Abstract
Lasso peptides are ribosomally synthesized and post-translationally modified peptides produced by bacteria. They are characterized by an unusual lariat-knot structure. Targeted genome scanning revealed a wide diversity of lasso peptides encoded in actinobacterial genomes, but cloning and heterologous expression of these clusters turned out to be problematic. To circumvent this, we developed an orthogonal expression system for heterologous production of actinobacterial lasso peptides in Streptomyces hosts based on a newly-identified regulatory circuit from Actinoalloteichus fjordicus. Six lasso peptide gene clusters, mainly originating from marine Actinobacteria, were chosen for proof-of-concept studies. By varying the Streptomyces expression hosts and a small set of culture conditions, three new lasso peptides were successfully produced and characterized by tandem MS. The newly developed expression system thus sets the stage to uncover and bioengineer the chemo-diversity of actinobacterial lasso peptides. Moreover, our data provide some considerations for future bioprospecting efforts for such peptides.
Similar content being viewed by others
Introduction
Microbial natural products have been the major source of human medicines and agrochemicals for hundreds of years. Recent advances in DNA sequencing and genomics revealed that microorganisms harbour a wealth of natural product biosynthetic gene clusters, far exceeding the number of known molecules identified via bioactivity-guided screenings. This discrepancy is attributed to the fact that a large number of such gene clusters are not expressed or yield low amounts of products that elude detection under standard laboratory conditions. To enable genomics-based discovery of new natural products, numerous strategies have been developed to activate otherwise cryptic gene clusters, which can be detected and classified using recently developed bioinformatics tools and databases1,2,3. One important approach is to express the pathways in heterologous hosts in combination with cluster engineering or co-expression of a specific regulator4.
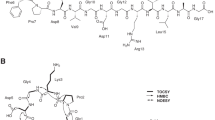
Lasso peptides are an emerging family of ribosomally synthesized and post-translationally modified peptides (RiPPs) produced by bacteria5. Composed of 15 to 24 amino acids, they are characterized by an interlocked structure formed by an N-terminal macrolactam ring and a C-terminal tail6,7,8. The 7- to 9-residue macrolactam ring is closed between the amino group of the first residue (Gly, Cys, rarely Ser or Ala) and the carboxyl side chain of a Glu or Asp. The tail threads through the ring; unthreading is prevented by steric hindrance from bulky residues and, in some cases, by additional disulphide linkages (Fig. 1). Lasso peptides are grouped into four classes: those without disulphide bonds form the dominant type II group, whereas the type I and III groups, defined by the presence of two or one disulphide bonds, respectively, comprise very few representatives (two for each type). They display interesting biological activities, such as enzyme inhibition and receptor antagonism, reflecting an advantageous binding of the constrained lasso scaffold to protein targets. Most of the bioactive lasso peptides are isolated from Actinobacteria7,9,10,11,12,13,14,15,16. Those displaying antibacterial activities are narrow-spectrum and frequently possess novel mode of action, including siamycins that attenuate the expression of virulence factors in Enterococcus faecalis by inhibiting a quorum sensing-related histidine kinase17,18, lassomycin that displays anti-mycobacterial activity via inhibition of the Clp protease11 and streptomonomicin, which potently inhibits the growth of Bacillus anthracis by targeting a vital two-component system12. These properties thus motivate the search for new lasso peptides from Actinobacteria19. In this regard, bioprospecting of untapped actinobacterial sources such as those from exotic environments seemed particularly promising in affording bioactive lasso peptides13,14.
Genomics-guided approaches greatly accelerated lasso peptides discovery in the recent years. Core lasso peptide biosynthetic genes encode a precursor peptide (A) that encompasses an N-terminal leader and a structural sequence, a cysteine protease (B), a macrolactam synthetase (C) and a RiPP recognition element (E) that binds to the leader sequence20. Many clusters alternatively encode fused E-B proteins. Precursor A-21 or enzyme B-focused mining19,22 of bacterial genomes as well as mass spectrometry-based peptidogenomics23 revealed a wide distribution of lasso peptide gene clusters in diverse phyla. Subsequently, a substantial amount of efforts has been made to explore the production potential of Proteobacteria, yielding more than 25 new lasso molecules since 2008.22 With the exception of microcin J25 (an archetype of lasso peptides produced by Escherichia coli), all proteobacterial lasso peptides were produced by expression of the corresponding gene clusters in E. coli. Heterologous production of lasso peptides is a preferred strategy, owing to the small size of their gene clusters that is amenable to manipulation and that these genes are frequently silent in the native strains under laboratory conditions. Several expression plasmids were developed to allow facile cloning of these clusters, in which transcription of core biosynthetic genes was under the control of an inducible promoter. In combination with genetic engineering and culture optimization, this strategy permitted the production of selected proteobacterial lasso peptides in E. coli at high yields24.
Exploration of Actinobacteria for bioactive lasso peptides using genomics-guided approach is emerging, with relatively few examples14,19,25,26,27. Among them, sviceucin is the only case in which high level production was achieved by expressing the cosmid-borne, native cluster in Streptomyces coelicolor26. Our previous work on other actinobacterial lasso peptides showed that simply transferring the native cluster to S. coelicolor did not reproducibly lead to peptide production7. There is a lack of a general strategy for heterologous expression of actinobacterial lasso peptides, in contrast to those originating from Proteobacteria. We report here an effective orthogonal expression system designed to circumvent toxicity problems encountered during cloning of actinobacetrial lasso peptides gene clusters in E. coli, and thus to produce new molecules in heterologous Streptomyces hosts. Our results also provide some guidelines in future bioprospecting efforts in Actinobacteria for lasso peptides.
Results
Identification of new lasso peptide gene clusters in the genomes of several actinomycetes
Recently-acquired genome sequences from several both known and newly isolated Actinobacteria were mined for putative lasso peptide biosynthetic gene clusters. In particular, draft genomes of the following strains were investigated: Streptomyces spp. ADI 94-01 and ADI 98-10 (isolated from the marine sponge Antho dichothoma, unpublished data), Actinoalloteichus fjordicus ADI 127-17 (DSM 46855 T)28, Streptomyces noursei ATCC 11455, and Streptomyces venezuelae ATCC 10712. Using a combination of antiSMASH 3.0429 and manual searches, lasso peptides gene clusters were identified in all genomes (ADI 94-01 contained two such clusters). They showed the conserved gene organization following the order A-C-E-B and some contained additional genes involved in the transport, regulation or putative further modifications (Fig. 2). For example, the Sven-LP gene cluster from S. venezuelae contained a gene encoding an acetyltransferase. The A127-LP gene cluster from A. fjordicus harboured genes encoding an oxidoreductase as well as a “Streptomyces antibiotic regulatory protein” (SARP) (Fig. 2). The primary sequences of the predicted lasso peptides were quite different and diverse, thus representing a considerable chemo-diversity (Table 1). Moreover, they appeared to display new features, including non-canonical residues involved in the macrolactam linkage (the first residue being Ala or Tyr instead of Gly or Cys commonly found in lasso peptides), the presence of one disulphide bond (rare type III lasso peptides) and post-translational modifications decorating the lasso scaffold (Table 1).
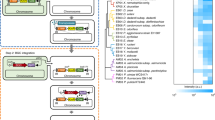
Gene organization of the selected lasso peptide clusters. The core biosynthetic genes (A/C/E/B) are labelled. The SARP and its cognate promoter in the A127-LP cluster used in this study are illustrated. SARP: Streptomyces antibiotic regulatory protein; RRE: RiPP recognition element. A127-LP is from Actinoalloteichus fjordicus ADI127-17; 9401-LP1 and 9401-LP2 are from Streptomyces sp. ADI94-01; 9810-LP is from Streptomyces sp. ADI98-10; Snou-LP is from Streptomyces noursei ATCC 11455; Sven-LP is from Streptomyces venezuelae ATCC 10712.
Construction of an orthogonal SARP-based expression system for lasso peptides
In an attempt to generate constructs for heterologous expression, the selected lasso peptide gene clusters were PCR-amplified from the respective genomic DNAs and subjected to Gibson assembly with the pSOK806 vector30, placing them under control of a strong constitutive promoter ermEp*. However, the analysis of recombinant constructs for all the clusters revealed multiple deletions. This suggests a potentially toxic effect of the lasso peptide products arising from gene expression in the cloning host, E. coli. These cloning attempts were repeated several times, with the same outcome. In order to determine whether the ermEp* promoter is active in E. coli, the plasmid pSOK80830 carrying an ermEp* promoter::glucuronidase-encoding gene (gusA) fusion was introduced into this bacterium, and the GusA activity was measured. A considerable GusA activity could be observed in the crude extracts of E. coli DH5a carrying pSOK808 (data not shown), confirming that the ermEp* promoter is indeed functional in E. coli.
To prevent expression of the selected lasso peptide gene clusters in E. coli, an alternative orthogonal two-plasmid system was constructed. We reasoned that promoters from a rare actinobacterium, such as Actinoalloteichus sp., will unlikely be recognized by the E. coli sigma factors. The A127-LP cluster from A. fjordicus encodes a SARP regulator immediately upstream of the precursor gene A (Fig. 2). It was thus postulated that this SARP controls the expression of A127-LP biosynthetic genes. Several SARPs and their promoters have been characterized in Streptomyces31,32. Currently it is still impossible to identity the SARP promoter consensus by in silico analysis, especially in an understudied genus such as Actionalloteichus. Therefore, the putative promoter region upstream of the precursor gene A was chosen for the experiment. We replaced the ermEp* promoter in pSOK808 with the above-mentioned A127-LPp promoter and tested the resulting construct, designated pSOK809, in E. coli for expression of GusA. As expected, almost no GusA activity was detected.
Next, we introduced the latter plasmid into S. venezuelae, where rather moderate (ca. 15 times lower compared to ermEp*-based pSOK808) level of GusA activity was observed (data not shown). In order to test our hypothesis about SARP-dependent upregulation of the A127-LPp promoter, a multi-copy plasmid pSARP carrying the A. fjordicus SARP-encoding gene under control of ermEp* was constructed and introduced into the S. venezuelae strain carrying pSOK809. Comparative analysis of the GusA activity in the S. venezuelae (pSOK809) strains with or without pSARP revealed ca. 5 fold higher expression of the gusA gene in the latter. Taken together, these results strongly suggested that the developed two-plasmid system (Fig. 3) can be useful for assembly and heterologous expression of lasso peptide biosynthetic gene clusters.
Heterologous production of lasso peptides in Streptomyces
Indeed, selected lasso peptide gene clusters were successfully assembled in E. coli under control of the A127-LPp promoter using Gibson ligation and pSOK809-based template. The resulting constructs were integrated into the genomes of various heterologous Streptomyces hosts via the phage VWB attachment site of pSOK809. Subsequently, the pSARP plasmid was introduced into these recombinant strains. We mainly used Streptomyces albus J1074 and Streptomyces lividans TK24 as hosts, unless otherwise stated. It was observed that conjugation efficiency for lasso peptide expression plasmids was generally low, implying a potential toxicity problem in Streptomyces even at the background expression level without SARP. In particular, no exoconjugants could be obtained for the Sven-LP cluster in both above-mentioned strains, as well as in S. coelicolor M1146. Conjugation of pA127-LP and pSnou-LP into S. albus also failed. On the contrary, pA127-LP could be introduced into S. coelicolor.
For lasso peptide production, recombinant Streptomyces strains were grown in four commonly-used rich media and one phosphate-limiting defined medium33 in liquid cultures. The latter choice was based on the observation that production of some antimicrobial peptides was triggered by phosphate starvation, e.g. in the case of microcin J2534. Liquid chromatography coupled to mass spectrometry (LC-MS) analyses of cell extracts indicated that the best culture condition was MYM or MYM-g medium, under which the expected masses of three peptides (9401-LP1, 9810-LP and Snou-LP) were detected in respective extracts, albeit in trace amount (Table 2).
In parallel, seven solid media were tested for lasso peptide production (Table 2). Three out of five expected peptides, 9401-LP1, 9810-LP and Snou-LP, were detected in extracts by LC-MS. They were produced in two or more culture conditions used for the respective strains. The best production medium was specific for each lasso peptide, and media preference did not show a general pattern. No significant differences among expression hosts for either peptide were noticed. It nevertheless appeared, that S. albus is a slightly better host (Table 2). On the contrary, the peptides A127-LP and 9401-LP2 could not be detected under any tested conditions. The scale-up production and purification was performed for Snou-LP. A yield of 0.8 mg/L solid SFM culture was obtained, a good starting point for further improvement of production.
Comprehensive characterisation of new lasso peptides by mass spectrometry
The lasso peptides 9401-LP1, 9810-LP and Snou-LP were eluted at retention times of 8.9, 4.5 and 5.4 min under used LC conditions, respectively (Figs 4–6). The measured m/z matched with calculated m/z with less than 2 ppm error (Table S4) and the MS/MS spectra matched with the peptide sequences. The MS/MS spectrum of 9401-LP1 revealed mostly fragmentation in the tail regions, yielding b- and y- product ions covalently attached through the disulphide bridge, together with their complementary internal product ions. Moreover, reduction of 9401-LP1 with DTT yielded a mass increase of 2 Da, confirming the presence of one disulphide bond (Figure S1). The peptide 9810-LP appeared more recalcitrant to fragmentation given its high number of basic residues (2 Arg and 1 Lys in the tail and macrolactam ring, respectively). The most abundant fragment ions were b15 and y7, observed together with product ions resulting from cleavages in the macrolactam ring. Finally, the MS/MS spectrum of Snou-LP appeared more complex, showing many complementary b- and y-type product ions generated by cleavages within the tail region, together with a few [2]rotaxane product ions diagnostic of the lasso structure35. The fact that only Snou-LP displayed a MS/MS spectrum characteristic of the lasso topology is not surprising when considering the general tendencies reported on a collection of lasso peptides36. For 9401-LP1, the disulphide bridge precludes the formation of the diagnostic [2]rotaxane product ions, while for 9810-LP, these ions may be disfavored due to the short loop region.
The lasso topology was further characterized by carboxypeptidase Y treatment. This exoprotease can cleave branched-cyclic peptides (i.e. non-lasso topoisomers) from the C-terminus up to the amino acid immediately next to the macrolactam ring (see positive control in Figure S2), while lasso peptides are resistant to proteolysis or are hydrolyzed up to the bulky amino acid acting as a plug maintaining the lasso topology37. The type III lasso peptide 9401-LP1 was resistant to proteolysis in non-reducing conditions, while it yielded two degradation products upon reduction: [A1-I17] and [A1-W10] (Figure S3). This suggests that in this peptide, the lasso structure is maintained by two factors: the disulphide bridge and a bulky amino acid located at the vicinity of I17 (most probably W16). When the disulphide bridge is reduced, 9401-LP1 appears to become partly unthreaded, thus generating [A1-W10] in the presence of carboxypeptidase Y, while a portion of the peptide still retains the lasso topology, generating [A1-I17]. Peptide 9810-LP yielded several hydrolysis products, namely [G1-I15], [G1-R14] and [G1-Y10] (Figure S4). This suggests that the peptide displays a lasso structure stabilized by amino acids R13-R14 (yielding [G1-I15] and [G1-R14]), but can partially unthread during incubation (yielding [G1-Y10]). Finally, Snou-LP was found to be almost completely resistant to carboxypeptidase Y hydrolysis (Figure S5). Only a minor degradation product [Y1-N9] was observed, suggesting a partial structure unthreading.
The lasso structure of peptides 9401-LP1 and Snou-LP was further confirmed by ion mobility mass spectrometry (IM-MS). This method permits differentiation of lasso peptides from their non-lasso topoisomers based on their difference in collision cross section (CCS, named Ω), especially at high charge states. It has revealed several indicators of the lasso structure, such as the ratio ΔΩ/Ω that measures the relative range of CCS covered by all charge states and the mean charge divided by the molecular mass (named ζ)38,39. The IM-MS experiments were carried out in the presence of the sulfolane supercharging agent, in order to favor high charge states. The indicators ΔΩ/Ω and ζ were calculated for 9401-LP1 and Snou-LP (Table S5), and compared with the values obtained for a collection of non-lasso and lasso peptides (including type I, type II and type III representatives). These two indicators were typical of those of lasso peptides for both 9401-LP1 and Snou-LP (Figs S6 and S7).
Antibacterial activity
None of the new lasso peptides showed activity against tested Gram-negative (E. coli, Salmonella enterica, Pseudomonas aeruginosa) and Gram-positive bacteria (Bacillus megaterium, Micrococcus luteus and Staphylococcus aureus). This is consistent with previous reports11,12,19, indicating that lasso peptides usually have a very narrow spectrum of activity and hence their target bacteria are difficult to identify.
Attempts to improve production level by genetic engineering
Although the production of the studied peptides was observed, the yields were unsatisfying. It has been observed that many lasso peptide gene clusters harbour an inverted repeat motif in the intergenic region between the precursor gene A and downstream biosynthetic genes40. This terminator-like motif may play a regulatory role in gene transcription. Indeed, modification of this region by an optimized RBS or promoter significantly increased production titers of proteobacterial lasso peptides in E. coli24. Consequently, we employed the same strategy in an attempt to improve the yield of 9401-LP1 as a proof of principle. The 216-bp intergenic region in the 9401-LP1 expression plasmid was replaced by a Kan-ermEp* cassette by PCR targeting41 so that the transcription of downstream genes was driven by an independent ermEp* promoter (Figure S8). This modification led to a 2–4 fold increase of 9401-LP1 production in S. albus, curiously without the SARP regulator (Figure S9). Results for S. lividans strains were highly variable and did not give a clear tendency.
Discussion
Here we report the development of a heterologous expression strategy that allowed the production of three new lasso peptides with a relatively small effort in culture screening. To our knowledge, Snou-LP is the first lasso peptide to have a bulky residue as the first residue (Tyr) and 9401-LP1 represents the third example of the type III group. To date, regulatory mechanisms and factors that trigger lasso peptide biosynthesis in the native producers remain largely unknown. The use of the SARP-responsive promoter A127-LPp to drive lasso peptide gene expression in Streptomyces bears the advantage of bypassing the native regulatory elements. This strategy is thus applicable to exploit lasso peptide production potential from rare actinobacteria and from metagenomics sources. It also opens a convenient way to generate lasso peptide variants by genetic engineering, facilitating structure-activity relationship studies37,42. In addition, our data reveal some aspects that need to be considered in future bioprospecting lasso peptides from Actinobacteria by heterologous expression (e.g. toxicity issue, choice of expression hosts etc.).
S. albus J1074 and S. lividans TK24 used in this study are well established hosts for heterologous production of secondary metabolites, each being more suitable than the other for certain metabolites43. Our data suggest that genotypes of these two hosts do not have a significant influence on lasso peptide production level, at least for the peptides and conditions examined. S. albus appeared slightly better because lasso peptide production in a wider range of conditions or higher yields were frequently observed for this strain (Table 2). In terms of growth media, solid cultures were clearly found to be superior to the liquid ones for lasso peptide biosynthesis, in consistence with previous studies23. However, the most suitable production medium is peptide-dependent and culture optimization of recombinant strains is still necessary. Since S. albus and S. lividans are common laboratory strains, the screening process can benefit from the abundant knowledge of their metabolism, and be simplified because of the availability of a range of tested media to choose from.
Modification of the native 9401-LP1 cluster led to a slight improvement of peptide production, showing the necessity of genetic engineering. The knowledge of how the native cluster is regulated will be required for efficient gene cluster refactoring.
Nevertheless, the strategy developed here failed to produce three other lasso peptides (A127-LP, 9401-LP2 and Sven-LP). The gene clusters of A127-LP and 9401-LP2 could be transferred to different Streptomyces hosts, and their gene transcription in heterologous hosts was unambiguously confirmed by RT-PCR (Supplementary methods and Figure S10). Therefore, we suspect that the toxicity of the produced peptides to the expression host is a major issue. A precedent example to support this hypothesis is that lariatin, a lasso peptide produced by Rhodococcus jostii, could not be produced by expressing the corresponding gene cluster in Rhodococcus strains that are closely related to the producer42. Several cellular targets of antibacterial lasso peptides have been identified11,18,44. In particular, the RNA polymerase has been shown to be a common target of several lasso peptides including microcin J25,44 siamycins and capistruin45, despite their different antibacterial spectra. The immunity mechanism of the native producer to its toxic lasso peptide is conferred by a dedicated ABC transporter, often encoded in the gene cluster. The striking difference in production between 9401-LP1 and 9401-LP2 clearly indicated that the lack of a resistance mechanism in the heterologous host could account for the failure of 9401-LP2 synthesis, as its gene cluster does not encode a pathway-specific transporter in contrast to that of 9401-LP1. For A127-LP, although transporter genes are present, it is likely that Streptomyces strains cannot install effectively the immunity mechanism originating from a phylogenetically distant actinobacterium. Similar phenomenon was observed when expressing the gene cluster of microbisporicin originating from Microbispora coralline in Streptomyces46. Worth of note, Snou-LP could be produced although no transporter genes were found in the cluster, suggesting that its cellular target might be different to those for 9401-LP2 and A127-LP. It is remarkable that the Sven-LP biosynthetic genes could not be conjugated into Streptomyces. One peculiarity of this cluster, in addition to the lack of transporter genes, is the presence of a gene whose product shows similarity to ribosomal protein serine acetyltansferases. A precedent example showed that genes encoding ribosomal proteins prevented the conjugation of the gene cluster of GE2270, a thiopeptide identified in Planobispora rosea, in Streptomyces47. It is therefore likely that this gene involved in protein acetylation plays a role in the conjugation process, or its expression is toxic to the host. An alternative explanation for failure to produce lasso peptides heterologously could be the absence in the new host of an unknown factor critical for maturation of the peptide. Overall, our data indicate that the success of heterologous expression is mainly determined by the intrinsic properties of the lasso peptide cluster of interest.
In conclusion, this study developed a SARP-controlled orthogonal expression system for the production of actinobacterial lasso peptides in Streptomyces. It is suitable for lasso peptide bioprospecting from yet untapped actinobacteria, and opens a convenient way to produce and engineer lasso peptides with new features that could hold promise for peptide-based drug discovery.
Methods
Bacterial strains, plasmids and growth conditions
Bacterial strains and plasmids are listed in Table S1. Streptomyces strains are routinely maintained on Soya-Flour-Mannitol (SFM) agar plates. For lasso peptide production, recombinant Streptomyces strains were screened in liquid media including GYM (4 g/L glucose, 4 g/L yeast extract and 10 g/L malt extract), MYM (4 g/L maltose, 4 g/L yeast extract, 10 g/L malt extract, 1.9 g/L MOPS), MYM-g in which maltose was replaced by 24 mL/L glycerol, MP5 (7 g/L yeast extract, 5 g/L NaCl, 1 g/L NaNO3, 36 ml/L glycerol, 20.9 g/L MOPS, pH 7.5) and phosphate-limited defined medium in which glucose was replaced with 24 mL/L glycerol33. Solid media used for lasso peptide production were GYM, SFM, ISP2 (BD Difco™), ISP4 (BD Difco™), marine broth (Pronadisa), R548 and Tryptic Soy Broth (TSB, BD Bacto™) agar. Conjugation of recombinant plasmids into Streptomyces was performed as previously described by Flett et al.49. Antibiotics used were apramycin (50 µg/mL) and thiostrepton (17 µg/mL) for selection of recombinant Streptomyces strains.
Construction and testing of SARP-based expression system
The pSOK809 reporter plasmid was constructed from PCR-amplified fragments using Gibson assembly50. DNA fragment encompassing the vector part with gusA gene was amplified from the pSOK808 template19 using primers 808GUS-F and 808GUS-R (Table S2). Putative SARP-dependent promoter region from the Actinoalloteichus sp. ADI127-17 lasso peptide gene cluster upstream of the A gene was amplified from the genomic DNA of the organism using primers SARp-F and SARp-R (Table S2). The pSARP plasmid for expression of the SARP regulator was constructed by a similar procedure. The vector part was amplified from the pUWLoriT template51 using primers UWLS-F and UWLS-R (Table S2). The SARP-encoding gene was amplified from the genomic DNA of the Actinoalloteichus sp. ADI127-17 using primers SARPg-F and SARPg-R (Table S2). Both plasmids were introduced in S. venezuelae ATCC 10712 via conjugation from E. coli ET12567/pUZ8002 either alone or in combination. The resulting strains were tested for GusA activity after 48 h of fermentation in the MYM liquid medium as described in Sekurova et al.30.
Gibson assembly was also used to construct all the plasmids for lasso peptide expression using the pSOK809 part without the gusA gene as a backbone and PCR-amplified genomic fragments encompassing respective gene clusters. Oligonucleotide primers used in construction of recombinant plasmids are listed in Table S2. Sequences of the lasso peptide gene clusters in this study were deposited in the GenBank. Accession numbers are KX379996 for 9401-LP1, KX379997 for 9401-LP2, KX379998 for 9810-LP, KX379999 for A127-LP, KX380000 for Snou-LP and KX380001 for Sven-LP (Table S3).
Oligonucleotide primers were designed using software j552 and Clone Manager 9 (Sci-Ed Software, USA). All constructs were sequenced to verify that no mutations were introduced during PCR amplification.
Production and extraction of lasso peptides
The SARP-based lasso peptide expression plasmids were introduced into Streptomyces strains by conjugation with E. coli ET12567/pUZ8002 as donor strain. Exconjugants were selected on SFM agar supplemented with 10 mM MgCl2 and apramycin. Integration of the gene cluster into the chromosome was confirmed by PCR. The pSARP plasmid was subsequently transferred to the resulting recombinant strain in a similar manner by conjugation. Exconjugants were selected with thiostrepton supplemented in the medium.
Growth on liquid medium for production: A seed culture of each recombinant Streptomyces strains harbouring the lasso peptide cluster was grown in the liquid medium of choice supplemented with apramycin and thiostrepton at 30 °C for 2 days. This was used to inoculate 50 mL of the same medium and the culture was grown in the presence of 5% XAD-16 resin at 30 °C for 7 days. The mycelia and the resin were extracted with MeOH at room temperature for 3 hours. The extracts were dried in a rotary evaporator and re-suspended in 60% acetonitrile before LC-MS analysis.
Growth on solid medium for production: A seed culture of each recombinant Streptomyces strains harbouring the lasso peptide cluster was grown in liquid GYM medium supplemented with apramycin and thiostrepton at 30 °C for 2 days. This seed culture was spread on the solid medium of choice. The plates were sealed and incubated at 30 °C for 7 days. Next, the agar with mycelia were cut into pieces and extracted in 100% MeOH at room temperature for 3 hours. The debris were removed by filtration; the resulting extracts were dried and resuspended in 60% acetonitrile. To scale up, selected strains were grown in square Petri dishes (10 cm × 10 cm) and extraction was performed as described above. Peptides were enriched from the crude extracts by solid phase extraction (SPE) using a Waters C18 SPE cartridge. 9401-LP1 and 9810-LP were produced in S. albus on ISP2 and ISP4 plates, respectively, and Snou-LP was produced in S. lividans on SFM agar.
LC-MS and MS/MS analysis
SPE fractions were analyzed by LC-MS and LC-MS/MS on an ultra-high performance LC system (Ultimate 3000 RSLC, Thermo Scientific) connected to a high-resolution electrospray ionization – quadrupole – time of flight (ESI-Q-TOF) mass spectrometer (Maxis II ETD, Bruker Daltonics). Separation was achieved on an Acclaim RSLC Polar Advantage II column (2.2 μm, 2.1 × 100 mm, Thermo Scientific) at a flow rate of 300 μL/min, using the following gradient of solvent A (ultra-pure water/0.1% formic acid) and solvent B (HPLC-MS grade acetonitrile/0.08% formic acid) over a total run time of 17.5 min: linear increase from 10% B to 60% B for 12 min, linear increase to 100% B for 0.2 min, decrease to 10% B for 0.5 min. The ESI-Q-TOF instrument was externally calibrated before each run using a sodium formate solution consisting of 10 mM sodium hydroxide in isopropanol/0.2% formic acid (1:1, v/v). The MS data were acquired in positive ion mode in the mass range m/z 60–2000. The source parameters were as follows: nebulizer gas 35 psi, dry gas 8 L/min, capillary voltage 3500 V, end plate offset 500 V, temperature 200 °C. LC-MS/MS data were acquired in the multiple reaction monitoring (MRM) mode with selection of the [M + 2 H]2+ or [M + 3 H]3+ ions of the lasso peptides. The data were treated with Data Analysis 4.3 (Bruker Daltonics). Relative peptide production level was measured by integrating mass peaks corresponding to all charge states.
The nomenclature introduced by Roepstorff and Fohlman53, further modified by Biemann54, was adopted to describe the product ions generated by MS/MS. The internal fragment ions were thus noted (brys)(r+s−t), where r, s, and t indicate the number of amino acids cleaved counting from the N-terminus, the number of amino acids cleaved from the C-terminus and the total number of amino acid residues in the peptide, respectively. The subscript (r + s − t) accounts for the number of residues in the internal fragment. When the b- and y-type product ions remained associated due to a covalent link (either the isopeptidic bond forming the macrolactam ring or a disulphide bridge), we used the nomenclature [brys]. As an example, [9401-LP1 - {FAGTAWI}] was denoted [b10y1] (Fig. 4). Finally, the b- and y- type ions associated non-covalently due to the steric entrapping of the tail within the ring were named using the nomenclature [(br)*(ys)] introduced previously by our group35. These [2]rotaxane product ions detected for certain lasso peptides constitute a signature of the lasso topology.
Carboxypeptidase Y treatment
The pre-purified peptides 9401-LP1, 9810-LP and Snou-LP were submitted to carboxypeptidase Y hydrolysis, as previously described22. Briefly, carboxypeptidase Y from Saccharomyces cerevisiae (Sigma) was added at 0.1 U/mL to the peptide solution in MES buffer (50 mM MES, 1 mM CaCl2, pH 6.7) and the solution was incubated for 3 h at 37 °C. Negative control consisted of the peptides with no enzyme. Positive control consisted of the non-lasso topoisomer of microcin J25. In case of reduction of 9401-LP1, 50 mM DTT was added before carboxypeptidase Y treatment.
Ion mobility mass spectrometry
IM-MS experiments were carried out on a hybrid quadrupole ESI-Q-TOF IM-MS instrument (Synapt G2, Waters). The pre-purified peptides 9401-LP1 and Snou-LP, as well as the lasso and non-lasso peptide controls, MccJ25 and MccJ25-lcm, were solubilized in H2O/CH3CN 1:1 + 0.1% formic acid +0.5% sulfolane. The solutions were introduced in the source by direct infusion using a syringe pump with a flow rate of 300 μL/h. Ionization was operated in positive ion mode under the experimental set-up as described39. The CCS calibration was carried out from the CCS values of doubly-protonated polyalanine ions determined with uniform field drift tube ion mobility with helium as drift gas55. The 9810-LP was in too low quantity/purity to provide information on the peptide topology by IM-MS.
Antibacterial activity assays
The antibacterial activity of the new lasso peptides were tested with pre-purified peptides using the soft-agar overlay technique.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Rutledge, P. J. & Challis, G. L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Micro 13, 509–523 (2015).
Blin, K., Medema, M. H., Kottmann, R., Lee, S. Y. & Weber, T. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res 45, D555–D559 (2017).
Medema, M. H. et al. Minimum information about a biosynthetic gene cluster. Nat Chem Biol 11, 625–631 (2015).
Wenzel, S. C. & Müller, R. Recent developments towards the heterologous expression of complex bacterial natural product biosynthetic pathways. Curr Opin in Biotech 16, 594–606 (2005).
Arnison, P. G. et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30, 108–160 (2013).
Maksimov, M. O., Pan, S. J. & James Link, A. Lasso peptides: structure, function, biosynthesis, and engineering. Nat Prod Rep 29, 996–1006 (2012).
Li, Y., Zirah, S. & Rebuffat, S. Bacterial Strategies to Make and Maintain Bioactive Entangled Scaffolds in SpringerBriefs in Microbiology (Springer-Verlag New York, 2015).
Hegemann, J. D., Zimmermann, M., Xie, X. & Marahiel, M. A. Lasso peptides: an intriguing class of bacterial natural products. Acc Chem Res 48, 1909–1919 (2015).
Iwatsuki, M. et al. Lariatins, antimycobacterial peptides produced by Rhodococcus sp. K01-B0171, have a lasso structure. J Am Chem Soc 128, 7486–7491 (2006).
Detlefsen, D. J. et al. Siamycins I and II, new anti-HIV-1peptides: II. Sequence analysis and structure determination of siamycin I. J Antibiot 48, 1515–1517 (1995).
Gavrish, E. et al. Lassomycin, a ribosomally synthesized cyclic peptide, kills mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem Biol 21, 509–518 (2014).
Metelev, M. et al. Structure, bioactivity, and resistance mechanism of streptomonomicin, an unusual lasso Peptide from an understudied halophilic actinomycete. Chem Biol 22, 241–250 (2015).
Um, S. et al. Sungsanpin, a lasso peptide from a deep-sea Streptomycete. J Nat Prod 76, 873–879 (2013).
Elsayed, S. S. et al. Chaxapeptin, a lasso peptide from extremotolerant Streptomyces leeuwenhoekii Strain C58 from the hyperarid atacama desert. J Org Chem 80, 10252–10260 (2015).
Weber, W., Fischli, W., Hochuli, E., Kupfer, E. & Weibel, E. K. Anantin–a peptide antagonist of the atrial natriuretic factor (ANF). I. Producing organism, fermentation, isolation and biological activity. J Antibiot 44, 164–171 (1991).
Potterat, O. et al. BI-32169, a bicyclic 19-peptide with strong glucagon receptor antagonist activity from Streptomyces sp. J Nat Prod 67, 1528–1531 (2004).
Nakayama, J. et al. Siamycin attenuates fsr quorum sensing mediated by a gelatinase biosynthesis-activating pheromone in Enterococcus faecalis. J Bacteriol 189, 1358–1365 (2007).
Ma, P. et al. Anti-HIV siamycin I directly inhibits autophosphorylation activity of the bacterial FsrC quorum sensor and other ATP-dependent enzyme activities. FEBS Lett 585, 2660–2664 (2011).
Tietz, J. I. et al. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat Chem Biol 13, 470–478 (2017).
Burkhart, B. J., Hudson, G. A., Dunbar, K. L. & Mitchell, D. A. A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat Chem Biol 11, 564–570 (2015).
Maksimov, M. O., Pelczer, I. & Link, A. J. Precursor-centric genome-mining approach for lasso peptide discovery. Proc Natl Acad Sci USA 109, 15223-8 (2012).
Hegemann, J. D., Zimmermann, M., Zhu, S., Klug, D. & Marahiel, M. A. Lasso peptides from proteobacteria: Genome mining employing heterologous expression and mass spectrometry. Biopolymers 100, 527–542 (2013).
Kersten, R. D. et al. A mass spectrometry–guided genome mining approach for natural product peptidogenomics. Nat Chem Biol 7, 794–802 (2011).
Pan, S. J., Rajniak, J., Maksimov, M. O. & Link, A. J. The role of a conserved threonine residue in the leader peptide of lasso peptide precursors. Chem Commun (Camb) 48, 1880–1882 (2012).
Takasaka, N., Kaweewan, I., Ohnishi-Kameyama, M. & Kodani, S. Isolation of a new antibacterial peptide actinokineosin from Actinokineospora spheciospongiae based on genome mining. Lett Appl Microbiol 64, 150–157 (2017).
Li, Y. et al. Characterization of sviceucin from Streptomyces provides insight into enzyme exchangeability and disulfide bond formation in lasso peptides. ACS Chem Biol 10, 2641–9 (2015).
Kaweewan, I., Ohnishi-Kameyama, M. & Kodani, S. Isolation of a new antibacterial peptide achromosin from Streptomyces achromogenes subsp. achromogenes based on genome mining. J Antibiot 70, 208–211 (2017).
Nouioui, I. et al. Actinoalloteichus fjordicus sp. nov. isolated from marine sponges: phenotypic, chemotaxonomic and genomic characterisation. Antonie van Leeuwenhoek 110, 1705–1717 (2017).
Weber, T. et al. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43, W237–43 (2015).
Sekurova, O. N., Zhang, J., Kristiansen, K. A. & Zotchev, S. B. Activation of chloramphenicol biosynthesis in Streptomyces venezuelae ATCC 10712 by ethanol shock: insights from the promoter fusion studies. Microb Cell Fact 15, 85 (2016).
Santos-Aberturas, J. et al. Hierarchical control on polyene macrolide biosynthesis: PimR modulates pimaricin production via the PAS-LuxR transcriptional activator PimM. PLoS ONE 7, e38536 (2012).
Garg, R. P. & Parry, R. J. Regulation of valanimycin biosynthesis in Streptomyces viridifaciens: characterization of VlmI as a Streptomyces antibiotic regulatory protein (SARP). Microbiology 156, 472–483 (2010).
Borodina, I. et al. Antibiotic overproduction in Streptomyces coelicolor A3(2) mediated by phosphofructokinase deletion. J Biol Chem 283, 25186–25199 (2008).
Chiuchiolo, M. J., Delgado, M. A., Farı́as, R. N. & Salomón, R. A. Growth-phase-dependent expression of the cyclopeptide antibiotic microcin J25. J Bacteriol 183, 1755–1764 (2001).
Zirah, S. et al. Topoisomer differentiation of molecular knots by FTICR MS: lessons from class II lasso peptides. J Am Soc Mass Spectrom 22, 467–479 (2011).
Jeanne Dit Fouque, K. et al. General rules of fragmentation evidencing lasso structures in CID and ETD. Analyst 143, 1157–1170 (2018).
Ducasse, R. et al. Sequence determinants governing the topology and biological activity of a lasso peptide, microcin J25. Chembiochem 13, 371–380 (2012).
Jeanne Dit Fouque, K. et al. Ion mobility–mass spectrometry of lasso peptides: signature of a rotaxane topology. Anal Chem 87, 1166–1172 (2015).
Jeanne Dit Fouque, K. et al. Signatures of mechanically interlocked topology of lasso peptides by ion mobility–mass spectrometry: lessons from a collection of representatives. J Am Soc Mass Spectrom 28, 315–322 (2017).
Severinov, K., Semenova, E., Kazakov, A., Kazakov, T. & Gelfand, M. S. Low-molecular-weight post-translationally modified microcins. Mol Microbiol 65, 1380–1394 (2007).
Gust, B., Challis, G. L., Fowler, K., Kieser, T. & Chater, K. F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA 100, 1541–1546 (2003).
Inokoshi, J., Koyama, N., Miyake, M., Shimizu, Y. & Tomoda, H. Structure-activity analysis of Gram-positive bacterium-producing lasso peptides with anti-mycobacterial activity. Sci Rep 6, 30375 (2016).
Baltz, R. H. Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J Ind Micro Biotech 43, 343–370 (2016).
Yuzenkova, J. et al. Mutations of bacterial RNA polymerase leading to resistance to microcin J25. J Biol Chem 277, 50867–50875 (2002).
Kuznedelov, K. et al. The antibacterial threaded-lasso peptide capistruin inhibits bacterial RNA polymerase. J Mol Biol 412, 842–848 (2011).
Foulston, L. & Bibb, M. Feed-forward regulation of microbisporicin biosynthesis in Microbispora corallina. J Bacteriol 193, 3064–3071 (2011).
Flinspach, K., Kapitzke, C., Tocchetti, A., Sosio, M. & Apel, A. K. Heterologous expression of the thiopeptide antibiotic GE2270 from Planobispora rosea ATCC 53733 in Streptomyces coelicolor requires deletion of ribosomal genes from the expression construct. PLoS ONE 9, e90499 (2014).
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. Practical Streptomyces Genetics. 613 pages (John Innes Foundation, 2000).
Flett, F., Mersinias, V. & Smith, C. P. High efficiency conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA restricting Streptomycetes. FEMS Microbiol Lett 155 (1997).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6, 343–345 (2009).
Luzhetskyy, A. et al. Generation of novel landomycins M and O through targeted gene disruption. Chembiochem 6, 675–678 (2005).
Hillson, N. J., Rosengarten, R. D. & Keasling, J. D. j5 DNA assembly design automation software. ACS Synth Biol 1, 14–21 (2012).
Roepstorff, P. & Fohlman, J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed Mass Spec 11, 601 (1984).
Biemann, K. Appendix 5. Nomenclature for peptide fragment ions (positive ions). Methods in Enzymol 193, 886–887 (1990).
Bush, M. F., Campuzano, I. D. G. & Robinson, C. V. Ion mobility mass spectrometry of peptide ions: effects of drift gas and calibration strategies. Anal Chem 84, 7124–7130 (2012).
Acknowledgements
Part of this work was supported by the European Commission under the 7th Framework Program through the “Collaborative Project” action “STREPSYNTH” Grant No. 613877, University of Vienna and NTNU. A “Research in Paris” postdoctoral fellowship from the City of Paris to HM is acknowledged. Soufiane Telhig is thanked for his help in constructing modified p9401-LP1 plasmid. Drs Hélène Lavanant, Isabelle Schmitz-Afonso and Marie Hubert from the COBRA laboratory (UMR 6014) are thanked for their help in IM-MS experiments. The personnel in the bioorganic analytical platform of MNHN is thanked for assistance in the mass spectrometry analyses.
Author information
Authors and Affiliations
Contributions
S.B.Z. and Y.L. designed the study. J.M., C.G., O.S., O.N.S., H.M., S.Z. and Y.L. performed the experiments. J.M., C.G., S.Z. and Y.L. analysed the mass spectrometry data. S.Z., S.R., S.B.Z. and Y.L. supervised the project. C.A. provided access to the ion mobility mass spectrometry and expertise. S.R., S.B.Z. and Y.L. wrote the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mevaere, J., Goulard, C., Schneider, O. et al. An orthogonal system for heterologous expression of actinobacterial lasso peptides in Streptomyces hosts. Sci Rep 8, 8232 (2018). https://doi.org/10.1038/s41598-018-26620-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-26620-0
This article is cited by
-
How to harness biosynthetic gene clusters of lasso peptides
Journal of Industrial Microbiology and Biotechnology (2020)
-
Lassomycin and lariatin lasso peptides as suitable antibiotics for combating mycobacterial infections: current state of biosynthesis and perspectives for production
Applied Microbiology and Biotechnology (2019)
-
Genome mining for lasso peptides: past, present, and future
Journal of Industrial Microbiology and Biotechnology (2019)
-
Discovery and characterization of a novel C-terminal peptide carboxyl methyltransferase in a lassomycin-like lasso peptide biosynthetic pathway
Applied Microbiology and Biotechnology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.