Abstract
Preliminary studies conducted on the human gastro-intestinal tract have revealed that enteric viral communities play a preponderant role in microbial homeostatis. However to date, such communities have never been investigated in the fish gut. Herein, we examined the main ecological traits of viruses in the digestive tract of a euryhaline fish, the tilapia Sarotherodon melanotheron. Individuals were collected at 8 different sites in Senegal covering a salinity gradient from 3 to 104‰, and showing large disparities in their organic pollutant concentrations. Results showed that the gut of S. melanotheron is home to a highly abundant viral community (0.2–10.7 × 109 viruses ml−1), distinct from the surrounding water, and essentially composed of phages of which a substantial proportion is temperate (the fraction of lysogenized cells-FLC ranging from 8.1 to 33.0%). Also, a positive and significant correlation was detected between FLC and the concentrations of polycyclic aromatic hydrocarbon in sediment, while no clear relationships were found between salinity and any of the microbial parameters considered. Finally, our data suggest that virus-bacteria interactions within the fish intestine are likely sensitive to the presence of particular xenobiotics, which may compromise the balance in the gut microbiota, and subsequently affect the health of their host.
Similar content being viewed by others
Introduction
The digestive tract of vertebrates houses an extraordinary number and variety of microorganisms including bacteria, viruses, fungi, and archaea which have co-evolved with their host, and are collectively referred to as the gut microbiota. Over the past two decades, researchers have tried to understand its implications for the host’s health, and its primary role in maintaining gut homeostasis. Commensal bacteria naturally present in the human intestinal tract are now well recognized to confer several beneficial functions to the host, not only for the digestion of food products but also for protecting from pathogenic microbial infections, and shaping the mucosal immune system1,2.
Most of the studies focusing on the gut microbiota of vertebrates have been conducted in humans, for which the medical challenge is of tremendous relevance3. However, little is known about the intestine microbiota of marine vertebrates, such as fish or mammals, despite their crucial ecological role and economical/societal values. The few information available suggests that the microbial network in fish gastrointestinal tract is very complex and probably plays a vital role in fish nutrition and disease prevention4,5. However, we still lack of clear information on how the microbial equilibrium is regulated within the intestine. Indeed, vertebrates need a stable, balanced gut bacteriome to be healthy6, and recently there has been increasing interest in understanding the factors that shape the activity and composition of enteric bacteria, as well as their subsequent role in host physiology and immunity.
Researchers discovered that viruses infecting bacteria (ie, phages) are highly abundant and diversified within the human gut, and forming key components of this specific microflora7,8,9. However to our knowledge, the occurrence, composition, and life traits of such enteric viruses have never been rarely explored in marine organisms for which they could also could play pivotal role, through lytic and/or lysogenic control of their bacterial hosts. For instance there is no information about viral abundances in the digestive tract of fishes, their infection strategies, nor their susceptibilty to ambient environmental conditions (salinity, temperature, xenobiotics, etc.). The only information available refers to the diversity of culturable phages in freshwater fishes (He & Yang 2015), which only represent a small fraction of the whole gastrointestinal community of viruses10.
In the present study, we examined the main ecological features of viruses present in the gut of the black-chinned tilapia (Sarotherodon melanotheron). This fish species was selected as a potential sentinel because of its abundance, physiological properties, and wide distribution in tropical coastal ecosystems. Indeed, this extreme euryhaline species, which is endemic to West Africa, is able to live in all types of water bodies, from freshwater to marine and even hypersaline waters11.
Individuals were collected from various sites in Senegal covering a salinity range from 3 to 104‰, and exhibiting contrasted concentrations of xenobiotics. This sampling strategy allowed us to (i) acquire basic information about the abundance, life strategies and morphological composition of viral communities found in the fish gut, and (ii) explore whether such viral communities are affected by salinity and/or the presence of persistent pollutants in their environment.
Material and Methods
Sampling sites
The study was conducted in Senegal (West Africa) during the dry season (May 2010) at 8 contrasted stations, namely Niayes 1 (NE1), Niayes 2 (NE2), Koular (KUE), Kaoloack (KOE), Koïlal (KOI), Hann Bay (BHE), Foundiougne (FE), and Missirah (ME) (Fig. 1). These sites are located in the Dakar peninsula and along the estuary of Sine-Saloum, covering a salinity gradient from 3 to 104‰ (see Table 1).
