, Q20, Q30, GC-content and sequence duplication level of the clean data were calculated. All the downstream analyses were based on clean data with high quality. The transcriptome de novo assembly was performed with Trinity (Grabherr et al., 2011) with min_kmer_cov set to 2 by default and all other parameters set default. After assembling, the unigenes were searched against protein databases, such as Nr, Swiss-Prot, KEGG, and GOG, using BLASTx with a cut-off E-value of 10−5. Gene Orthology (GO) and Cluster of Orthologous Groups (COG) were determined using Blast2GO program.
Identification of candidate genes involved in chemoreception
To identify putative OBP, CSP, NPC2, OR, IR, GR and SNMP genes, we searched the transcriptome data set with keywords (odorant-binding protein, chemosensory protein, NPC2, odorant receptor, ionotropic receptor, gustatory receptor and sensory neuron membrane protein). The open reading frames (ORFs) of each unigene was predicted by ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/). The signal peptides of candidate OBP and CSP genes were predicted using signalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/). In addition, transmembrane domains in proteins (OR and IR) were predicted using TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). To obtain a more reliable sequence, we performed PCR reaction to amplify the intact or partial sequences of each gene. Gene-specific primers were designed online by Primer3 (version 0.4.0) (http://bioinfo.ut.ee/primer3-0.4.0/) based on the transcriptome data (Supplementary Table S4). PCR products were sequenced by a commercial company (Sunny, China).
Phylogenetic analysis
The phylogenetic analysis was performed based on the amino sequences of the C. lividipennis and other insect species olfaction genes. GenBank accession numbers of genes were listed in Supplementary Table S5. The putative amino acid sequences from C. lividipennis OBPs, CSPs (without signal peptide sequences) and ORs were aligned using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). We constructed phylogenetic trees using the maximum likelihood analysis with MEGA 7 (JTT model, 1000 bootstrap replications)65.
Relative transcript accumulation of chemosensory genes in female and male antennae
To compare the expression patterns of chemosensory genes in male and female antennae of C. lividipennis, qRT-PCR was performed using RNA (3 replicates) extracted from female, male antennae and legs (male and female mixture). Legs were used as the control. PrimerScript RT reagent Kit with gDNA Eraser (Takara, Japan) was used to synthesize cDNA. All the primers used in qRT-PCR were designed online (http://bioinfo.ut.ee/primer3-0.4.0/), and sequences were listed in Supplementary Table S6. The 18S rRNA gene and ribosomal protein S15 (RPS15) were used as reference genes. SYBR Premix Ex Taq II was used in qRT-PCR according to the manufacturer’s protocol. The reaction program was (1) 95 °C, 30 s; (2) 95 °C, 5 s; (3) 60 °C, 30 s; (4) go to (2), 40 cycles in the CFX96 machine (Bio-Rad, Japan). Relative transcript accumulation of different samples were measured by the 2−ΔΔCt method66.
RNA interference (RNAi) targeting orco
We performed RNAi to explore the role of orco in the foraging behavior of the predator C. lividipennis. The MEGAscript T7 High Yield Transcription Kit (Ambion, Austin, TX, USA) was used to synthesize dsRNA. dsRNA primers (Supplementary Table S6) were designed by SnapDragon (http://www.flyrnai.org/cgi-bin/RNAi_find_primers.pl#userconsent#). Aequorea victoria green fluorescent protein (GFP) was used as the control. We injected 100 fifth instar (day 1) with about 150 ng dsRNA (dsorco and dsGFP) according a method reported in N. lugens67,68. Each treatment was replicated three times. Two days after the injections, RNA was extracted from 5 nymphs to examine the gene silencing efficiency by qRT-PCR. The remaining nymphs were used for the olfactory response experiments.
