Abstract
Complete tyrosine kinase 2 (TYK2) deficiency has been previously described in patients with primary immunodeficiency diseases. The patients were infected with various pathogens, including mycobacteria and/or viruses, and one of the patients developed hyper-IgE syndrome. A detailed immunological investigation of these patients revealed impaired responses to type I IFN, IL-10, IL-12 and IL-23, which are associated with increased susceptibility to mycobacterial and/or viral infections. Herein, we report a recessive partial TYK2 deficiency in two siblings who presented with T-cell lymphopenia characterized by low naïve CD4+ T-cell counts and who developed Epstein-Barr virus (EBV)-associated B-cell lymphoma. Targeted exome-sequencing of the siblings’ genomes demonstrated that both patients carried novel compound heterozygous mutations (c.209_212delGCTT/c.691C > T, p.Cys70Serfs*21/p.Arg231Trp) in the TYK2. The TYK2 protein levels were reduced by 35% in the T cells of the patient. Unlike the response under complete TYK2 deficiency, the patient’s T cells responded normally to type I IFN, IL-6, IL-10 and IL-12, whereas the cells displayed an impaired response to IL-23. Furthermore, the level of STAT1 was low in the cells of the patient. These studies reveal a new clinical entity of a primary immunodeficiency with T-cell lymphopenia that is associated with compound heterozygous TYK2 mutations in the patients.
Similar content being viewed by others
Introduction
Interferons (IFN) and other cytokines, which play important roles in multiple innate and adaptive immune responses, transduce signals via the JAK-STAT pathway. When the cytokines bind and induce the dimerization of their receptors, receptor-associated Janus kinases (JAKs) become phosphorylated and activated. The activated JAKs then phosphorylate downstream substrates, the signal transducers and activators of transcription (STAT) molecules, which subsequently dimerize and translocate to the nucleus to activate the transcription of specific genes. Mutations of the genes encoding components of the JAK-STAT pathway cause various immunological disorders, including increased susceptibility to infection, such as in growth hormone insensitivity syndrome, severe combined immunodeficiency, and others1,2,3,4,5,6,7,8,9,10,11.
One of the JAKs, tyrosine kinase 2 (TYK2), which is associated with the receptors of type I IFN, interleukin (IL)-6, IL-10, IL-12 and IL-23, plays a central role in the signal transduction of these cytokines12,13. TYK2 deficiency was first described in a 22-year-old Japanese male patient who developed symptoms of hyper-IgE syndrome (HIES) with susceptibility to various pathogens, including Staphylococcus, mycobacteria and herpes simplex virus2. Genomic DNA sequencing analysis in the patient has revealed a homozygous frame-shift mutation in the TYK2 gene, which resulted in a frameshift at codon 90 with the premature termination of translation. Therefore, the patient’s cells expressed no functional TYK2 protein that could be detected via immunoblot analysis. The cells derived from the TYK2-deficient patient displayed nearly abolished responses to type I IFN, IL-12, IL-23, IL-6 and IL-10. More recently, the comprehensive immunological investigation of seven other TYK2-deficient patients has been reported14. Unlike the first TYK2-deficient patient, cells from these TYK2-deficient patients displayed an impaired but not abolished response to type I IFN, IL-12, IL-23 and IL-10. The study suggested that the susceptibility to intracellular bacterial and/or viral infections identified in all the TYK2-deficient patients was caused by impaired responses to IL-12 and type I IFN14. All of these accumulating reports have elucidated the functional impacts of a complete TYK2-deficiency on clinical outcomes. However, little is known regarding the functional impact of other TYK2 variants (e.g., insertion, deletion and substitution).
In this study, we present two cases of patients who had immunodeficiency associated with novel heterozygous mutations in the four-point-one, ezrin, radixin, moesin (FERM) domain region of TYK2. Unlike previous reports on TYK2 deficiency, the patients exhibited severe T-cell lymphopenia characterized by low naïve CD4+ T-cell counts. Furthermore, both patients developed Epstein-Barr virus (EBV)-associated B-cell lymphoma. We analyzed the responses to cytokines in the cells of the patient. The cellular functions partially resemble but are less defective than those previously described in the TYK2-deficient patient.