At each station, water salinity was measured with a refractometer. Surface sediment samples were also collected using a sediment corer (core tube 70 mm inner diameter; maximum sediment penetration depth of 6 cm), which allows collecting samples hermetically sealed. Visual inspection of the overlying waters and sediment surfaces revealed very limited resuspension effects during sampling. Sediment samples were then stored in aluminum containers and frozen at −20 °C for further chemical analysis of persistent organic pollutants such as polychlorinated biphenyls (PCB), organochlorine pesticides (OCP), and polycyclic aromatic hydrocarbons (PAH)12.
Extraction of fish gut
At each sampling site, three adult tilapia Sarotherodon melanotheron heudelotii of similar size (130–150 mm fork length) were caught using a cast net and were anaesthetized progressively in icy water. Fish were then killed by cervical dislocation (following the European directive 2010/63/UE), as there is no regulation or guidelines in Senegal where experiments were carried out. Captured fish were individually placed in plastic bags and immediately stored in ice before dissection (within 5-h after sampling)13. Briefly, the digestive tract was extracted from each individual and cut from below the stomach to the rectum. Each gut was squeezed to expel content, which represented a minimum volume of 3 ml.
At each stations, triplicate raw gut contents (RGC) were processed for the different analyses as follows: (i) 1 ml was immediately processed for estimating the proportion of bacterial cells under lysogenic infections, and (ii) 2–3 ml were fixed with formaldehyde (final concentration f.c. 3% v/v), flash-frozen in liquid nitrogen and stored at −80 °C until measurement of viral and bacterial abundances, and distribution of the viral morphotypes. Triplicate subsurface water samples (20 ml) were also collected at each station, fixed with formaldehyde (f.c. 2% v/v) and stored at −80 °C in liquid nitrogen for determination of the parameters described above.
Enumeration of viruses and prokaryotes
For each replicate, 1 mL of the fixed RGC was suspended in 1 mL of a 9‰ NaCl sterile saline solution complemented with 0.2 mL pyrophosphate (0.1 M) to extract bacterial and viral particles from gut matrix13. After vortexing for 2 min at room temperature, the supernatant was immediately transferred into a sterile tube containing 10 mL of 9‰ NaCl. The number of viruses and prokaryotes contained in each replicate samples was determined after retention of the particles on 0.02 µm pore-size membranes (Anodisc) and staining with SYBR Gold14. On each slide, 300–500 bacteria and viruses per replicate were counted with an Olympus Provis-AX70 epifluorescence microscope, under blue light excitation (488 nm). For planktonic viruses and bacteria, since the extraction procedure was not necessary, we only applied the staining protocol, as described above, by using a volume of 300–500 µL of seawater.
Fraction of lysogenic bacteria
We used the method of Jiang and Paul15 to initiate prophage induction in gut and planktonic bacteria. Mitomycin C (final concentration: 1 mg mL−1, Sigma-Aldrich) was added to triplicate 1-mL volumes of unfixed and fresh RGC and water. Triplicate untreated samples served as control. All sub-samples were formalin fixed (f.c., 3% [vol/vol]) after being incubated for 12 h in the dark, at in situ temperatures (eg between 25.3–29.3 °C). Prophage induction was calculated as the difference in viral abundance (epifluorescence counts, see above) between mitomycin C-treated (Vm) and control incubations (Vc). The fraction of lysogenic bacterial cells (FLC) was calculated as follows:
where BS is the burst size (number of viruses per bacterial cell) and BAt0 is the bacterial abundance at the start of the experiment, i.e., before adding mitomycin C16. Since no burst size from infected gut bacteria has been published so far, we used a burst size of 24, which is an average value calculated from the range for different studied marine environments17.
Examination of viral morphotypes
The distribution of the different viral morphotypes in gut and water samples was examined using transmission electron microscopy (TEM)18. Viruses from 500 µL aliquots of formalin fixed and pyrophospate-treated samples (see above) were harvested by repeated ultracentrifugation of 50 µL onto grids (400 mesh Cu electron microscope grids with carbon coated Formvar film) using an A-100/30 rotor in an air-driven ultracentrifuge (Airfuge®, Beckman) at 105,000 × g for 70 min. The grids were then stained for 30 s with uranyl acetate (2%, w/w), and viruses were examined and measured using a JEOL 1200EX TEM operated at 80 kV and magnification from x20,000 to x100,000. Between 300 and 400 viruses were examined per grid. Three morphotypes were distinguished for shape classification of tailed viruses (Caudovirales). Tailed viruses with isometric heads and long non-contractile tails were considered to belong to the family Siphoviridae. Tailed viruses with isometric heads and contractile tails (presence of a neck) to the family Myoviridae. Tailed viruses with isometric heads and short tails to the Podoviridae. The proportion of tailless icosahedral viruses was also evaluated.
Data analysis
Data were log transformed to satisfy the requirements of normality and homogeneity of variance necessary for parametric analyses. Simple relationships between original data sets were tested using Pearson correlation analysis. Statistical comparisons of mean values were performed using one-way Anova and Tukey post hoc tests. All statistical analyses were performed using SIGMASTAT software.
Results and Discussion
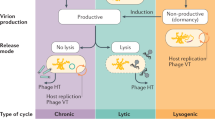
The digestive tract of S. melanotheron harbored a highly concentrated community of viruses and bacteria, ranging respectively from 0.2 to 10.7 × 109 viruses ml−1, and from 0.1 to 6.8 × 109 bacterial cells ml−1 of gut content (Table 2). This is to our knowledge, the first report of viral abundances in the fish gut. Such abundances fall within the same order of magnitude of those commonly found in human gastrointestinal tracts (e.g., around 109 viruses g−1)8,19, but are much greater than those reported in natural environments such as marine or even soil ecosystems, which rarely surpass 109 viruses per milliliter of water or gram of soil20,21. Whilst the main information available on gut-associated viruses refers to those examined in humans9,22,23, the positive and highly significant correlation between viral and bacterial concentrations suggested that the fish gut viral communities were dominated by phages (Pearson correlation r = 0.90, p < 0.05, Table 3). This was also corroborated by the important proportion of tailed viruses of the Caudovirales families (Sipho-, Myo- and Podoviridae), which was significantly higher in the fish gut than in the surrounding water (Table 3). Given that the Firmicutes, Proteobacteria and Bacteroidetes typically represent up to 90% of the fish intestinal microbiota4, one may suspect that these abundant tailed and temperate phages are specific of these particular bacterial groups. Interestingly, similar bacteria prevalence were reported in the human gastrointestinal tract1,24 strongly supporting the paradigm of a core gut microbiome comprised of common bacterial members ensuring the primary functions of this vital organ (e.g., defense against pathogen, etc.). By analogy, one may also suspect the existence of a core viral diversity in vertebrate gut. The rest of the microbial community being rather dedicated to the specific digestion processes and diet, which are highly species-specific.
The mitomycin-C inductions revealed a high proportion of lysogenic cells in the fish gut (min-max, 8.1–33.0%), which was significantly higher than that was found in their planktonic counterparts (min-max, 0.3–8.8%) (Table 2). However, incubations were performed ex vivo and therefore the results should be taken with caution. At first sight, such results are surprising as lysogeny is usually considered as a refuge strategy for viruses in habitats where the conditions for their reproduction is threatened by either low abundance and/or compromised host metabolism25,26. Yet, the gut of S. melanotheron contained a dense population of bacteria (eg, 0.1–6.8 × 109 cells ml−1), resulting in a rather low virus-to-bacteria ratio (VBR) (mgut = 1.5 ± 0.8) compared to that measured in the ambient water (meanwat = 3.9 ± 1.4) (Table 2). These results clearly contrast with the refuge theory that predicts rather low prevalence of lysogeny in highly ‘bacterialized’ habitats, like the fish gut. Interestingly, such high proportions of lysogens were also found in the coral mucus which is a nutrient-rich medium, and which typically hosts an abundant community of bacteria27,28. Recently a new model, the piggyback-the-winner model postulates that lysogeny can dominate in certain biomes hosting abundant and fast growing bacteria29. This may be the case in animal microbiomes where commensal bacteria are abundant, functionally crucial for their host, and to which lysogeny could represent an advantageous strategy, providing cells immunity against viral surinfections together with acquisition of genes allowing optimal survival in stressing conditions30,31. Temperate phages have also been long shown to prevail in the human gut9,32, suggesting that the digestive tract may harbor a cosmopolitan lysogenic phageome capable of controlling the bacterial homeostastis in vertebrates at large. The metagenomic research of prophages sequences in a large variety of terrestrial and marine vertebrates will be necessary to further explore this sensitive question.
We did not observe any significant correlation between salinity and any of the studied microbial parameters. Overall, given the euryhaline nature of S. melanotheron heudelotii12 and because the fish intestine is a major osmoregulatory organ, it was not surprising to see that gut-associated bacterial and viral communities were not impacted by external osmolarity.
One of the most striking findings here was the positive correlation between the fraction of lysogenized cells and the concentrations of polycyclic aromatic hydrocarbons (PAH). We know that the gut bacteriome of fishes (and of human) is affected by external environmental conditions33, including the presence of xenobiotics34,35. However the potential relationships between gut-associated viruses and environmental pollutants have only received little attention so far. PAHs are ubiquitous environmental pollutants, generated primarily during the incomplete combustion of organic materials (e.g. coal, oil, petrol, and wood), which exhibit toxic, mutagenic and/or carcinogenic properties to many aquatic organisms36,37. Previous reports have shown that PAHs can accumulate into the sediment38, and since they are highly lipid soluble, they can be readily absorbed from the intestinal tract of marine vertebrates39. Herein, the positive relationship between the PAH concentrations and FLC may indicate that phage-bacteria interactions within the fish gut are also strongly susceptible to this specific persistent pollutants, contrary the other POPs investigated here (e.g. PCB and OCP). Since the lysis-lysogenic decision usually depends upon host cell metabolism40, one may suspect that the physiological stress caused by the presence of PAH could have triggered lysogenic pathways. This may impact the fragile balance between temperate and virulent phages within the gut microbiota, with potentially severe implications for fish health.
References
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Sommer, F. & Backhed, F. The gut microbiota - masters of host development and physiology. Nature Review Microbiology 11, 227–238 (2013).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Ghanbari, M., Kneifel, W. & Domig, K. J. A new view of the fish gut microbiome: Advances from next-generation sequencing. Aquaculture 448, 464–475 (2015).
Escalas, A. et al. Functional diversity and redundancy across fish gut, sediment and water bacterial communities. Environmental Microbiology 19, 3268–3282 (2017).
Maurice, C. F., Haiser, H. J. & Turnbaugh, P. J. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152, 39–50 (2013).
Reyes, A., Semenkovich, N. P., Whiteson, K., Rohwer, F. & Gordon, J. I. Going viral: next-generation sequencing applied to phage populations in the human gut. Nature Review Microbiology 10, 607–617 (2012).
Ogilvie, L. A. & Jones, B. V. The human gut virome: a multifaceted majority. Frontiers in Microbiology 6, 918 (2015).
Mirzaei, M. K. & Maurice, C. F. Ménage à trois in the human gut: interactions between host, bacteria and phages. Nature Review Microbiology 15, 397–408 (2017).
He, Y. & Yang, H. The gastrointestinal phage communities of the cultivated freshwater fishes. FEMS Microbiology Letters 362, fnu027 (2015).
Panfili, J. et al. Influence of salinity on the life-history traits of the West African black-chinned tilapia (Sarotherodon melanotheron): comparison between the Gambia and Saloum estuaries. Aquatic Living Resources 17, 65–74 (2004).
Ndiaye, A. et al. Multiparametric approach for assessing environmental quality variations in West African aquatic ecosystems using the black-chinned tilapia (Sarotherodon melanotheron) as a sentinel species. Environment Science and Pollution Research 19, 4133–4147 (2012).
Mouchet, M. A. et al. Genetic difference but functional similarity among fish gut bacterial communities through molecular and biochemical fingerprints. FEMS Microbiology Ecology 79, 568–80 (2012).
Patel, A. et al. Virus and prokaryote enumeration from planktonic aquatic environments by epifluorescence microscopy with SYBR Green I. Nature Protocols 2, 269–276 (2007).
Jiang, S. C. & Paul, J. H. Occurrence of lysogenic bacteria in marine microbial communities as determined by prophage induction. Marine Ecology Progress Series 142, 27–38 (1996).
Bettarel, Y. et al. Corallivory and the microbial debacle in two branching scleratinians. The ISME Journal (in press, https://doi.org/10.1038/s41396-017-0033-5) (2018).
Parada, V., Herndl, G. J. & Weinbauer, M. G. Viral burst size of heterotrophic prokaryotes in aquatic systems. Journal of Marine Biology Association UK 86, 613–621 (2006).
Bettarel, Y. et al. Viral distribution and life strategies in the tropical Bach Dang estuary (Vietnam). Microbial Ecology 62, 143–154 (2011).
Kim, M. S., Park, E. J., Roh, S. W. & Bae, J. W. Diversity and abundance of single-stranded DNA viruses in human feces. Applied and Environmental Microbiology 77, 8062–8070 (2011).
Suttle, C. A. Marine viruses-major players in the global ecosystem. Nature Review Microbiology 5, 801–812 (2007).
Reavy, B., Swanson, M. M. & Taliansky, M. Viruses in Soil. In: Dighton J., Krumins J. (eds) Interactions in Soil: Promoting Plant Growth. Biodiversity, Community and Ecosystems, vol 1. Springer, Dordrecht (2014).
Minot, S. et al. Rapid evolution of the human gut virome. Proceedings of the National Academy of Sciences USA 110, 12450–12455 (2013).
Norman, J. M. et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160, 447–460 (2015).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Paul, J. H. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? The ISME Journal 2, 579–589 (2008).
Howard-Varona, C., Hargreaves, K. R., Abedon, S. T. & Sullivan, M. B. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. The ISME Journal 11, 1511–1520 (2017).
Garren, M. & Azam, F. New method for counting bacteria associated with coral mucus. Applied and Environmental Microbiology 76, 6128–6133 (2010).
Nguyen-Kim, H. et al. Coral mucus is a hot spot for viral infections. Applied and Environmental Microbiology 81, 5773–5783 (2015).
Knowles, B. et al. Lytic to temperate switching of viral communities. Nature 531, 466–470 (2016).
Bettarel, Y., Bouvier, T., Nguyen-Kim, H. & Pham, T. T. The versatile nature of coral-associated viruses. Environmental Microbiology 17, 3433–3439 (2015).
Silveira, C. B. & Rohwer, F. Piggyback-the-Winner in host-associated microbial communities. NPJ Biofilms Microbiomes 2, 16010 (2016).
Manrique, P. et al. Healthy human gut phageome. Proceedings of the National Academy of Sciences USA 113, 10400–10405 (2016).
Barnerjee, G. & Ray, A. K. Bacterial symbiosis in the fish gut and its role in health and metabolism. Symbiosis 72, 1–11 (2017).
Taguer, M. & Maurice, C. F. The complex interplay of diet, xenobiotics, and microbial metabolism in the gut: implications for clinical outcomes. Clinical Pharmacology and Therapeutics 99, 588–599 (2016).
Claus, S. P., Guillou, H. & Ellero-Simatos, S. The gut microbiota: a major player in the toxicity of environmental pollutants? NPJ Biofilms Microbiomes 2, 16003 (2016).
Menzie, C. A., Potocki, B. B. & Santodonato, J. Exposure to carcinogenic PAHs in the environment. Environmental Science and Technology 26, 1278 (1992).
Abdel Shafi, H. I. & Mansour, M. S. M. A review on polycyclic aromatic hydrocarbons: Source, environmental impact, effect on human health and remediation. Egyptian Journal of Petroleum 25, 107–123 (2016).
Tavakoly Sany, S. B. et al. Polycyclic aromatic hydrocarbons in coastal sediment of Klang Strait, Malaysia: distribution pattern, risk assessment and sources. PLoS ONE 9, e94907 (2014).
Beyer, J., Jonsson, G., Porte, C., Krahn, M. M. & Ariese, F. Analytical methods for determining metabolites of polycyclic aromatic hydrocarbon (PAH) pollutants in fish bile: A review. Environmental Toxicology and Pharmacology 30, 224–244 (2010).
Erez, Z. et al. Communication between viruses guides lysis–lysogeny decisions. Nature 451, 488–493 (2017).
Acknowledgements
This research was supported by the ECLIPSE Project (EC2CO INSU, France), the IRD DPF, and the Montpellier University of Excellence (MUSE) program. We wish to thank Khady Diop (IRD Dakar), Waly Ndiaye (PhD student, UCAD), Corinne Bouvier (CNRS Montpellier), and the fishermen for their technical help and assistance during sampling.
Author information
Authors and Affiliations
Contributions
J.D.D. conceived the study. J.D.D., J.P., Y.B., T.B. and A.N. collected samples. M.C. and A.A. performed epifluorescence and transmission electron microscopy analyses. Y.B. drafted the manuscript, and all the authors reviewed the final version.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bettarel, Y., Combe, M., Adingra, A. et al. Hordes of Phages in the Gut of the Tilapia Sarotherodon melanotheron. Sci Rep 8, 11311 (2018). https://doi.org/10.1038/s41598-018-29643-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-29643-9
This article is cited by
-
Bacteriophage-based techniques for elucidating the function of zebrafish gut microbiota
Applied Microbiology and Biotechnology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.