We tested the responses of C. lividipennis treated with dsorco and dsGFP to different odor sources (healthy plants and plant-BPH gravid female complex) in a two-choice H-shaped olfactometer (Fig. 6), which was similar to the method described by Khan and Saxena6,69. The plants used were 40-day-old TN1 rice seedlings. Six rice seedlings were infested by 120 gravid BPH females for 6 h before an assay. 25 fifth instar mirid nymphs (two days after injection) that had been starved for 12 h were introduced into the H-shaped olfactometer through A (1 cm diameter). Two hours later, the number of the predators in the B, C and D area of the glass tube was recorded. The predators that in the C area were regarded as no response mirids. The experiments were conducted in a separate dark room at 26 °C ± 2 °C and 70–80% relative humidity with five biological replicates.
Statistical analysis
Statistical analysis was performed using Data Processing System (DPS) software v9.570. Data was represented as mean ± SE. Means were compared using two-samples t test in choice test of C. lividipennis. Relative transcript accumulation of chemosensory genes in female and male antennae was measured by one -way analysis of variance (ANOVA) with the least significant difference (LSD).
Data availability statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
References
Frago, E. et al. Symbionts protect aphids from parasitic wasps by attenuating herbivore-induced plant volatiles. Nat. Commun. 8, 1860, https://doi.org/10.1038/s41467-017-01935-0 (2017).
Ponzio, C. et al. Volatile-mediated foraging behaviour of three parasitoid species under conditions of dual insect herbivore attack. Anim. Behav. 111, 197–206 (2016).
Heong, K. L., Bleih, S. & Lazaro, A. A. Predation of of Cyrtorhinus lividipennis Reuter on eggs of the green leafhopper and brown planthopper in rice. Res. Popul. Ecol. 32, 255–262 (1990).
Qiao, F. et al. Reciprocal intraguild predation between two mirid predators, Cyrtorhinus lividipennis and Tytthus chinensis (Hemiptera: Miridae). Biocontrol Sci. Techn. 26, 1267–1284 (2016).
Wang, G. Y. et al. Identification of candidate odorant-binding protein and chemosensory protein genes in Cyrtorhinus lividipennis (Hemiptera: Miridae), a key predator of the rice planthoppers in Asia. Environ. Entomol. 46, 654–662 (2017).
Lou, Y. G. & Cheng, J. A. Role of rice volatiles in the foraging behaviour of the predator Cyrtorhinus lividipennis for the rice brown planthopper Nilaparvata lugens. Biocontrol 48, 73–86 (2003).
Rapusas, H. R., Bottrell, D. G. & Coll, M. Intraspecific variation in chemical attraction of rice to insect predators. Biol. Control 6, 394–400 (1996).
An, X. K. et al. Identification and expression analysis of an olfactory receptor gene family in green plant bug Apolygus lucorum (Meyer-Dur). Sci. Rep. 6, 37870, https://doi.org/10.1038/srep37870 (2016).
Song, Y. Q., Sun, H. Z. & Du, J. Identification and tissue distribution of chemosensory protein and odorant binding protein genes in Tropidothorax elegans Distant (Hemiptera: Lygaeidae). Sci. Rep. 8, 7803, https://doi.org/10.1038/s41598-018-26137-6 (2018).
Zhou, Y. L. et al. Silencing in Apolygus lucorum of the olfactory coreceptor Orco gene by RNA interference induces EAG response declining to two putative semiochemicals. J. Insect Physiol. 60, 31–39 (2014).
Wang, B., Liu, Y. & Wang, G. R. Chemosensory genes in the antennal transcriptome of two syrphid species, Episyrphus balteatus and Eupeodes corollae (Diptera: Syrphidae). BMC Genomics 18, 586, https://doi.org/10.1186/s12864-017-3939-4 (2017).
Zhang, J. et al. Antennal transcriptome analysis and comparison of chemosensory gene families in two closely related noctuidae moths, Helicoverpa armigera and H. assulta. Plos One 10, e0117054, https://doi.org/10.1371/journal.pone.0117054 (2015).
Wang, J., Hu, P., Gao, P., Tao, J. & Luo, Y. Antennal transcriptome analysis and expression profiles of olfactory genes in Anoplophora chinensis. Sci. Rep. 7, 15470, https://doi.org/10.1038/s41598-017-15425-2 (2017).
Fleischer, J., Pregitzer, P., Breer, H. & Krieger, J. Access to the odor world: olfactory receptors and their role for signal transduction in insects. Cell. Mol. Life Sci. 75, 485–508 (2017).
Leal, W. S. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391 (2013).
Clyne, P. J., Warr, C. G. & Carlson, J. R. Candidate taste receptors in Drosophila. Science 287, 1830–1834 (2000).
Marchant, A. et al. Under-expression of chemosensory genes in domiciliary bugs of the chagas disease vector Triatoma brasiliensis. Plos Neglect. Trop. D. 10, e0005067, https://doi.org/10.1371/journal.pntd.0005067 (2016).
Vogt, R. G. et al. The insect SNMP gene family. Insect Biochem. Molec. 39, 448–456 (2009).
Zhu, J. et al. Niemann-Pick C2 proteins: a new function for an old family. Front. Physiol. 9, 52, https://doi.org/10.3389/fphys.2018.00052 (2018).
Pelosi, P., Zhou, J. J., Ban, L. P. & Calvello, M. Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 63, 1658–1676 (2006).
Liu, S. et al. Identification of candidate chemosensory genes in the antennal transcriptome of Tenebrio molitor (Coleoptera: Tenebrionidae). Comp. Biochem. Phys. D 13, 44–51 (2015).
Andersson, M. N. et al. Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae). BMC Genomics 14, 198, https://doi.org/10.1186/1471-2164-14-198 (2013).
Laughlin, J. D., Ha, T. S., Jones, D. N. M. & Smith, D. P. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell 133, 1255–1265 (2008).
Zhang, J. et al. Identification and expression profiles of novel odorant binding proteins and functional analysis of OBP99a in Bactrocera dorsalis. Arch. Insect Biochem. 98, e21452, https://doi.org/10.1002/arch.21452 (2018).
Xu, X. Z. et al. H-1, N-15, and C-13 chemical shift assignments of the mosquito odorant binding protein-1 (CquiOBP1) bound to the mosquito oviposition pheromone. Biomol. Nmr Assigm. 3, 195–197 (2009).
Yang, K. et al. Identification of novel odorant binding protein genes and functional characterization of OBP8 in Chilo suppressalis (Walker). Gene 591, 425–432 (2016).
Zhou, J. J. Odorant-binding proteins in insects. Vitam. Horm. 83, 241–272 (2010).
Pelosi, P., Iovinella, I., Felicioli, A. & Dani, F. R. Soluble proteins of chemical communication: an overview across arthropods. Front. Physiol. 5, 320, https://doi.org/10.3389/fphys.2014.00320 (2014).
Jin, X. et al. Expression and immunolocalisation of odorant-binding and chemosensory proteins in locusts. Cell. Mol. Life Sci. 62, 1156–1166 (2005).
Kitabayashi, A. N., Arai, T., Kubo, T. & Natori, S. Molecular cloning of cDNA forp10, a novel protein that increases in the regenerating legs of Periplaneta americana (American cockroach). Insect Biochem. Molec. 28, 785–790 (1998).
Wanner, K. W., Isman, M. B., Feng, Q., Plettner, E. & Theilmann, D. A. Developmental expression patterns of four chemosensory protein genes from the Eastern spruce budworm, Chroistoneura fumiferana. Insect Mol. Biol. 14, 289–300 (2005).
Liu, Y. L., Guo, H., Huang, L. Q., Pelosi, P. & Wang, C. Z. Unique function of a chemosensory protein in the proboscis of two Helicoverpa species. J. Exp. Biol. 217, 1821–1826 (2014).
Zhu, J. et al. Conserved chemosensory proteins in the proboscis and eyes of Lepidoptera. Int. J. Biol. Sci. 12, 1394–1404 (2016).
Pelosi, P., Iovinella, I., Zhu, J., Wang, G. & Dani, F. R. Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects. Biol. Rev. 93, 184–200 (2018).
Yang, S., Cao, D., Wang, G. & Liu, Y. Identification of genes involved in chemoreception in Plutella xyllostella by antennal transcriptome analysis. Sci. Rep. 7, 11941, https://doi.org/10.1038/s41598-017-11646-7 (2017).
Li, Z. Q. et al. Identification and expression pattern of candidate olfactory genes in Chrysoperla sinica by antennal transcriptome analysis. Comp. Biochem. Phys. D 15, 28–38 (2015).
Su, C. Y., Menuz, K. & Carlson, J. R. Olfactory perception: receptors, cells, and circuits. Cell 139, 45–59 (2009).
Trible, W. et al. orco mutagenesis causes loss of antennal lobe glomeruli and impaired social behavior in ants. Cell 170, 727–735 (2017).
Nakagawa, T., Pellegrino, M., Sato, K., Vosshall, L. B. & Touhara, K. Amino acid residues contributing to function of the heteromeric insect olfactory receptor complex. Plos One 7, e32372, https://doi.org/10.1371/journal.pone.0032372 (2012).
Sato, K. et al. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452, 1002–U1009 (2008).
Wang, Q. et al. Characterization and comparative analysis of olfactory receptor co-receptor Orco orthologs among five mirid bug species. Front. Physiol. 9, 158, https://doi.org/10.3389/fphys.2018.00158 (2018).
Li, Y. et al. CRISPR/Cas9 in locusts: Successful establishment of an olfactory deficiency line by targeting the mutagenesis of an odorant receptor co-receptor (Orco). Insect Biochem. Molec. 79, 27–35 (2016).
Yan, H. et al. An engineered orco mutation produces aberrant social behavior and defective neural development in ants. Cell 170, 736–747 (2017).
Benton, R., Vannice, K. S., Gomez-Diaz, C. & Vosshall, L. B. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162 (2009).
Abuin, L. et al. Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69, 44–60 (2011).
Wu, Z. Z. et al. Transcriptome sequencing of Tessaratoma papillosa antennae to identify and analyze expression patterns of putative olfaction genes. Sci. Rep. 7, 3070, https://doi.org/10.1038/s41598-017-03306-7 (2017).
Croset, V. et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. Plos Genet. 6, e1001064, https://doi.org/10.1371/journal.pgen.1001064 (2010).
Kwon, J. Y., Dahanukar, A., Weiss, L. A. & Carlson, J. R. The molecular basis of CO2 reception in Drosophila. P. Natl. Acad. Sci. USA 104, 3574–3578 (2007).
Benton, R., Vannice, K. S. & Vosshall, L. B. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature 450, 289–U213 (2007).
Zhou, J. J. et al. Genome annotation and comparative analyses of the odorant-binding proteins and chemosensory proteins in the pea aphid Acyrthosiphon pisum. Insect Mol. Biol. 19, 113–122 (2010).
Xue, W. et al. Identification and expression analysis of candidate odorant-binding protein and chemosensory protein genes by antennal transcriptome of Sitobion avenae. Plos One 11, e0161839, https://doi.org/10.1371/journal.pone.0161839 (2016).
Xue, J. et al. Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. Genome Biol. 15, 521, https://doi.org/10.1186/s13059-014-0521-0 (2014).
Zhou, W. W. et al. Identification and expression profiling of putative chemosensory protein genes in two rice planthoppers, Laodelphax striatellus (Fallen) and Sogatella furcifera (Horvath). J. Asia-Pac. Entomol. 18, 771–778 (2015).
Bin, S. Y., Qu, M. Q., Pu, X. H., Wu, Z. Z. & Lin, J. T. Antennal transcriptome and expression analyses of olfactory genes in the sweetpotato weevil Cylas formicarius. Sci. Rep. 7, 11073, https://doi.org/10.1038/s41598-017-11456-x (2017).
Ge, X., Zhang, T., Wang, Z., He, K. & Bai, S. Identification of putative chemosensory receptor genes from yellow peach moth Conogethes punctiferalis (Guenee) antennae transcriptome. Sci. Rep. 6, 32636, https://doi.org/10.1038/srep32636 (2016).
Zhao, Y. et al. Transcriptome and expression patterns of chemosensory genes in antennae of the parasitoid wasp Chouioia cunea. Plos One 11, e0148159, https://doi.org/10.1371/journal.pone.0148159 (2016).
Li, X. et al. Chemosensory gene families in adult antennae of Anomala corpulenta Motschulsky (Coleoptera: Scarabaeidae: Rutelinae). Plos One 10, e0121504, https://doi.org/10.1371/journal.pone.0121504 (2015).
Chang, H. et al. A pheromone antagonist regulates optimal mating time in the moth Helicoverpa armigera. Curr. Biol. 27, 1610–1615 (2017).
Liu, N. Y., He, P. & Dong, S. L. Binding properties of pheromone-binding protein 1 from the common cutworm Spodoptera litura. Comp. Biochem. Phys. B 161, 295–302 (2012).
Larsson, M. C. et al. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43, 703–714 (2004).
Zheng, W., Zhu, C., Peng, T. & Zhang, H. Odorant receptor co-receptor Orco is upregulated by methyl eugenol in male Bactrocera dorsalis (Diptera: Tephritidae). J. Insect Physiol. 58, 1122–1127 (2012).
Zhang, R. et al. Molecular basis of alarm pheromone detection in aphids. Curr. Biol. 27, 55–61 (2017).
Yang, B., Fujii, T., Ishikawa, Y. & Matsuo, T. Targeted mutagenesis of an odorant receptor co-receptor using TALEN in Ostrinia furnacalis. Insect Biochem. Molec. 70, 53–59 (2016).
Zhang, R., Gao, G. & Chen, H. Silencing of the olfactory co-receptor gene in Dendroctonus armandi leads to EAG response declining to major host volatiles. Sci. Rep. 6, 23136, https://doi.org/10.1038/srep23136 (2016).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2-approximately maximum-likelihood trees for large alignments. Plos One 5, e9490, https://doi.org/10.1371/journal.pone.0009490 (2010).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408 (2001).
Xu, H. J. et al. Genome-wide screening for components of small interfering RNA (siRNA) and micro-RNA (miRNA) pathways in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Mol. Biol. 22, 635–647 (2013).
Xu, H. J. et al. Two insulin receptors determine alternative wing morphs in planthoppers. Nature 519, 464–467 (2015).
Khan, Z. R. & Saxena, R. C. Effect of steam distillate extracts of resistant and susceptible rice cultivars on behavior of Sogatella furcifera (Homoptera, Delphacidae). J. Econ. Entomol. 79, 928–935 (1986).
Tang, Q. Y. & Zhang, C. X. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 20, 254–260 (2013).
Acknowledgements
This work was supported by the Industry Project of the Ministry of Agriculture of China (201403030), the National Natural Science Foundation of China (31371935). We thank XX Shi, P Qian, MJ Zhang, FQ Li and other lab mates for their kind cooperation during the research.
Author information
Authors and Affiliations
Contributions
Z.R.Z., W.W.Z. and G.Y.W. designed the experiments. G.Y.W. and J.L.Z. performed the experiments. G.Y.W. analyzed the sequence data. W.W.Z., S.L., Q.M.K., A.N.A. and Z.R.Z. revised the manuscript. G.Y.W. wrote the paper.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, GY., Zhu, JL., Zhou, WW. et al. Identification and expression analysis of putative chemoreception genes from Cyrtorhinus lividipennis (Hemiptera: Miridae) antennal transcriptome. Sci Rep 8, 12981 (2018). https://doi.org/10.1038/s41598-018-31294-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-31294-9
This article is cited by
-
Comparative transcriptomic analysis of salivary glands between the zoophytophagous Cyrtorhinus lividipennis and the phytozoophagous Apolygus lucorum
BMC Genomics (2024)
-
Reliable reference genes for qPCR normalization in females of the mirid predator, Cyrtorhinus lividipennis (Hemiptera: Miridae)
Biologia (2022)
-
Nitrogen fertilization of rice plants improves ecological fitness of an entomophagous predator but dampens its impact on prey, the rice brown planthopper, Nilaparvata lugens
Journal of Pest Science (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.