Results
Characteristics of Patients
Patient 1 (P1) was referred as a 15-year-old boy born to non-consanguineous parents (Fig. 1a). He had a history of varicella and recurrent parotitis. He had received the following vaccinations: bacille Calmette-Guérin (BCG); diphtheria-pertussis-tetanus (DPT); polio; influenza; measles-rubella (MR) and Japanese encephalitis. A parotid biopsy was performed at 12 years of age and exhibited diffuse medium- to large-sized B-cell proliferation (Fig. 2a–c). Furthermore, a lymphoepithelial lesion was detected (Fig. 2a). The lesion was diagnosed as mucosa-associated lymphoid tissue (MALT)-type B-cell lymphoma (stage III). He was treated with chemotherapy according to the reference standard, and he initially obtained remission. At the age of 16 years, EBV-associated diffuse large B-cell lymphoma (stage III) was diagnosed by parotid gland biopsy. Large B cells were expanded monoclonally (Fig. 2e–g). These cells as well as a biopsy sample at the age of 12 years were positive for EBV-encoded RNAs (EBERs) (Fig. 2d,h). Positivity for anti-EBV viral capsid antigen (VCA) IgG was observed without anti-VCA IgM antibodies, and the EBV DNA copy number in the peripheral blood was 5,200 copies/mL (Table 1). The patient died at the age of 16 years due to a severe graft-versus-host disease (GVHD) after allogeneic bone marrow transplantation (BMT).
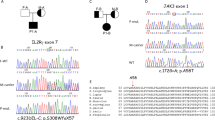
Identification of TYK2 compound heterozygous mutations in siblings with primary immunodeficiency. (a) Pedigree of a family in which compound heterozygous mutations in TYK2 were identified. Squares and circles denote males and females, respectively. Closed boxes indicate affected individuals, and a diagonal bar represents a deceased individual. (b) Validation by Sanger sequencing of the TYK2 mutations in the patients and their parents. (c) Schematic representation of the TYK2 protein. (d) western blot analysis of TYK2 protein expression in EBV-BCLs established from the PBMCs of a healthy donor and the TYK2-mutated patient P2. ß-tubulin served as a loading control. n = 3 EBV-BCLs per genotype. Data represent mean ± SEM. **P < 0.01. P values were derived from 2-tailed Student’s t-test. Full-length immunoblots are presented in Supplementary Fig. S6.
Histologic analysis of biopsy specimens from the patients. A biopsy specimen from the parotid gland of P1 at 12 years of age (a–d) and at 16 years of age (e–h). The parotid gland of P2 at 13 years of age (i–l); the left nasopharynx of P2 at 16 years of age (m–p). Staining with hematoxylin and eosin (HE) and immunostaining with anti-CD20 antibody demonstrates the infiltration of CD20+ lymphocytes. An in situ hybridization study demonstrated EBV-encoded RNAs (EBERs). The cell clonality was assessed by in situ hybridization for κ and λ mRNAs.
Patient 2 (P2), the 14-year-old sister of P1 exhibited similar symptoms. She had a history of varicella and recurrent otitis media with effusion. She had received vaccines of BCG, DPT, polio, influenza and MR. At the age of 13 years, a biopsy of the right parotid gland was histologically diagnosed as EBV-associated B-cell lymphoma (stage II). The biopsy exhibited a diffuse lymphoid infiltrate composed of small to medium lymphocytes. The lymphoid cells included monocytoid B cells and centrocyte-like cells, reminiscent of MALT-type B-cell lymphoma (Fig. 2i–k). The cells were positive for EBERs (Fig. 2l). P2 received chemotherapy and achieved provisional remission. At the age of 16 years, she developed EBV-associated B-cell lymphoma (stage II) in the left nasopharynx (Fig. 2m–p). Immunostaining for light chains revealed predominant staining of the κ light chain, indicating monoclonal proliferation of the tumor cells (Fig. 2o). Similar to P1, the patient was positive for anti-EBV VCA IgG in the absence of anti-VCA IgM antibodies. A PCR analysis revealed EBV infection with a viral load of 4,467 copies/mL in the peripheral blood (Table 1). P2 underwent another round of chemotherapy and obtained remission. Currently, P2 is being followed up in Japan and exhibited persistent increase in EBV viral load (1.4 × 104 copies/mL) even 15 months after the final chemotherapy. The results of serologic screening for HIV and PCR screening for cytomegalovirus and human herpes virus-6 were negative in both patients.
Immunophenotyping of the peripheral blood mononuclear cells (PBMCs) revealed that both patients had significant T-cell lymphopenia characterized by low naïve CD4+ T-cell (CD4+CD31+CD45RA+, recent thymic emigrant cells) counts (Table 2). The frequencies of B cells and NK cells were close to those of age-matched controls. High EBV antibody titers indicated intact B-cell function in the patients (Table 1). The NK cell function could not be successfully evaluated because of inadequate cellular materials. Both patients had impaired T-cell proliferative responses following phytohemagglutinin(PHA) stimulation (Table 2). The immunoglobulin levels in both patients’ sera were normal with the exception of elevated IgA and slightly reduced IgE levels in P2.
To evaluate the lymphocyte proliferation response in P2, T cells were stimulated with anti-CD3 monoclonal antibody (mAb) plus anti-CD28 mAb-coated beads and expanded in culture with IL-2. The result showed that normal T-cell proliferation was restored in P2 compared with the cells derived from her parents and a control donor (Supplementary Fig. S1).
Identification of causative mutations
Initially, an autosomal recessive inheritance of primary immunodeficiency was suspected because the parents were non-consanguineous and had no symptoms related to immune disorders. Therefore, we performed targeted exome sequencing for P1, P2 and their parents to identify causative or candidate genes. In total, 7 Mbp of exome sequences (targeting approximately 2,761 genes registered as disease-implicated in the Human Genome Mutation Database) were enriched and analyzed with the Illumina MiSeq. The sequencing resulted in 1.1–1.3 Gbp of raw sequence data per patient. In total, 87.8–90.0% of the targeted bases were sequenced with at least 20-fold coverage depth. A genetic variant analysis resulted in the detection of 4,816 (P1) and 4,829 (P2) variants. These variants were further filtered using the following criteria: (1) linked to missense, frameshift, loss or gain of a stop codon, initiator codon, in-frame insertion/deletion, or splice-site mutations; (2) detected as uncommon variants (a frequency of less than 5%, which was set according to previous reports)15,16 in the Asian population; (3) inherited in an autosomal recessive pattern; (4) shared by both of the patients. The results revealed that 16 variants in four genes, TYK2, ANK2, RYR1 and DSPP fulfilled the above criteria (Supplementary Table S1). Of these variants, seven synonymous and four non-synonymous mutations were identified in two genes, RYR1 and DSPP. As biological functions of the two genes are likely irrelevant to primary immunodeficiency, we narrowed down to five non-synonymous variants in TYK2 and ANK2 to identify the causative mutations. Of note, our analysis of structural variants (SVs) linked to the T-cell lymphopenia displayed 22 rare SVs that are observed with the expected frequency of less than 5% in either the 1000 Genome Project data or the Exome Aggregation Consortium (ExAC) data17,18. However, any of these SVs were not associated with a recessive inheritance (Supplementary Table S2).
In addition, since our patients developed aggressive EBV-associated B-cell lymphoma that is rarely observed in children, we further validated exome sequencing data for the genes of which mutations have been reported in the patients suffering from EBV-associated lymphoproliferative diseases, including EBV-associated lymphoma; IL-2 inducible tyrosine kinase (ITK)19, CD2720, SH2 domain protein 1A (SH2D1A)21, X-linked inhibitor-of-apoptosis protein (XIAP)22, Wiskott-Aldrich syndrome protein (WASP)23, coronin, actin-binding protein 1A (CORO1A)24,25, mammalian sterile 20-like kinase-1 (MST1)26 and magnesium transporter 1 (MAGT1)27. All of these genes are involved in the survival and/or differentiation of T cells. The results showed that no nonsense mutations were detected in these genes. Nevertheless, further investigations are required for verifying an alternative possibility that unknown other genetic mutations might be associated with aggressive development of EBV-associated B-cell lymphoma under low TYK2 activity.
Our verification of the five mutations by Sanger sequencing indicated that two variants of the TYK2 gene were validated whereas three mutations of the ANK2 were false positive on the MiSeq analysis. As shown in Fig. 1b, both parents are heterozygous for the TYK2 gene localized on chromosome 19: c.209_212 del GCTT in the mother and c.691C > T in the father. Their children, the two patients, carry compound heterozygous mutations in TYK2 (Fig. 1b). The first TYK2 mutation, c.209_212 deletion, resulted in a frame-shift mutation and generated a premature stop codon at amino acid residue 90. The homozygous form of this mutation was reported previously2 and registered in the dbSNP database (rsID: rs770927552) with a minor allele frequency of 0.00006. The second TYK2 mutation is a c.691C > T transition and resulted in a p.Arg231Trp substitution in the FERM domain region (Fig. 1c). This transition mutation has been registered in the dbSNP database (rsID: rs201917359) with a minor allele frequency of 0.0008.
These results suggest that the causative mutations are strongly linked to the compound heterozygous mutations, namely the c.209_212 del GCTT and the c.691C > T in the TYK2 genes of these two patients.
Assessment of type I IFN response
To probe possible association between the TYK2 variants and cellular functions, we measured the TYK2 mRNA and protein levels in EBV-transformed human B-cell lines (EBV-BCLs) prepared from PBMCs of healthy donors and P2. The result indicated that no significant reduction of the mRNA level was observed in P2 compared with healthy donor controls (Supplementary Fig. S2). In contrast, the immunoblot analysis showed that the TYK2 protein level (Fig. 1d, Supplementary Fig. S2) was low in P2 (47–51% relative to the control donors). The reduction was similarly observed in primary CD4+ T cells derived from P2, whereas the TYK2 protein levels in CD4+ T cells from her parents were similar to the levels in the healthy control (Supplementary Fig. S3). These results suggest no significant impact of single allelic variation for the c.209_212 del GCTT or the c.691C > T on the TYK2 expression.
Next, to assess the function of TYK2 in the patient, the levels of STAT1 (Tyr 701), STAT2 (Tyr 690) and STAT3 (Tyr 705) phosphorylation in patient-derived EBV-BCLs and T cells upon stimulation with IFN-α and IFN-ß were analyzed by immunoblotting (Fig. 3a) and compared by quantifying the gel images (Fig. 3c,e and g). The relative expression level of STAT1 was less than 50% with or without IFN stimulation in the patient-derived cells compared with the control cells (Fig. 3b). However, the ratio of phosphorylated STAT1 (pSTAT1) to STAT1 was 2-fold higher in the P2-derived cells than in the control cells, suggesting that the patient-derived cells responded to type I IFN normally or even better than the healthy donor cells (Fig. 3c). In contrast, the relative expression levels of STAT2 (Fig. 3d) and the IFN-induced phosphorylated STAT2 (Fig. 3e) were similar between the P2 cells and the control cells. The levels of STAT3 exhibited a pattern similar to that of STAT2 (Fig. 3f,g).
Type I IFN-induced STAT activation in EBV-BCLs from a healthy donor and the patient. (a) Total STAT and tyrosine-phosphorylated STAT (pSTAT) protein levels were analyzed by western blotting. Cells were stimulated with IFN-α (1000 U/mL) or IFN-ß (500 U/mL) for 15 min. (b–g) Based on three independent analyses, the expression levels of STAT proteins (b,d,f) and pSTAT proteins (c,e,g) in the patient-derived EBV-BCLs were compared with those in untreated healthy donor-derived cells (n = 3 EBV-BCLs per genotype). Data represent mean ± SEM. **P < 0.01, *P < 0.05. P values were derived from 2-tailed Student’s t-test. Full-length immunoblots are presented in Supplementary Fig. S7.
Furthermore, the transcriptional upregulation of type I IFN-inducible genes, including IRF1, SOCS1, SOCS3, STAT1 and TAP1, were analyzed. One of the JAK-STAT pathway components, STAT1, is itself an IFN-inducible gene product28. The transcriptional upregulation of type I IFN-inducible genes in response to type I IFN was comparable to that of the control cells (Fig. 4 and Supplementary Fig. S4). Among the five genes, only low STAT1 expression significantly differed between the patient’s and healthy donor cells (Fig. 4). These results suggest that the TYK2 compound heterozygous mutations are associated with a low level of constitutive STAT1 mRNA expression.
Analysis of type I IFN-inducible gene induction in EBV-BCLs from a healthy donor and the patient.Cells were stimulated with IFN-α (1000 U/mL), IFN-ß (500 U/mL) or without any IFN for 2 h. The cDNAs generated from the total RNA were quantified with qPCR assays. The expression levels of IRF1, SOCS1, SOCS3, STAT1 and TAP1 mRNAs were determined by normalizing each with cyclophilin B levels. The induction level is presented as n-fold expression that in the untreated healthy donor cell control, which was set as 1. The data are derived from three independent experiments using 3 EBV-BCLs per genotype. Data represent mean ± SEM. **P < 0.01. P values were derived from 2-tailed Student’s t-test.
Assessment of response to IL-6, IL-10, IL-12 and IL-23
In addition to the type I IFN signaling pathway, TYK2 is also involved in the IL-6 and IL-10 signaling pathways, which regulate a broad range of physiological responses through the activation of STAT3. To assess the IL-6 and IL-10 signaling pathways in the patient, the levels of phosphorylated STAT3 (Tyr 705) proteins in the patient-derived T cells stimulated with IL-6 or IL-10 were analyzed. As depicted in Fig. 5a, the patient’s cells responded similarly or even more efficiently to IL-6 and IL-10 compared to the control cells.
STAT activation in response to IL-6, IL-10, IL-12 and IL-23 in T cells from a healthy donor and the patient.Total STAT and tyrosine-phosphorylated STAT (pSTAT) protein levels were analyzed by western blotting. (a) The cells were stimulated with IL-6 (10 ng/mL) or IL-10 (10 ng/mL) for 15 min. (b) The cells were stimulated with IL-12 (10 U/mL) or IFN-α (1000 U/mL) for 15 min. (c) The cells were stimulated with IL-23 (500 ng/mL) or IFN-α (1000 U/mL) for 15 min. Full-length immunoblots are presented in Supplementary Fig. S8–10. Based on three independent analyses using same batch of CD4+ T cells, STAT proteins and pSTAT proteins expression levels were calculated relative to those of untreated healthy donor-derived cells. Data represent mean ± SEM. *P < 0.05. P values were derived from 2-tailed Student’s t-test.
Finally, we tested the effects of the TYK2 variants on the IL-12 and IL-23 signaling pathways because previous studies using TYK2-deficient cells from an immunodeficient patient or TYK2-knockout mice have shown that a lack of TYK2 affects the response to IL-12 and IL-232,14,29,30. IL-12 and IL-23 are pro-inflammatory cytokines that mediate signaling via STAT4 and STAT3, respectively. A comparative analysis of STAT4 phosphorylation (Tyr 693) after IL-12 stimulation revealed a slight reduction in phosphorylation in the P2 cells compared with the control cells (Fig. 5b); however, the difference was not significant. In contrast, the IL-23-dependent phosphorylation of STAT3 (Tyr 705) was reduced in the P2-derived cells (41%) relative to that in the control cells (Fig. 5c). These observations demonstrated that the signaling pathway of IL-23 but not that of IL-12 was significantly impaired in the patient’s cells.
Discussion
The present study investigated two patients with primary immunodeficiency who developed a type of EBV-associated B-cell lymphoma that is rare in children. Through the targeted exome sequencing analysis of the patients’ genomic DNA, we identified compound heterozygous mutations (c.209_212 del GCTT /c.691 C > T, p.Cys70Serfs*21/p.Arg231Trp) in the TYK2-encoding gene. The four-base deletion locus of the TYK2 gene (c.209_212 del GCTT) was previously reported in the first TYK2-deficient patient2. The missense mutation (c.691 C > T), which causes an Arg231Trp mutation, was previously registered in the dbSNP database; however, the mutational effect was unknown.
Our immunoblot analysis revealed that the patient-derived cells expressed nearly 50% (47–51%) of the TYK2 protein compared with healthy donor cells (Fig. 1d and Supplementary Fig. S2). In contrast, cells isolated from the parents carrying either of the TYK2 mutations (c.209_212 del GCTT or c.691 C > T) exhibited a level of TYK2 protein similar to that of the cells with wild-type TYK2 alleles. These data indicated that these TYK2 mutations might affect the TYK2 protein expression level, and the mutations in the parent’s cells may be compensated by the wild-type allele. The levels of TYK2 mRNA in the patient were similar to those in the healthy donors (Supplementary Fig. S2). Interestingly, the second mutation (c.691 C > T) that we identified in this study results in the substitution of the Arg231 within the FERM domain of TYK2 for a tryptophan. Mapping of the residue in the domain structure (PDB 4PO6) showed that it is located at positively charged TYK2 surface that is critical for maintaining the FERM F2 conformation stability as well as interacting with plasma/nuclear membrane (Supplementary Fig. S5)31. Therefore, the missense mutation (c.691 C > T) may lead to disruption of intracellular localization and/or intracellular stability of TYK2 protein. This mechanistic scenario is strongly supported by a previous finding that alanine substitutions of Arg231-233-235 impaired TYK2 nuclear localization32.
The first Japanese patient with TYK2 deficiency reported by Minegishi et al.2 was characterized by atopic dermatitis with highly elevated serum IgE (2100 IU/mL) and diagnosed with HIES. In contrast, serum IgE concentrations of 23 IU/mL and 5 IU/mL for P1 (at the age of 16 years) and P2 (at the age of 11 years), respectively, were noted in this study; these levels were even lower than those for the healthy control. More recently, another report of seven TYK2-deficient patients has shown that the core clinical phenotype of TYK2 deficiency is due to mycobacterial and/or viral infections and not to HIES14.
In addition, the immunological investigation of T cells derived from these patients displayed impaired responses to IFN-α/ß, IL-12, IL-23 and IL-1014. Upon IFN-α/ß stimulation, a lower level of STAT1 phosphorylation and no STAT3 phosphorylation was detected in the TYK2-deficient patients14. In contrast, our patient-derived cells displayed similar levels of phosphorylated STAT1 and STAT3 proteins in response to IFN-α/ß compared with the TYK2-proficient control cells. Additionally, the transcriptional induction of type I IFN-inducible genes in response to IFN-α/ß was normal in the patient’s PBMCs. The reduced expression of only STAT1 mRNA and protein was observed in the patient’s cells compared to the control cells. Such STAT1 protein reduction has also been observed in the complete TYK2-deficient patients and in TYK2-/- mice as previously reported2,14,29. Because basal expression levels of IFN-inducible genes, including STAT1, are maintained by low amounts of constitutively secreted IFN-ß in immune cells28,33, this minimal constitutive activation of STAT signaling might enable immune cells to maintain a rapid immune response to infections. Therefore, the reduced STAT1 expression observed in this study could be attributed to the absence or decreased basal level of TYK2 protein expression that regulates the IFN signaling pathway. Previous reports have suggested apparently inconsistent mechanisms in immunodeficient patients with low levels of STAT1 expression caused by an exonic splicing enhancer mutation in STAT14,5 or by insufficiently stable STAT1 protein folding6. Consequently, those patients exhibited susceptibility to mycobacterial and viral infections and/or displayed progressive losses in lymphocyte numbers and functions. Further studies are warranted to better understand whether the clinical features of the patients in the present study can be attributed to the effects of reduced STAT1 levels or other unidentified factors associated with TYK2 mutation.
In contrast to the aforementioned immunological features that are in common between our patient and the previously reported patients with complete TYK2 deficiency, there are two immunological signatures that are different. First, STAT3 activation is noted upon IL-6/IL-10 stimulation. Kreins et al. have shown that STAT3 phosphorylation after IL-10 stimulation was impaired in cells from complete TYK2-deficient patients, whereas STAT3 phosphorylation upon IL-6 stimulation was normal in those cells, suggesting that TYK2 is indispensable for IL-10 signaling but not for IL-6 signaling14. However, we observed no significant impairment in the response to either IL-10 or IL-6 in our patient’s T cells. Residual TYK2 activity might account for the normal IL-6/IL-10 signaling in our patient. Second, approximately normal responses to IL-12 were observed in our patient-derived cells regarding STAT4 phosphorylation, whereas a drastic impairment of IL-12 signaling was detected in the complete TYK2-deficient patients2,14. Disorders involving IL-12-dependent signal transduction may result in mycobacterial diseases, such as BCG infection or Salmonella infection34. Indeed, BCG and Salmonella infections that are likely due to impaired IL-12 responses have been previously observed in TYK2-deficient patients2,3,14. However, the patients in our present study did not have any episodes of BCG or Salmonella infection. This clinical evidence strongly indicates that residual TYK2 activity involving the IL-12 signaling pathway was sufficiently maintained to induce protective immunity against those bacteria.
An intriguing observation of this study was that the patients presented atypical clinical features that have not been observed in TYK2-deficient patients. In addition, as a prominent clinical feature of our patients, T-cell lymphopenia that is characterized by low naïve CD4+ T-cell counts was similarly observed in our two patients. The reduction in the T-cell count observed in our patients was not found in previous reports of TYK2-deficient patients or TYK2-deficient mice2,3,14,29,30. There are several reports that mutations in a variety of genes cause primary immunodeficiency associated with T-cell lymphopenia6,24,26,35,36 and that these T-cell lymphopenias are attributed mainly to an impaired proliferative capacity of T cells26,35,36 or a high expression of apoptosis-related proteins6,26. In the present study, we obtained inconsistent results for the proliferative capacity of T cells in the patient. The proliferative capacity of PBMCs after PHA stimulation were impaired (Table 2), whereas T cells isolated from the patients proliferated in response to anti-CD3 and anti-CD28 antibodies and exhibited no apparent signs of apoptosis (Supplementary Fig. S1). Interestingly, a previous in vitro study has shown that the inhibitor that primarily targets TYK2 altered the development and proliferation of CD4+ T cells, suggesting an involvement of TYK2 in T-cell development37. Hence, low expression and/or assumingly altered membrane localization of TYK2 due to Arg231Trp mutation in the patients’ cells may give a negative effect on T-cell development. Still, we cannot eliminate an alternative possibility that lymphopenia observed in the patient could be due to the EBV infection38.
Several patients displayed either decreased NK cell activity or a reduced number of natural killer T (NKT) cells, suggesting that low NK/NKT-cell activities are correlated with a susceptibility to EBV-associated lymphoproliferative disease19,20,22,23,24. A recent study using mice has suggested a tumor-surveillance function of TYK2 independent of the kinase activity of this protein39. The decreased cytotoxic activities of NK and NKT cells in TYK2-/- mice are considered to be involved in virus-induced tumor development40. Further assessment of the function of NK/NKT cells in patients will be required to gain a better understanding of the pathogenic mechanisms of EBV-associated B-cell lymphoma. Notably, other studies reported finding gain-of-function mutations of TYK2 in cells derived from a patient with CD30-positive lymphoproliferative disorders41 and T-cell acute lymphoblastic leukemia (T-ALL)-derived cell lines42. These observations suggest that in T-ALL patients, the survival of aberrant cells is attained through increased STAT1 phosphorylation followed by the upregulation of the anti-apoptotic BCL2 protein42. To clarify the tumorigenic mechanism in these patient’s cells, further detailed analysis of the tumor tissue is necessary.
In conclusion, we identified novel compound heterozygous mutations in TYK2 associated with primary immunodeficiency characterized by a rare type of EBV-associated B-cell lymphoma and T-cell lymphopenia. Complete TYK2 deletion genotypes have been previously reported; however, this is the first report of mutations that are associated with reduced TYK2 expression. The results of cytokine response study using patient’s cells demonstrated partial loss of function of TYK2 which might be involved in the unique clinical phenotypes in the patients. The combination of a rare type of EBV-associated B-cell lymphoma with severe T-cell lymphopenia associated with compound heterozygous TYK2 mutations represents a distinct disease entity.
Methods
Subjects
We investigated two siblings (P1 and P2) from non-consanguineous parents originating from and living in Japan. Both patients were chemotherapy-free at the time of the immunophenotyping. The immunophenotyping was conducted 3 years after the last chemotherapy of P1 that was treated with JPLSG B-NHL03 Group 343 and 4 months after the last chemotherapy of P2 that was treated with FAB/LMB96 Group B44. Given that cells from P1, who died from GVHD, were no longer available, immunological characterization was only performed with the cells from P2. P2 was not treated with any immunosuppressive drugs at the time of the experiments. Twice blood collections were required for (A) making growth curves and generation of EBV-BCLs which is used for IFN responsiveness assay and (B) performing IL-6, IL-10, IL-12, IL-23 responsiveness assay. Experiment (A) was conducted 2 years and 10 months after the first chemotherapy with cyclophosphamide, vincristine, prednisolone, methotrexate and cytarabine. Experiment (B) was conducted 6 months after the second chemotherapy with 4 courses of rituximab, etoposide, prednisolone, vincristine, cyclophosphamide and doxorubicin.
Histologic examination
Biopsies of primary and relapsed parotid lesions and those of primary parotid and nasopharynx lesions were collected from P1 and P2, respectively. Each biopsy was embedded in paraffin for conventional histological and immunohistochemical analyses and was examined for morphological characteristics based on the WHO classification. Immunophenotyping were based primarily on CD20-positive expression. In situ hybridization analysis was performed to determine the EBERs and κ/λ expressions. The INFORM Kappa, Lambda and EBER Probes were purchased from Ventana Medical Systems.
Mutation screening
Genomic DNA of PBMCs isolated from EDTA-anticoagulated blood was obtained using the QIAamp DNA Blood Mini Kit (QIAGEN). Total RNA was isolated with the RNeasy Plus Mini Kit (QIAGEN), and used to synthesize cDNA with the SuperScript III First-Strand Synthesis System (Life Technologies) and random hexamers, according to the manufacturer’s protocol. Targeted exome sequencing was performed using the genomic DNA isolated from P1, P2 and their parents. We followed the manufacturer’s protocol for the Illumina TruSight Exome Kit to build a genomic library starting from 50 ng of the genomic DNA samples. The enriched libraries were sequenced to obtain 75-bp paired-end reads on an Illumina MiSeq. All the sequence data were aligned to the human reference genome sequence (GRCh37/hg19) with the Burrows-Wheeler Aligner (BWA) program (http://bio-bwa.sourceforge.net). Genetic variants were identified with the Genome Analysis Toolkit (https://software.broadinstitute.org/gatk/), followed by data annotation using the Illumina VariantStudio software. Analyses of the SVs from the exome sequencing data were performed using the programs, Pindel45, Delly46 and gridss (https://github.com/PapenfussLab/gridss), at their default settings. For Sanger DNA sequencing, coding regions of the TYK2 gene were amplified from the synthesized cDNA using the following primer set: forward (5′-TTTGAATTCTTGCTTGAGTTGACACAGGGAGCT-3′) and reverse (5′-TTTGCGGCCGCTCTCTAGACAGGAGTAAGGCACAC-3′). The resulting DNA fragment was cloned into the pCR-Blunt II TOPO vector (Life Technologies).
Antibodies and cytokines
The anti-CD19 and anti-CD27 mAbs were purchased from Sigma-Aldrich and the anti-CD20 mAb from Ventana Medical Systems. The antibodies against human TYK2, STAT1, STAT3 and STAT4, and their phosphorylated forms were purchased from Cell Signaling Technology. The antibodies directed against ß-tubulin, STAT2, phosphorylated STAT2 and horseradish peroxidase-conjugated rabbit polyclonal antibody were obtained from Abcam. The human IL-2, IL-12 and IL-23 recombinant proteins were purchased from Cell Signaling Technology, while IFN-α was obtained from Sigma-Aldrich, and IFN-ß, IL-6 and IL-10 were purchased from Peprotech.
Cell culture and stimulation
CD4+ T cells were prepared from the heparinized blood of patients and healthy donors as previously described47. Before stimulation with cytokines, the expanded CD4+ T cells were washed and serum-starved for 18 h in RPMI-1640 supplemented with 1% FBS. The cells were then stimulated with IFN-α (1000 U/mL), IFN-ß (500 U/mL), IL-6 (10 ng/mL), IL-10 (10 ng/mL), IL-12 (10 U/mL) or IL-23 (500 ng/mL) at 37 °C for 15 min.
Western blot analysis
After stimulation with cytokines, cells were harvested and lysed with lysis buffer [50 mM Tris (pH 8.0), 1% Triton X-100, 150 mM NaCl, 2 mM EDTA, 2 μg/mL aprotinin, 1 mM sodium orthovanadate, 100 μg/mL PMSF and 1 mM NaF], followed by sonication. The protein samples were subjected to 10% SDS-PAGE and transferred onto PVDF membranes (Millipore). The membranes were immunoblotted with primary antibodies followed by the appropriate secondary antibodies. The intensity of the immunoblotted bands was quantified using ImageQuant (GE Healthcare).
Gene expression analysis
For the analysis of IFN-inducible genes, EBV-BCLs and freshly isolated PBMCs were stimulated with IFN-α (1000 U/mL) or IFN-ß (500 U/mL) for 2 h. Quantification of the cDNA generated by reverse transcription from total RNA was performed using a real-time PCR assay in a Thermal Cycler Dice Real-Time System TP800 (Takara Bio) according to the manufacturer’s protocol for SYBR Premix DimerEraser (Takara Bio). Real-time PCR assays were employed to analyze the levels of cyclophilin B, TYK2, IRF1, SOCS1, SOCS3, STAT1 and TAP1 mRNA using each primer set listed in Supplementary Table S3. The expression level of each gene was determined using the standard curve method.
Statistics
All data are expressed as mean ± SEM. Statistical significance was determined using 2-tailed Student’s t-test. For all studies, values of P < 0.05 were considered statistically significant.
Study approval
Written informed consent for the genetic and functional investigations was obtained from the patients and their parents in accordance with local regulations, with approval by the ethics committee of Nagoya Medical Center (registration #2013-679). The experiments were conducted at the Clinical Research Center, National Hospital Organization Nagoya Medical Center, Japan.
Data Availability
Exome sequencing data were deposited in the Japanese Genotype-phenotype Archive (JGA)48 under accession number JGAS00000000098 under Type I security.
References
Casanova, J. L., Holland, S. M. & Notarangelo, L. D. Inborn errors of human JAKs and STATs. Immunity 36, 515–528, https://doi.org/10.1016/j.immuni.2012.03.016 (2012).
Minegishi, Y. et al. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity 25, 745–755, https://doi.org/10.1016/j.immuni.2006.09.009 (2006).
Kilic, S. S. et al. A patient with tyrosine kinase 2 deficiency without hyper-IgE syndrome. J Pediatr 160, 1055–1057, https://doi.org/10.1016/j.jpeds.2012.01.056 (2012).
Chapgier, A. et al. A partial form of recessive STAT1 deficiency in humans. J Clin Invest 119, 1502–1514, https://doi.org/10.1172/JCI37083 (2009).
Kong, X. F. et al. A novel form of human STAT1 deficiency impairing early but not late responses to interferons. Blood 116, 5895–5906, https://doi.org/10.1182/blood-2010-04-280586 (2010).
Sharfe, N. et al. Fatal combined immunodeficiency associated with heterozygous mutation in STAT1. J Allergy Clin Immunol 133, 807–817, https://doi.org/10.1016/j.jaci.2013.09.032 (2014).
Candotti, F. et al. Structural and functional basis for JAK3-deficient severe combined immunodeficiency. Blood 90, 3996–4003 (1997).
Notarangelo, L. D. et al. Of genes and phenotypes: the immunological and molecular spectrum of combined immune deficiency. Defects of the gamma(c)-JAK3 signaling pathway as a model. Immunological Reviews 178, 39–48, https://doi.org/10.1034/j.1600-065X.2000.17812.x (2000).
Bernasconi, A. et al. Characterization of immunodeficiency in a patient with growth hormone insensitivity secondary to a novel STAT5b gene mutation. Pediatrics 118, E1584–E1592, https://doi.org/10.1542/peds.2005-2882 (2006).
Kofoed, E. M. et al. Growth hormone insensitivity associated with a STAT5b mutation. New England Journal of Medicine 349, 1139–1147, https://doi.org/10.1056/NEJMoa022926 (2003).
Minegishi, Y. et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature 448, 1058–U1010, https://doi.org/10.1038/nature06096 (2007).
Velazquez, L., Fellous, M., Stark, G. R. & Pellegrini, S. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell 70, 313–322 (1992).
Strobl, B., Stoiber, D., Sexl, V. & Mueller, M. Tyrosine kinase 2 (TYK2) in cytokine signalling and host immunity. Front Biosci (Landmark Ed) 16, 3214–3232 (2011).
Kreins, A. Y. et al. Human TYK2 deficiency: Mycobacterial and viral infections without hyper-IgE syndrome. J Exp Med 212, 1641–1662, https://doi.org/10.1084/jem.20140280 (2015).
Altshuler, D. M. et al. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58, https://doi.org/10.1038/nature09298 (2010).
Ku, C. S., Loy, E. Y., Pawitan, Y. & Chia, K. S. The pursuit of genome-wide association studies: where are we now? Journal of Human Genetics 55, 195–206, https://doi.org/10.1038/jhg.2010.19 (2010).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285, https://doi.org/10.1038/nature19057 (2016).
Sudmant, P. H. et al. An integrated map of structural variation in 2,504 human genomes. