Abstract
The Schizosaccharomyces pombe sirtuin Hst4, functions in the maintenance of genome stability by regulating histone H3 lysine56 acetylation (H3K56ac) and promoting cell survival during replicative stress. However, its molecular function in DNA damage survival is unclear. Here, we show that hst4 deficiency in the fission yeast causes S phase delay and DNA synthesis defects. We identified a novel functional link between hst4 and the replisome component mcl1 in a suppressor screen aimed to identify genes that could restore the slow growth and Methyl methanesulphonate (MMS) sensitivity phenotypes of the hst4Δ mutant. Expression of the replisome component Mcl1 rescues hst4Δ phenotypes. Interestingly, hst4 and mcl1 show an epistatic interaction and suppression of hst4Δ phenotypes by mcl1 is H3K56 acetylation dependent. Furthermore, Hst4 was found to regulate the expression of mcl1. Finally, we show that hSIRT2 depletion results in decreased levels of And-1 (human orthologue of Mcl1), establishing the conservation of this mechanism. Moreover, on induction of replication stress (MMS treatment), Mcl1 levels decrease upon Hst4 down regulation. Our results identify a novel function of Hst4 in regulation of DNA replication that is dependent on H3K56 acetylation. Both SIRT2 and And-1 are deregulated in cancers. Therefore, these findings could be of therapeutic importance in future.
Similar content being viewed by others
Introduction
The concerted action of histone acetyltransferases (HATs) and histone deacetylases (HDACs) regulate key DNA metabolic processes such as replication, transcription and repair by modulating the acetylation status of histone and non-histone proteins1,2,3. HDACs are classified into four classes based on their sequence homology4. The class III HDACs, also called sirtuins require nicotinamide adenine nucleotide (NAD+) as a co-factor for deacetylating their substrates5,6. Sirtuins function in several important cellular processes including gene expression, heterochromatin maintenance, genome stability and replicative life span7,8,9.
The S. pombe sirtuin family comprises three sirtuins, hst2, sir2 and hst410. Fission yeast lacking either hst2 or sir2 do not display phenotypes such as slow growth and sensitivity to DNA damaging agents11. However, deletion of hst4 alone results in characteristic phenotypes such as elongated cell morphology (30%), fragmented DNA and sensitivity to DNA damaging agents like ultra-violet (UV) radiation, methyl methane sulphonate (MMS), hydroxy urea (HU) and camptothecin (CPT)12,13. Hst4 deacetylates lysine 56 on histone H313. Originally discovered in S. cerevisiae, H3K56 acetylation is a histone core domain modification, which peaks during the S phase of the cell cycle and is conserved from yeast to higher eukaryotes13,14,15,16,17. Acetylation of the H3K56 residue on newly synthesized histone H3 is essential for its deposition on chromatin, while reduction in H3K56 acetylation leads to genomic instability18,19. In addition, induction of H3K56 acetylation upon DNA damage facilitates repair, chromatin reassembly and checkpoint recovery3,19,20. In S. pombe, Hst4 down regulation is accompanied by increased H3K56 acetylation during cell cycle, DNA damage and oxidative stress13,21. Hst4 has been shown to interact with Myh1, a protein involved in base excision repair and cell cycle checkpoint clamp 9-1-1 complex21. However, the exact molecular mechanism by which Hst4 functions in DNA transactions is not understood.
Mcl1 is the S. pombe homologue of human And-1 and the budding yeast Ctf4. These proteins are characterized by the presence of WD-repeat domains, which aid in protein-protein interactions. Originally identified in a genetic screen for mutants affecting chromosome transmission fidelity22, Ctf4 functions in sister chromatid cohesion. It also couples Mcm2-7 helicase to DNA polymerase alpha (Polα) within the replisome complex and facilitates replication by binding to Mcm1023,24. Studies using Xenopus egg extracts showed the conserved function of Ctf4 in Polα recruitment during DNA replication and cell cycle progression25. Reports on human And-1 also indicate its importance in DNA replication through its involvement in the formation of CDC45-MCM2-7-GINS complex (CMG helicase complex)26. And-1 participates in several important cellular processes such as checkpoint activation, sister chromatid cohesion and DNA repair27. Studies in fission yeast show that Mcl1 is a multifunctional protein that associates with Polα and is required for genome stability, telomere replication, chromosome segregation and DNA repair28,29,30,31. Deletion of either hst4 or mcl1 show similar phenotypes such as elongated cell morphology and sensitivity to DNA damaging agents12,13,29. Additionally, these mutants exhibit elevated chromosome loss12,32.
In this study, we identified sirtuin Hst4 as a regulator of Mcl1, a S. pombe orthologue of Ctf4/And-1. We show that the deletion of hst4 causes S phase delay and DNA synthesis defects, which are partially suppressed by overexpression of mcl1. The sensitivity of hst4Δ mutant cells to agents that cause replication stress (MMS, HU), with an exception to CPT, are completely rescued by expression of mcl1. Our genetic analysis reveals that mcl1 and hst4 function in same genetic pathways to preserve genomic integrity. Further, we discovered that during replicative stress Mcl1 levels are altered via Hst4 to maintain genome integrity. Our results indicate that the role of hst4 in DNA replication is dependent on H3K56 acetylation. Finally, we demonstrate that the human SIRT2 regulates the levels of human Mcl1 orthologue, And-1, revealing conservation of this sirtuin dependent regulatory mechanism in humans.
Results
Deletion of hst4 causes S phase delay
In fission yeast, deficiency of hst4 results in slow growth and DNA fragmentation phenotypes in the absence of external genotoxic agents12,13. Earlier studies have indicated that S. pombe Hst4 functions in DNA damage response pathways. However, the molecular functions of Hst4 in DNA metabolic pathways are not clear. Cells either arrest or progress slowly through the cell cycle in response to DNA damage33,34. To determine the effect of hst4 deficiency on cell cycle progression, we constructed wild type and hst4Δ mutant strains bearing cdc25-22 mutation. We synchronized these wild type and hst4Δ mutant strains at G2/M interface using the temperature sensitive cdc25-22 allele. Following their arrest cells were released into cell cycle by lowering the temperature from 36 °C to 25 °C, aliquots of cells were collected at indicated time points and the progression through the cell cycle was monitored using flow cytometry. The results presented in Fig. 1A show that the hst4∆ cells display 30 minutes delay in completing S phase compared to the wild type. To further confirm this S phase delay, septation index analysis was carried out which shows synchrony and cell cycle position. Figure 1B shows that wild type cells reached the peak of septation index in 90 minutes after release from arrest, whereas there is a delay in peak of septation index by 1 hour in the hst4∆ cells. Next, in order to confirm that the delay of S phase in the hst4Δ mutant cells is not due to G2/M delay, we constructed wild type and hst4Δ mutant strains bearing cdc10-v50 mutation. We synchronized these wild type and hst4Δ mutant strains at the G1/S phase boundary of cell cycle using the temperature sensitive cdc10-v50 allele. Following their arrest cells were released into cell cycle by lowering the temperature from 36 °C to 25 °C, aliquots of cells were collected at indicated time points and the progression through cell cycle was monitored using flow cytometry. Figure 1C shows that the wild type cells complete S- phase within 120 minutes after release from the arrest, however, the hst4Δ cells progress through the S phase slowly, entering the G2 phase one hour later (180 minutes post-release) than the wild type cells. Altogether, these results suggest that Hst4 is required for progression through the S phase, indicating that it may play a role in DNA replication.
Deletion of hst4 causes S phase delay. (A) Flow cytometry profile showing cell cycle progression of wild type (WT) and hst4Δ mutants synchronized at G2/M phase.The cdc25-22 mutant strain (FY4225) and cdc25-22 hst4Δ (DHP56) were grown in YES medium to log phase at permissive temperature (25 °C) and shifted to restrictive temperature (36 °C) for 4 hr, inducing G2/M arrest. Cells were shifted to permissive temperature (25 °C) after 4 hr, cells were collected every 30 minutes and cell cycle profile was analyzed by flow cytometry. (B) Septation index for corresponding cell cycle arrest in (A) showing percentage of cells with septa at each time point after release from G2 arrest. Calcofluor was used to stain the septa and DAPI was used to localize the nucleus. (C) Flow cytometry profile showing cell cycle progression of wild type (WT) and hst4Δ mutants synchronized at G1/S phase. The cdc10-v50 mutant strain (FY563) and cdc10-v50 hst4Δ (DHP91) were grown in YES medium to log phase at permissive temperature (25 °C) and shifted to restrictive temperature (36 °C) for 4 hr, inducing G1/S arrest. Cells were shifted to permissive temperature (25 °C) after 4 hr, cells were collected every 30 minutes and cell cycle profile was analyzed by flow cytometry.
Deficiency of hst4 leads to replication defects
We hypothesized that the delayed S phase progression phenotype of hst4Δ mutant cells may be due to replication defects. To examine this possibility, we generated wildtype and hst4Δ mutant cells capable of bromodeoxyuridine (BrdU) uptake and incorporation into DNA35. The strains were grown for an hour in the presence of BrdU, to label the nascent DNA, and harvested. We determined the BrdU positive cells by immunofluorescence and quantitated them. Figure 2A,B show a two-fold reduction of the BrdU positive population in hst4 deficient cells compared to wild type cells (47% BrdU positive cells in wild-type versus 23% in hst4∆ mutant cells). Further, we quantified the amount of BrdU incorporated into DNA by slot blot, using anti-BrdU antibodies, and observed reduced BrdU incorporation in hst4Δ cells (Fig. 2C,D). These results suggest that DNA synthesis is defective in hst4 deficient cells. Many studies directed towards understanding the mechanisms of the S phase delay indicate that problem in replication origin firing and fork progression contribute to reducing the rate of DNA replication36,37. The slowing of S phase should result in more number of S phase cells (BrdU positive cells), however, as all S phase cells in the mutant are not actively incorporating BrdU, the number of BrdU incorporating mutant cells decrease. This situation may arise in cases where S phase slowing occurs due to DNA synthesis defects, there are cells which entered S phase but are not actively incorporating BrdU (this situation could be due to replication fork stalling). Although these cells are in S phase, they do not incorporate BrdU. This results in BrdU negative S phase cells. The size of this cell population varies with the severity of DNA synthesis defects. The DNA replication defects generate ssDNA, which could result in DNA damage. Rad22 is a marker for DNA damage as it binds to ssDNA and double strand breaks arising due to replication defects or other exogenous damage38,39,40. Since hst4Δ cells are known to accumulate spontaneous DNA damage, we hypothesized that this could be due to replication defects. Therefore, we generated hst4 deficient strains expressing Rad22-YFP from its genomic loci. A striking increase in Rad22 foci were observed in hst4Δ mutants compared to wild type (40% ± 4.54 in hst4Δ as compared to 8% ± 1.02 in the wild type, Fig. 2E). We have also observed multiple Rad22 foci appearing only in hst4Δ mutants (Fig. 2E,F). Cell cycle position analysis of the Rad22 foci containing cells was also performed to examine whether accumulation of Rad22 foci was occurring in the S phase cells. The S/early G2 phase hst4Δ cells exhibited significant accumulation of Rad22 foci (Fig. 2G). Overall, these results confirm that deletion of hst4 causes defects in DNA replication.
Deficiency of hst4 leads to replication defects. (A) Asynchronously growing wild type (FY4225) and hst4Δ (DHP56) strains were grown in presence of BrdU (150 µg/ml) for 30 minutes. Cells were fixed and stained with anti-BrdU antibody and visualized under fluorescent microscope. Bar = 2 µm. (B) Quantification of the immunofluorescence data presented in Fig. 2A. The percentage of S-phase cells were determined by counting BrdU positive cells in wild-type and hst4Δ mutants. Plotted are the mean values from three independent experiments. Student’s t test was used for statistical analysis. Error bars indicate mean ± S.D whereas three asterisks represent extremely significant difference, p-value < 0.001. (C) Slot blot showing BrdU incorporation in genomic DNA of wild type (FY4225) and hst4Δ (DHP56) strains using anti-BrdU antibody. Equal amounts of genomic DNA was loaded for wild type and hst4Δ mutants. (D) Quantification of BrdU incorporation of slot blot analysis. Average and standard deviations from samples of three independent experiments were plotted. Statistical significance p < 0.05 between wild-type and hst4Δ strains is indicated with a single asterisk. (E) Rad22-YFP foci formation by life cell microscopy. Wild-type (ENY0670) and hst4Δ mutant (DHP63) containing genomically tagged Rad22-YFP were grown to mid-log phase and percentage of nuclei with at least one Rad22 Foci is shown. Bar = 2 µm. (F) Graph showing percent cells with single and multiple Rad22 foci in wild type and hst4Δ mutants. Mean values from three independent experiments were used to calculate standard deviation. (G) Graph showing percent Rad22 foci according to different cell cycle stages of wild-type and hst4Δ mutants. SD was calculated from three independent experiments.
Overexpression of replisome component Mcl1 suppresses the phenotypes of hst4 deficient cells
To further understand the pathways functioning aberrantly in the absence of hst4, we performed a genetic screen to identify high copy suppressors of the slow growth and DNA damage (MMS) sensitivity phenotypes of hst4Δ mutant cells. The S. pombe genomic library used for this study was generated as described earlier41. We have transformed this genomic library in hst4∆ mutant cells and selected colonies which were growing like wild type and insensitive to MMS. By isolating plasmids from these cells and sequencing them, we identified multiple classes of genes involved in RNA metabolism, protein transport, and DNA replication as suppressors of hst4 mutant phenotypes. A clone containing the full length mcl1 gene was among the suppressors of hst4Δ mutant phenotypes. The fission yeast Mcl1, a homologue of S. cerevisiae Ctf4, is a WD domain containing protein that interacts with many proteins involved in DNA replication and act as a hub for replication factors32,42. It also couples the CMG helicase and Polα43. To validate Mcl1 as a candidate suppressor of hst4 mutant phenotypes, mcl1 gene was cloned into a high copy plasmid (pRO314) and transformed into cells lacking hst4. The slow growth phenotype of hst4Δ cells was rescued in the presence of mcl1 as monitored by spot assay (Fig. 3A). As reported previously, expression of mcl1 alone in the wild type cells leads to slow growth32. Thus, the rescue of slow growth of hst4Δ cells by mcl1 expression is specific to its function in hst4Δ mutant cells. The elongated cell morphology of cells lacking hst4 was also rescued by mcl1 expression and the percentage of elongated cells in hst4Δ mutant culture increases in stationary phase. We observed reduction in the percentage of elongated cells (from 28% to 3.5%) in hst4 deficient cells expressing mcl1 as compared to hst4Δ cells (Fig. 3B,C). In S. cerevisiae, Ctf4, which is an orthologue of Mcl1, has been shown to couple the CMG helicase complex to Polα. Therefore, we sought to investigate whether hst4 genetically interact with MCM complex and GINS complex components. We expressed mcm4 and sld5 in hst4Δ mutants and monitored growth rate by spot assay. Interestingly, expression of mcm4 and sld5 had no effect on the slow growth phenotype of hst4Δ mutant as compared to mcl1 (Fig. 3D). The expression levels of Mcl1, Mcm4 and Sld5 in hst4 mutant cells are shown in Supplementary Fig. 1. These results indicate that hst4 specifically interacts with mcl1.
Mcl1 expression rescues the slow growth and elongated morphology phenotypes of hst4Δ mutants. (A) Wild type strain (ROP191) or hst4∆ (ROP58) expressing hst4, mcl1 or pRO314 (empty vector) were grown to OD 1, five-fold serially diluted, spotted onto EMM-Ura plates and incubated at 30 °C to assay growth rate. (B) Morphology of strains indicated in (A) were grown to saturation in liquid EMM medium at 30 °C and observed under phase-contrast microscope. (C) Plot showing quantification of elongated cells. Percentage elongated cells were determined by counting at least 100 cells from three independent experiments. Elongated cells were identified as cells that were greater than the size of a normal dividing cell (>14 µm). Error bars represent standard deviation of the mean from three independent experiments. (D) hst4∆ mutant strain (ROP58) expressing mcm4, sld5 or harboring pSLF272 (empty vector) were grown to OD 1, five-fold serially diluted, spotted onto EMM-Ura plates and incubated at 30 °C to assay growth rate. (E) hst4Δ mutant cells expressing indicated genes were grown in EMM-URA medium at 30 °C and observed under microscope for examining morphology. (F) Plot showing quantification of elongated cells in indicated strains. Percentage elongated cells were determined by counting at least 100 cells from three independent experiments. Average and standard deviations were calculated from three independent experiments.
Mcl1 expression partially rescues the S-phase delay phenotype of hst4 deficient cells
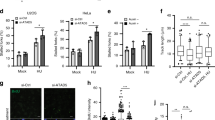
The S phase delay and decreased BrdU incorporation observed in cells lacking hst4 indicate that its deficiency could be resulting in defective DNA replication. Since Ctf4 has been shown to play a crucial role in coupling DNA unwinding and DNA synthesis machineries, we examined whether mcl1 can rescue the S-phase delay phenotype of the hst4Δ mutant cells. Wild type and hst4Δ mutant strains were arrested in G2 phase and progression through the cell cycle was monitored using flow cytometry. Our results show that expression of mcl1 could partially rescue the S-phase delay phenotype of hst4Δ deletion mutants (Fig. 4A). However, the rate of S phase progression through S phase was slower in hst4Δ cells than the wild type cells (Fig. 4A). To check whether expression of mcl1 rescues the DNA synthesis defect of hst4 deletion mutants, hst4Δ mutants were grown in the presence of BrdU and the cells which incorporated BrdU were detected by immunofluorescence. The defects in DNA synthesis in hst4∆ mutants was rescued by expression of mcl1 (Fig. 4B,C).
The rescue of S phase delay phenotypes of hst4Δ mutants is partly dependent on Mcl1. (A) Flow cytometry showing cell cycle progression of wild type (WT) and hst4Δ mutants synchronized at G2/M phase. The cdc25-22 mutant strain (FY4225) and cdc25-22 hst4Δ (DHP56) expressing hst4, mcl1 or pSLF272 (empty vector) were grown in EMM-URA medium to log phase at permissive temperature (25 °C) and shifted to restrictive temperature (36 °C) for 4 hr inducing G2/M arrest, cells were collected every 30 minutes and cell cycle profile was analyzed by flow cytometry. (B) Asynchronously growing wild type (FY4225) and hst4Δ (DHP56) expressing hst4, mcl1 or pSLF272 (empty vector) were grown in presence of BrdU (150 μg/ml) for 30 minutes. Cells were fixed and stained with anti-BrdU antibody and visualized under fluorescent microscope. Bar = 2 μm. (C) Quantification of the immunofluorescence data presented in (B). The percentage of S-phase cells was determined by counting BrdU positive cells. Plotted are the mean values from three independent experiments. Error bars indicate mean ± S.D.
Mcl1 expression rescues hst4 deficient cells from sensitivity to genotoxic agents
To determine whether expression of mcl1 rescues the DNA fragmentation phenotype of hst4 deficient cells, we determined nuclear morphology of the hst4Δ cells by staining live cells with Hoechst. The results presented in (Fig. 5A) shows that the mutant cells acquired the normal cell and nuclear morphology on expression of mcl1. To check whether mcl1 can suppress MMS sensitivity of hst4Δ mutant cells, five-fold serially diluted wild type or hst4Δ mutant cells with indicated plasmids were spotted in the presence or absence of MMS on minimal medium. The MMS sensitivity of the hst4Δ cells was rescued by expression of mcl1 (Fig. 5B). The hst4Δ mutants are also sensitive to other DNA damaging agents such as camptothecin (CPT) (DNA topoisomerase I inhibitor) and hydroxyurea (HU). To test whether the suppressor mcl1 can rescue the sensitivity of hst4Δ cells to other DNA damaging agents, five-fold serially diluted indicated strains were spotted in the presence or absence of 10 mM HU or 10 µM camptothecin. Mcl1 expression could partially rescued HU sensitivity but not CPT sensitivity of hst4∆ mutant cells (Fig. 5B). Overall, these results suggest that Hst4 and Mcl1 function together in cell survival following replicative stress, and these genes specifically interact in certain DNA damage response pathways.
Mcl1 and Hst4 cooperate to promote cell survival following replicative stress. (A) Wild type strain (ROP191) or hst4∆ (ROP58) expressing hst4, mcl1 or pSLF272 (empty vector) were grown to stationary phase, DNA was stained with Hoechst and visualized under confocal microscope. Bar = 2 µm. (B) Wild type strain (ROP191) or hst4∆ (ROP58) or rad3∆ (ROP266) expressing hst4, mcl1 or pSLF272 (empty vector) were five-fold serially diluted and spotted on EMM-Ura plates in the presence or absence of 0.01% MMS or 10 mM HU or 10 µM camptothecin for five days at 30 °C. (C) Double deletion mutants of hst4 and mcl1 were generated by crossing hst4∆ (ROP58) and mcl1∆ (NYSPE19) strains followed by tetrad dissection. Wild type strain (ROP191), hst4∆ (ROP58), mcl1∆ (NYSPE19), hst4∆mcl1∆ (DHP69) and rad3∆ (ROP266) strains were five-fold serially diluted, grown in YES medium in the presence or absence of 0.005, 0.01 and 0.02% MMS and incubated for three days at 30 °C.
Mcl1 and Hst4 function in an epistatic manner to maintain genome integrity
To further characterize the genetic interaction between hst4 and mcl1, hst4Δ and mcl1Δ mutant cells were crossed to generate a double mutant. The results presented in Fig. 5C shows that the double mutants are viable and have phenotypes similar to hst4Δ cells, suggesting an epistatic interaction between hst4 and mcl1. However, deletion of hst4 in mcl1Δ cells partially rescues its MMS sensitivity, suggesting a harmful role of hst4 in the absence of mcl1. This effect could be partially due to hyperacetylation of histone H3K56 which may aid in survival and may also be attributed to the function of these proteins in replication and/or cohesion pathways.
Analysis of H3K56 acetylation on phenotype suppression by Mcl1
The histone H3 lysine 56 (H3K56) is deacetylated by Hst4. Interestingly, the phenotypes of H3K56R and H3K56Q mutants, which mimic constitutive deacetylated and acetylated states respectively, are similar to hst4Δ cells13. Therefore, we examined whether expression of mcl1 could rescue the slow growth and MMS sensitivity phenotypes of hst4Δ cells. The results presented in Fig. 6A show that mcl1 expression could not suppress the phenotypes of these mutants, indicating H3K56ac may play an important role in this pathway. To further confirm the function of H3K56ac, we have investigated whether expression of mcl1 can rescue the slow growth and MMS sensitivity phenotypes of H3K56Qhst4Δ and H3K56Rhst4Δ double mutant cells. The results presented in Fig. 6B show that the slow growth phenotype of H3K56Rhst4Δ double mutant cells was partially recovered by mcl1 expression. However, it could not rescue the MMS sensitivity phenotype. Furthermore, expression of mcl1 could not rescue the slow growth and MMS sensitivity phenotypes of H3K56Qhst4Δ double mutant cells (Fig. 6C). Since the phenotypes of hst4Δ mutant cells are mainly attributed to increased H3K56ac levels, we analyzed the status of H3K56ac in hst4Δ mutant cells on expression of Mcl1 by immunoblotting. The levels of acetylation in hst4Δ mutant cells remain unchanged on expression of mcl1 indicating mcl1 functions downstream to H3K56ac (Fig. 6D). In summary, these results suggest that the suppression of hst4Δ mutant phenotypes by expression of mcl1 is dependent on H3K56ac pathway.
Suppression of hst4Δ phenotypes by Mcl1 is dependent of H3K56ac. (A) hst4Δ (ROP58) expressing hst4, mcl1 or pSLF272 (empty vector), h3K56Q (ROP247) or h3K56R (ROP253) expressing mcl1 or pSLF272 (empty vector) and rad3Δ (ROP266) were grown to OD 1, five-fold serial dilutions were prepared and spotted on to EMM-Ura or EMM-Ura + 0.005% MMS plates. (B) hst4Δ (ROP58) expressing hst4, mcl1 or pSLF272 (empty vector), h3K56R (ROP253), h3K56Rhst4Δ (ROP275) expressing mcl1 or pSLF272 (empty vector) grown to OD 1, five-fold serial dilutions were prepared and spotted on to EMM-Ura or EMM-Ura + 0.005% MMS, or EMM-Ura + 0.01% MMS plates. (C) hst4Δ (ROP58) expressing hst4, mcl1 or pSLF272 (empty vector), h3K56Q (ROP247), h3K56Qhst4Δ (ROP276) expressing mcl1 or pSLF272 (empty vector) grown to OD 1, five-fold serial dilutions were prepared and spotted on to EMM-Ura or EMM-Ura + 0.005% MMS, or EMM-Ura + 0.01% MMS plates. (D) Indicated strains were grown to mid log phase at 30 °C, whole cell extracts were prepared and Western blot was performed using antibodies against H3K56ac and histone H3.
Hst4 regulates the expression of replisome component Mcl1
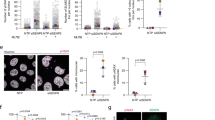
DNA replication is a tightly regulated process. The coupling between CMG helicase and DNA polymerases is a crucial determinant for DNA replication. In S. cerevisiae, it has been shown that Mcl1 homologue Ctf4 is a major target of H3K56ac pathway44. Our data showed that Mcl1 expression could suppress hst4Δ mutant phenotypes. Therefore, we hypothesized that Mcl1 levels might be low in hst4Δ mutants resulting in the slow S phase progression. And-1 is the human orthologue of fission yeast Mcl1. The anti-And-1 antibody raised against the conserved C-terminal region of And-1 was used to examine the protein levels of Mcl1. The C-terminal region of And-1 consists of multiple sepB domains, which are highly conserved amongst the members of And-1/Mcl1 family (Supplementary Fig. 2). To verify the specificity of And-1 antibody in yeast, extracts from wild type and mcl1Δ strains were prepared and analyzed by immunoblotting using anti-And-1 antibody. The 93 kDa Mcl1 protein was not recognized in mcl1 deletion mutants confirming the specificity of antibody (Fig. 7A). To examine whether Mcl1 expression is altered in hst4Δ mutant cells, we analyzed Mcl1 levels in wild type and hst4Δ mutant cells at the RNA and protein level. We observed a two-fold reduction in the protein levels of Mcl1 by Western blot in hst4Δ mutant cells as compared to wild type (Fig. 7B,C). We also observed corresponding two-fold reduction inMcl1 transcript levels in hst4Δ mutant cells compared to wild type cells, by quantitative RT-PCR (Fig. 7D). Next, the expression of Mcl1 in wild type and hst4Δ mutant strains bearing endogenous GFP-tagged mcl1 gene was analyzed using fluorescence microscopy. Our results confirmed a decrease of Mcl1 expression in hst4Δ strains (Fig. 7E,F). To further confirm the regulation of Mcl1 by Hst4, we checked whether overexpression of Hst4 can rescue Mcl1 expression. Overexpression of hst4 or mcl1 in hst4Δ mutant cells reveal that Hst4 is required for the expression of Mcl1(Fig. 7G). To test whether hyperacetylation of H3K56 in hst4Δ mutant cells affect Mcl1 expression, we analyzed mcl1 levels in h3K56Q and h3K56R mutants. Mcl1 expression was unaffected in these mutants (Fig. 7H). This result suggests that functional acetyl group at H3K56 might be required for down regulation of Mcl1. In order to check whether deletion of hst4 affect the expression of other replication proteins, we analyzed the expression of other replication proteins such as Pol1(Polα that binds to Mcl1), MCM complex (helicase component), and PCNA (clamp loader) in hst4Δ mutant cells. We did not observe any significant differences in the expression of other replication proteins (Fig. 7I). Collectively, these results reveal that Hst4 specifically regulates Mcl1 transcriptionally.
Hst4 regulates mcl1 expression. (A,B) Wild type (ROP191), hst4∆ (ROP58) and mcl1∆ (NYSPE19) strains were grown asynchronously to mid log phase in YES medium. Whole cell lysates were prepared and the levels of Mcl1 were monitored by Western blotting using anti-And-1 antibody. (C) The Mcl1 levels in WT and hst4Δ mutants were quantified relative to tubulin levels by densitometric analysis using Image J software. Error bars: standard deviation of the mean of densitometry values (three independent experiments). Statistical significance p < 0.05 between WT and hst4Δ strains is indicated with a single asterisk. (D) qPCR quantification of mcl1 levels in WT and hst4Δ mutants normalized with actin levels. Error bars represent standard deviation. (E) Live cells of wild type (YAP50) and hst4Δ mutant cells bearing endogenous GFP tagged mcl1 gene (DHP57) were visualized under confocal microscope. Bar = 2 µm. (F) Quantification of nuclear GFP signal in wild type versus hst4Δ described in (D). The intensity of Mcl1-GFP foci was analyzed using Image J software. Approximately 50 cells from three independent experiments against each genotype is plotted. Statistical significances between WT and hst4Δ indicated with single asterisk, p < 0.05. (G) Cell lysates from wild type strain (ROP191) or hst4∆ (ROP58) expressing hst4, mcl1 or pSLF272 (empty vector) were prepared and Mcl1 levels were analyzed by Western blotting using anti-And-1 antibody. (H) Logarithmically grown cultures of WT (ROP191), hst4Δ (ROP58), h3K56Q (ROP247) and h3K56R (ROP253) strains were lysed and Western blot was performed to check the levels of Mcl1. (I) WT(ROP191) and hst4∆ (ROP58) strains were grown asynchronously. Whole cell lysates were prepared and the levels of Mcl1, Pol1, PCNA, MCMs were monitored by Western blotting using individual antibodies. (J) Cell lysates from WT strains untreated or treated with MMS were analyzed by Western blot using anti-And-1antibody. (K) Assessment of Mcl1 and Hst4 levels by Western blot in WT (ROP191) and hst4∆ (ROP58) cells untreated or treated with either CPT or bleomycin or HU. For all the Western blots, tubulin was used as loading control. The amount of Mcl1 was quantitated relative to tubulin. The average relative intensity of bands was calculated from three independent experiments.
Mcl1 expression is reduced upon MMS treatment
It has been reported that expression of Hst4 is down regulated on MMS treatment13. To investigate the regulation of Mcl1 by Hst4, the expression of Mcl1 and Hst4 was analyzed in wild type cells treated with different DNA damaging agents, by immunoblotting. Interestingly, Hst4 levels were down regulated on MMS, bleomycin and HU treatment but not on CPT treatment13. We observed that the Mcl1 levels were also down regulated on MMS treatment (Fig. 7J). However, when cells were exposed to other DNA damaging agents, we did not observe any significant changes in Mcl1 expression (Fig. 7K).
Conservation of the regulation of Ctf4/Mcl1/And-1 by sirtuins from yeast to human cells
The functions of Mcl1 in replication and sister chromatid cohesion are conserved from budding yeast to vertebrates. We investigated whether the expression of human And-1(Mcl1 orthologue) is regulated by sirtuins in mammals. Mammals have seven sirtuins12. Mammalian SIRT1 and SIRT2 are localized in both cytoplasm and nucleus5,45,46, SIRT3 is mitochondrial and SIRT6 is a predominant nuclear protein47,48. We transfected HeLa cells with siRNA to knockdown SIRT1, SIRT2, SIRT3, and SIRT6 proteins and checked the levels of And-1 by immunoblotting. Depletion of SIRT2 reduced And-1 expression significantly compared to other sirtuins (Fig. 8A,B). To further confirm the regulation of And-1 by SIRT2, we transfected SIRT2 shRNA construct targeted to 3′ end of endogenous mRNA in HeLa cells and overexpressed with FLAG-SIRT2. We observed rescued And-1 expression on overexpression of FLAG-SIRT2 (Fig. 8C). To examine whether human And-1 is down regulated on MMS treatment and on treatment with other DNA damaging agents, cell extracts were prepared from either untreated or treated HeLa cells and analyzed by Western blot using anti-And-1 antibody (Fig. 8D). We observed significant decrease in And-1 level upon MMS, bleomycin, and HU treatments (Fig. 8E). These results indicate that SIRT2 depletion leads to decreased And-1 expression in mammals suggesting that this regulatory mechanism is evolutionarily conserved.
Human Sirtuin2 regulates Mcl1/And-1 expression. (A) HeLa cells were transfected with scramble, SIRT1, SIRT2, SIRT3 and SIRT6 siRNA. At 48 h post transfection, whole cell extracts were prepared and And-1 expression was detected by Western blot. The expression of indicated sirtuins was detected by respective antibodies. (B) Western blot showing depletion of hSIRT2 reduces And-1 expression. (C) HeLa cells were transfected with SIRT2 shRNA targeted to 3′ end of endogenous mRNA for 48 h and then transfected with Flag-SIRT2 for 24 h. Whole cell lysates were analyzed for indicated proteins by Western blot (D) Western blot showing And-1 expression in cell lysates from HeLa cells untreated or treated with 0.005% MMS. (E) Western blot showing And-1 expression in HeLa cells treated with indicated damaging agents. The amount of And-1 was quantitated relative to tubulin. The average relative intensity of bands is from three independent experiments. (F) Model showing how decrease in Mcl1 level mediated by down regulation of Hst4 may cause stalling of replication fork to preserve genomic integrity. In wild-type cells, Mcl1 couples replicative helicase and polymerase leading to efficient DNA synthesis. Under replicative stress such as MMS, Hst4 is down regulated resulting in decrease in level of Mcl1 causing uncoupling and stalling of replication fork to prevent DNA synthesis.
Discussion
We have identified a novel regulatory role of sirtuin family HDAC Hst4 in the regulation of Mcl1, which acts as a hub for replication proteins42. In the absence of hst4, cells progress through S phase slowly as their DNA is damaged (Fig. 1). This slowing of S phase could be mediated via activation of intra S phase checkpoint, which provides cells time to repair the damaged DNA before its replication37,49,50. Our earlier work has shown synthetic lethal interaction between the mediator of intra S phase checkpoint cds1 and hst413. Many studies directed towards understanding the mechanisms of the S phase delay indicate that replication origin firing and fork progression contribute to reducing the rate of DNA replication. Here, we present data showing reduction of DNA synthesis on deletion of hst4, indicating it is required for DNA replication (Fig. 2). Interestingly, in fission yeast, similar phenotype results from reduced levels of a component of replicative MCM helicase Mcm2, which stalls replication fork progression and also cause S phase delay33,36,37. Studies towards understanding the cause of S phase delay phenotype in S. pombe and other eukaryotes indicate that it could be either defect in origin firing or fork progression or both33,36,51. However, not much is known about the role of sirtuins in slowing of DNA replication. In budding yeast, Sir2, a sirtuin family HDAC, has been implicated in negative regulation of DNA replication52. On contrary, two other studies on HDACs showed that Rpd3, Sir2, and Hst1 promote replication initiation at many origins53,54. In humans, SIRT1 has been shown to function in initiation of DNA replication55,56. Although these reports indicate the role of sirtuins in DNA replication, the mechanisms are not very clear. In order to uncover novel functions of Hst4, we performed a high-copy suppressor screen and identified Mcl1 (Fig. 3A), a protein which interacts with Polα and also functions in establishing sister chromatid cohesion. In Schizosaccharomyces pombe, mcl1 was first reported as an essential gene required for maintaining genome stability and deletion of which caused chromosome segregation defects32. However, another report showed that cells lacking mcl1 were viable but displayed sick phenotype29. The budding yeast orthologue of Mcl1, Ctf4 is a multifunctional protein involved in maintaining genomic integrity but not an essential gene22. Recent reports have shown that Ctf4 is involved in coupling replicative CMG helicase to Polα43. Here we show expression of mcl1 partially suppresses the S- phase delay phenotype observed in hst4Δ mutant cells. We report reduced expression of mcl1 in hst4 deletion mutants (Fig. 7). Our results have shown for the first time that Hst4 is required for efficient DNA replication. Reduction in the expression of replication proteins affect the replication process36. Limited mcl1 expression may lead to accumulation of ssDNA and uncoupling of lagging and leading strand replication. The presence of increased Rad22 foci in the hst4Δ cells especially in the S/early G2 phase (Fig. 2) also indicate the accumulation of high amount of ssDNA. The total levels of Polα and Mcm remained unchanged in hst4Δ mutants (Fig. 7H). Budding yeast CTF4 mutants fail to stabilize the helicase complex thereby resulting in defective DNA synthesis57. Human And-1 depletion also leads to replication defects26,27. We demonstrated that expression of Mcl1 suppresses the growth defects and restores MMS and HU sensitivity of hst4Δ mutants but not CPT sensitivity of hst4Δ cells. DNA damage by MMS causes stalling of replication forks and leads to activation of Cds1 dependent replication checkpoint, while CPT traps Topoisomerase I on DNA and leads to collapse of replication forks due to replisome run off58. Suppression of sensitivity of hst4Δ cells to MMS and HU by Mcl1 suggests that Hst4 may function in replication fork stabilization and recovery. The differential sensitivity to various damaging agents also points towards distinct roles for Hst4 in these repair pathways. We propose that recovery of replication forks after CPT treatment may require Hst4 but is independent of Mcl1 function.
Hst4 deacetylates the histone H3K56 after its acetylation dependent incorporation into chromatin3,16. The dynamic regulation of H3K56ac is required for cell survival on replication stress as H3K56R or H3K56Q mutants, which mimic constitutive deacetylated and acetylated states respectively, are sensitive to DNA damage. The phenotype of cells lacking hst4 are attributed to hyperacetylated chromatin and are similar to H3K56R and H3K56Q mutants13. Here, we showed that expression of mcl1 could rescue the phenotypes of hst4Δ mutants. However, our data showed the sensitivity of these mimics to DNA damaging agents, are not recovered by expression of mcl1, indicating functional acetyl group might be required for phenotype suppression. Previous reports have shown that acetyl lysine and glutamine are not perfect mimic due to their structural differences59. The expression of mcl1 does not alter the H3K56ac levels, suggesting that mcl1 functions downstream of H3K56ac. This study depicts that Mcl1 may also function in H3K56 acetylation pathway as in the case of S. cerevisiae44,60,61. Additionally, work in S. cerevisiae shows that suppression of hst3Δ hst4Δ is brought about by deletion of ctf4Δ, which is contrary to our observation in fission yeast where deletion of hst4 increase survival of mcl1Δ cells on replication stress59. This difference could be partly explained by the fact that in budding yeast, ctf4Δ mutants have milder phenotypes compared to hst3Δ hst4Δ, whereas S. pombe mcl1Δ mutants are more sensitive compared to hst4Δ mutants suggesting additional roles played by Mcl1.
The suppression of hst4Δ mutant phenotypes by expression of mcl1 is further correlated to its low cellular levels (Fig. 7). Additionally, Mcl1 is regulated by Hst4 in response to replication stress generated by MMS treatment. In S. pombe, it has been shown that upon replication stress, cell signals for degradation of replisome components to maintain genomic integrity62. Genetic analyses in S. cerevisiae suggest that in the presence of replicative stress H3K56 acetylation uncouples the Cdc45–Mcm2-7–GINS DNA helicase complex and DNA polymerases through the replisome component Ctf4. However, they could not detect significant decrease in Ctf4 level under replication stress44. This pathway is dependent on regulation of H3K56ac by Hst3p and Hst4p and represents a key mechanism for maintenance of genome stability61. However, our results have indicated that there could be indeed a direct role of Hst4 in the regulation of Mcl1 at the transcriptional level. Future studies towards understanding the molecular players that are involved in transcriptional regulation of Mcl1, along with Hst4, needs to be carried out. Although currently there is no evidence, it is tempting to speculate how reduction of mcl1 expression may affect DNA replication process on replicative stress. In S. pombe, Mcl1 has been shown to interact with Polα. However, whether Mcl1 interacts with Mcm complex and GINS complex has not been studied. We propose that Mcl1 couples replicative helicase to DNA polymerase in S. pombe, and on replicative stress reduction of Hst4 levels leads to decreased mcl1 expression (Fig. 7), causing uncoupling of replicative CMG helicase from polymerase to stall DNA synthesis, thus maintaining genomic integrity (Fig. 8F). Sirtuins are conserved in higher eukaryotes. Mammalian sirtuin SIRT2 plays a major role in maintaining genomic integrity63. Human SIRT2 deacetylates histone H3 on lysine 56, a signature chromatin mark involved in DNA replication and repair64. Our study shows that depletion of SIRT2 leads to reduced And-1 expression, a conserved regulatory mechanism of replication protein regulation by sirtuins. These data points out that SIRT2 might be the functional human homologue of fission yeast Hst4.
We have uncovered the novel Hst4-Mcl1 axis for regulation of DNA replication on replicative stress. Our results indicate that this pathway might be conserved in mammalian cells. The knowledge of such regulatory mechanism involving sirtuins during replicative stress will be useful in designing therapeutics against diseases, such as cancer where sirtuins and And-1 are deregulated.
Materials and Methods
Yeast Strains, media and growth conditions
Yeast strains used in this study are listed in Table 1. Standard techniques were used for growth, transformation and genetic manipulations65. S. pombe strains were grown in yeast extract plus supplements (YES) or Edinburgh minimal media (EMM) at 32 °C on plate or in liquid media. Transformations were done using lithium acetate protocol. 10 millilitres of culture were grown to an optical density OD600 = 1. The cells were washed with 10 ml of sterile water once followed with 5 ml of Tris-EDTA (TE) plus 0.1 M lithium acetate. Cells were resuspended in 0.1 ml of TE plus 0.1 M lithium acetate and incubated for 1 h on a roller drum at 32 °C. 5 µl of 10 mg/ml carrier DNA (salmon sperm DNA) and 1 µg of plasmid DNA was added to 0.1 mL of cells and incubated at 32 °C for 30 min. Then, 0.7 ml of polyethylene glycol solution (40% polyethylene glycol) was added to the cells and incubated at 32 °C for 1 h. The cells were heat shocked for 5 min at 42 °C, resuspended in 0.2 ml of water and plated on EMM plates supplemented without uracil.
Cloning of replication proteins
The mcl1 gene was amplified and cloned in the BamH1 and Xba1 sites of the pRO314 plasmid. The mcl1 was also cloned into the Xho1 and Not1 sites of pSLF272 plasmid. The mcm4 and sld5 genes were cloned in the Xho1 and Not1 sites of pSLF272 plasmid.
Cell lines
HeLa cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 U/mL penicillin and streptomycin in a humidified 5% CO2 incubator at 37 °C.
DNA damage treatment of yeast cells
Cells were grown to OD600 = 1. Cultures were subjected to fivefold serial dilution and spotted on YES or EMM plates containing different concentrations of 0.01% MMS or 10 mM HU or 10 µM CPT. The plates were placed in 30 °C incubator for 4–5 days.
Protein Preparation and Western blotting
Total cell lysates were prepared from 10 mL culture of S. pombe. Cells were re-suspended in 200 µL of lysis buffer containing 50 mM HEPES, 500 mM NaCl, 5 mM EDTA, 0.1% NP-40, 10% glycerol, 1% protease inhibitor cocktail. Cells were lysed by glass beads using bead beater. Crude extracts were clarified by centrifugation and proteins were estimated through bradford method. Samples were prepared by pre-heating proteins in SDS sample buffer (50 mM Tris pH 7.5, 5 mM EDTA, 5% SDS, 10% glycerol, 0.5% β-mercaptoethanol, 0.05% bromophenol blue) and resolved by SDS-polyacrylamide gel electrophoresis (PAGE) followed by Western blotting to detect specific proteins.
Live cell Microscopy
Cells expressing Mc1l-GFP and Rad22-YFP were grown to mid-log phase. Cells were pelleted and resuspended in PBS. Live cell miscroscopy was performed on a Confocal microscope (Zeiss, LSM700). Quantification of foci was done by three independent experiments and atleast 300 cells were counted individually. The cell cycle position analysis was performed as earlier66. Briefly, Calcofluor was used to stain the septa and hoechst staining was performed to count the nucleus.
Immunofluorescence
Immunofluorescence was performed as described previously35. Briefly, logarithmically growing cells (50 ml) were labelled for 30 min at 25 °C in media containing 150 µg/ml BrdU [Sigma]. Cells were fixed in methanol/paraformaldehyde fixative for 30 min, washed in PBS and then treated with 0.5 mg/ml Zymolyase 20 T and 1 mg/mL lysing enzymes in PEMS for 10 min. After washing in PBS, cells were resuspended in 1 ml of 4 M HCl and incubated for 10 min to denature the DNA. Cells were washed extensively in PBS then blocked in PBS with 10% foetal calf serum for 1 h. Cells were incubated overnight in BrdU antibody [BD Biosciences] at 1:50 in PBS with 10% foetal calf serum and 0.05% Tween-20. Cells were then washed in PBS and incubated with α-mouse-AlexaFluor 488 at 1:500 in PBS with 10% foetal calf serum and 0.05% Tween-20 for 2 h. Cells were washed and resuspended in PBS then put on cover slips previously treated with poly-l-lysine. DNA was detected with 4–6′ diamidino-2-phenylindole (DAPI). Cells were visualized under confocal microscope.
Measurement of DNA content by flow cytometry
Yeast strains bearing a temperature sensitive cdc25-22 mutation were used for synchronization. The wild type cdc25-22 and cdc25-22 hst4Δ temperature-sensitive cells transformed with either hst4 or mcl1 were arrested in G2 at 36 °C for 4 h and then released at 25 °C for 4 hours and samples were collected every 30 minutes. For G1 arrest, yeast strains bearing a temperature sensitive cdc10-v50 mutation were used for synchronization. The wild type cdc10-v50 and cdc10-v50 hst4Δ temperature-sensitive cells arrested in G1 at 36 °C for 4 h and then released at 25 °C for 4 hours and samples were collected every 30 minutes. Cells were fixed with 70% ethanol and stained with propidium iodide (PI). Flow cytometry was performed on FACS Scan instrument using Cell Quest software. Histograms were generated using Flow JO (7.6.5).
Septation Index
The cdc25-22 (ROP204) and cdc25-22 hst4Δ (ROP216) strains were grown at 25 °C to log phase and synchronized in G2 by shifting the cells to 36 °C for 4 h. Cells were then shifted to 25 °C and mitotic progression was determined by 4′,6′-diamidino-2-phenylindole (DAPI) and calcofluor (50 µg/ml) staining. Three hundred cells from each time point were counted and septation index was determined by calculating the percentage of septated cells.
Slot blot analysis
The slot blot analysis was performed as described previously67. Genomic DNA from BrdU incorporated yeast cells was isolated with a commercial kit (zymoresearch). The amount of purified DNA was determined with a nanodrop-spectrophotometer. 500 ng of genomic DNA was made single-stranded by incubating with 10 volumes of 0.4 N NaOH solution for 30 min at room temperature. The denatured DNA was placed on ice and neutralized by an equal volume of 1 M Tris·HCl (pH 6.8). The single-stranded neutralized DNA was diluted in 100 µL water to obtain a series of concentrations of DNA (50 ng, 25 ng and 12.5 ng) and then slot dot blotted onto a nitrocellulose membrane (Amersham) and fixed by ultraviolet cross-linker Stratalinker (Stratagene, La Jolla, CA). To visualize the BrdU signal, the membrane was blocked in 5% non-fat milk and incubated with mouse anti-BrdU monoclonal antibody and the signal was detected by chemiluminiscence. Signal intensities were quantified by densitometric analysis to determine the fold change in BrdU incorporation in the hst4Δ mutants compared to wild type.
RNA isolation and quantitative real-time polymerase chain reaction
RNA was isolated by acid phenol method68. Total RNA was used for cDNA synthesis using Superscript III reverse transcriptase (Invitrogen, USA) and the above prepared cDNA was used for RT-qPCR using EvaGreen qPCR Mastermix (GBiosciences, USA). Each sample was run in triplicate and amplification was detected using 7500 Real Time PCR system (Applied Biosystems, USA). Transcripts were normalized to Actin by using −∆∆CT method69. Primer sequences used in this assay for Mcl1 are F: AGCTAGTGATGAAACAGCAG and R: GATTCTGCCTCTAAAGAGGC.
AND-1 rescue experiment
SIRT2 shRNA targeted to 3′ end of SIRT2 mRNA was generated by cloning double stranded oligonucleotides in pGFP-V-RS plasmid, using BamHI and HindIII. The following oligonucleotides were ordered with overhangs: 5′GATCCGCTTATTGGAGACAAATTAAAAACATGTGCTGTCTGTTTTTAATTTGTCTCCAATAAGCTTTTTA 3′complementary strand 5′AGCTTAAAAAGCTTATTGGAGACAAATTAAAAACAGACAGCACATGTTTTTAATTTGTCTCCAATAAGC-3′. HeLa cells were transfected with both vector control and SIRT2 shRNA vector using Lipofectamine 2000 and media change was done after 12 h. Cells were incubated for 48 h before transfecting cells with Flag-SIRT2 for another 24 h.
References
Li, B., Carey, M. & Workman, J. L. The role of chromatin during transcription. Cell 128, 707–19 (2007).
Peterson, C. L. & Laniel, M. A. Histones and histone modifications. Curr Biol 14, R546–51 (2004).
Chen, C. C. et al. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 134, 231–43 (2008).
Yang, X. J. & Seto, E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol 9, 206–18 (2008).
Sauve, A. A., Wolberger, C., Schramm, V. L. & Boeke, J. D. The biochemistry of sirtuins. Annu Rev Biochem 75, 435–65 (2006).
Yuan, H. & Marmorstein, R. Structural basis for sirtuin activity and inhibition. J Biol Chem 287, 42428–35 (2012).
Bosch-Presegue, L. & Vaquero, A. Sirtuin-dependent epigenetic regulation in the maintenance of genome integrity. FEBS J 282, 1745–67 (2015).
Dai, Y. & Faller, D. V. Transcription Regulation by Class III Histone Deacetylases (HDACs)-Sirtuins. Transl Oncogenomics 3, 53–65 (2008).
Wierman, M. B. & Smith, J. S. Yeast sirtuins and the regulation of aging. FEMS Yeast Res 14, 73–88 (2014).
Blander, G. & Guarente, L. The Sir2 family of protein deacetylases. Annu Rev Biochem 73, 417–35 (2004).
Durand-Dubief, M. et al. Specific functions for the fission yeast Sirtuins Hst2 and Hst4 in gene regulation and retrotransposon silencing. EMBO J 26, 2477–88 (2007).
Freeman-Cook, L. L. et al. The Schizosaccharomyces pombe hst4(+) gene is a SIR2 homologue with silencing and centromeric functions. Mol Biol Cell 10, 3171–86 (1999).
Haldar, D. & Kamakaka, R. T. Schizosaccharomyces pombe Hst4 functions in DNA damage response by regulating histone H3 K56 acetylation. Eukaryot Cell 7, 800–13 (2008).
Adkins, M. W., Carson, J. J., English, C. M., Ramey, C. J. & Tyler, J. K. The histone chaperone anti-silencing function 1 stimulates the acetylation of newly synthesized histone H3 in S-phase. J Biol Chem 282, 1334–40 (2007).
Maas, N. L., Miller, K. M., DeFazio, L. G. & Toczyski, D. P. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol Cell 23, 109–19 (2006).
Masumoto, H., Hawke, D., Kobayashi, R. & Verreault, A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436, 294–8 (2005).
Vempati, R. K. et al. p300-mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals. J Biol Chem 285, 28553–64 (2010).
Clemente-Ruiz, M., Gonzalez-Prieto, R. & Prado, F. Histone H3K56 acetylation, CAF1, and Rtt106 coordinate nucleosome assembly and stability of advancing replication forks. PLoS Genet 7, e1002376 (2011).
Li, Q. et al. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134, 244–55 (2008).
Wurtele, H. et al. Histone H3 lysine 56 acetylation and the response to DNA replication fork damage. Mol Cell Biol 32, 154–72 (2012).
Chang, D. Y., Shi, G., Durand-Dubief, M., Ekwall, K. & Lu, A. L. The role of MutY homolog (Myh1) in controlling the histone deacetylase Hst4 in the fission yeast Schizosaccharomyces pombe. J Mol Biol 405, 653–65 (2011).
Kouprina, N. et al. CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Mol Cell Biol 12, 5736–47 (1992).
Gambus, A. et al. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J28, 2992–3004 (2009).
Wang, J., Wu, R., Lu, Y. & Liang, C. Ctf4p facilitates Mcm10p to promote DNA replication in budding yeast. Biochem Biophys Res Commun 395, 336–41 (2010).
Zhu, W. et al. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev 21, 2288–99 (2007).
Im, J. S. et al. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc Natl Acad Sci USA 106, 15628–32 (2009).
Yoshizawa-Sugata, N. & Masai, H. Roles of human AND-1 in chromosome transactions in S phase. J Biol Chem 284, 20718–28 (2009).
Natsume, T. et al. A DNA polymerase alpha accessory protein, Mcl1, is required for propagation of centromere structures in fission yeast. PLoS One 3, e2221 (2008).
Tsutsui, Y. et al. Genetic and physical interactions between Schizosaccharomyces pombe Mcl1 and Rad2, Dna2 and DNA polymerase alpha: evidence for a multifunctional role of Mcl1 in DNA replication and repair. Curr Genet 48, 34–43 (2005).
Williams, D. R. & McIntosh, J. R. Mcl1p is a polymerase alpha replication accessory factor important for S-phase DNA damage survival. Eukaryot Cell 4, 166–77 (2005).
Mamnun, Y. M., Katayama, S. & Toda, T. Fission yeast Mcl1 interacts with SCF(Pof3) and is required for centromere formation. Biochem Biophys Res Commun 350, 125–30 (2006).
Williams, D. R. & McIntosh, J. R. mcl1+, the Schizosaccharomyces pombe homologue of CTF4, is important for chromosome replication, cohesion, and segregation. Eukaryot Cell 1, 758–73 (2002).
Willis, N. & Rhind, N. Regulation of DNA replication by the S-phase DNA damage checkpoint. Cell Div 4, 13 (2009).
Tercero, J. A. & Diffley, J. F. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412, 553–7 (2001).
Hodson, J. A., Bailis, J. M. & Forsburg, S. L. Efficient labeling of fission yeast Schizosaccharomyces pombe with thymidine and BUdR. Nucleic Acids Res 31, e134 (2003).
Liang, D. T., Hodson, J. A. & Forsburg, S. L. Reduced dosage of a single fission yeast MCM protein causes genetic instability and S phase delay. J Cell Sci 112(Pt 4), 559–67 (1999).
Kumar, S. & Huberman, J. A. On the slowing of S phase in response to DNA damage in fission yeast. J Biol Chem 279, 43574–80 (2004).
Ostermann, K., Lorentz, A. & Schmidt, H. The fission yeast rad22 gene, having a function in mating-type switching and repair of DNA damages, encodes a protein homolog to Rad52 of Saccharomyces cerevisiae. Nucleic Acids Res 21, 5940–4 (1993).
Kim, W. J. et al. Rad22 protein, a rad52 homologue in Schizosaccharomyces pombe, binds to DNA double-strand breaks. J Biol Chem 275, 35607–11 (2000).
Noguchi, E., Noguchi, C., Du, L. L. & Russell, P. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol Cell Biol 23, 7861–74 (2003).
Adams, C., Haldar, D. & Kamakaka, R. T. Construction and characterization of a series of vectors for Schizosaccharomyces pombe. Yeast 22, 1307–14 (2005).
Villa, F. et al. Ctf4 Is a Hub in the Eukaryotic Replisome that Links Multiple CIP-Box Proteins to the CMG Helicase. Mol Cell 63, 385–96 (2016).
Simon, A. C. et al. A Ctf4 trimer couples the CMG helicase to DNA polymerase alpha in the eukaryotic replisome. Nature 510, 293–7 (2014).
Luciano, P., Dehe, P. M., Audebert, S., Geli, V. & Corda, Y. Replisome function during replicative stress is modulated by histone h3 lysine 56 acetylation through Ctf4. Genetics 199, 1047–63 (2015).
North, B. J. & Verdin, E. Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS One 2, e784 (2007).
Tanno, M., Sakamoto, J., Miura, T., Shimamoto, K. & Horio, Y. Nucleocytoplasmic shuttling of the NAD+ -dependent histone deacetylase SIRT1. J Biol Chem 282, 6823–32 (2007).
Onyango, P., Celic, I., McCaffery, J. M., Boeke, J. D. & Feinberg, A. P. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci USA 99, 13653–8 (2002).
Nakagawa, T. & Guarente, L. Sirtuins at a glance. J Cell Sci 124, 833–8 (2011).
Cobb, J. A., Shimada, K. & Gasser, S. M. Redundancy, insult-specific sensors and thresholds: unlocking the S-phase checkpoint response. Curr Opin Genet Dev 14, 292–300 (2004).
Willis, N. A. et al. Identification of S-phase DNA damage-response targets in fission yeast reveals conservation of damage-response networks. Proc Natl Acad Sci U S A 113, E3676–85 (2016).
Kumar, S. & Huberman, J. A. Checkpoint-dependent regulation of origin firing and replication fork movement in response to DNA damage in fission yeast. Mol Cell Biol 29, 602–11 (2009).
Pappas, D. L. Jr., Frisch, R. & Weinreich, M. The NAD(+)-dependent Sir2p histone deacetylase is a negative regulator of chromosomal DNA replication. Genes Dev 18, 769–81 (2004).
Yoshida, K. et al. The histone deacetylases sir2 and rpd3 act on ribosomal DNA to control the replication program in budding yeast. Mol Cell 54, 691–7 (2014).
Weber, J. M., Irlbacher, H. & Ehrenhofer-Murray, A. E. Control of replication initiation by the Sum1/Rfm1/Hst1 histone deacetylase. BMC Mol Biol 9, 100 (2008).
Fatoba, S. T. et al. Human SIRT1 regulates DNA binding and stability of the Mcm10 DNA replication factor via deacetylation. Nucleic Acids Res 41, 4065–79 (2013).
He, H., Yu, F. X., Sun, C. & Luo, Y. CBP/p300 and SIRT1 are involved in transcriptional regulation of S-phase specific histone genes. PLoS One 6, e22088 (2011).
Tanaka, H. et al. Ctf4 coordinates the progression of helicase and DNA polymerase alpha. Genes Cells 14, 807–20 (2009).
Xu, Y. & Her, C. Inhibition of Topoisomerase (DNA) I (TOP1): DNA Damage Repair and Anticancer Therapy. Biomolecules 5, 1652–70 (2015).
Kamieniarz, K. & Schneider, R. Tools to tackle protein acetylation. Chem Biol 16, 1027–9 (2009).
Collins, S. R. et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446, 806–10 (2007).
Celic, I., Verreault, A. & Boeke, J. D. Histone H3 K56 hyperacetylation perturbs replisomes and causes DNA damage. Genetics 179, 1769–84 (2008).
Roseaulin, L. C. et al. Coordinated degradation of replisome components ensures genome stability upon replication stress in the absence of the replication fork protection complex. PLoS Genet 9, e1003213 (2013).
Kim, H. S. et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell 20, 487–99 (2011).
Das, C., Lucia, M. S., Hansen, K. C. & Tyler, J. K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 459, 113–7 (2009).
Moreno, S., Klar, A. & Nurse, P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194, 795–823 (1991).
Ansbach, A. B. et al. RFCCtf18 and the Swi1-Swi3 complex function in separate and redundant pathways required for the stabilization of replication forks to facilitate sister chromatid cohesion in Schizosaccharomyces pombe. Mol Biol Cell 19, 595–607 (2008).
Bhaskara, S. et al. Histone deacetylases 1 and 2 maintain S-phase chromatin and DNA replication fork progression. Epigenetics Chromatin 6, 27 (2013).
Jang, Y. K. et al. A simple and efficient method for the isolation of total RNA from the fission yeast Schizosaccharomyces pombe. Biochem Mol Biol Int 37, 339–44 (1995).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–8 (2001).
Acknowledgements
We thank Susan Forsburg, Yashuhiro Tsutsui, Takashi Toda and A-Lien Lu for generously providing strains. Annapurna and Bala for the technical assistance in microscopy and FACS respectively. We thank Rohinton Kamakaka for supporting the pilot suppressor screen experiment. We thank Ms Lakshmi Manasa for proofreading this manuscript. Sincere thanks to members of LCBE. This study was supported by a grant from the Department of Biotechnology (DBT), Ministry of Science and Technology, India (Grant BT/PR13489/GBD/27/271/2010). The research was also partially funded by grant from Centre for DNA Fingerprinting and Diagnostics (CDFD), India. http://www.dbtindia.nic.in/. LK, RV supported by CSIR-Research Fellowships and CDFD Core Funding. SA is supported by UGC fellowship.
Author information
Authors and Affiliations
Contributions
D.H. conceived, designed the study and analyzed data from most of the experiments. L.K. performed and analyzed the experiments presented in Figures 1(A), 2(A–D), 3(A–C), 4(A), 5(B–C), 6(A,D) and 7. S.A. performed and analyzed the experiments presented in Figures 1(B–D), 2(E–G), 3(D–F), 4(B), 5(A), 6(B–C). L.K. and R.V. designed, performed and analyzed the experiments presented in Figure 8. D.H., L.K. and S.A. wrote the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Konada, L., Aricthota, S., Vadla, R. et al. Fission Yeast Sirtuin Hst4 Functions in Preserving Genomic Integrity by Regulating Replisome Component Mcl1. Sci Rep 8, 8496 (2018). https://doi.org/10.1038/s41598-018-26476-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-26476-4
This article is cited by
-
Effects of sirtuins on the riboflavin production in Ashbya gossypii
Applied Microbiology and Biotechnology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.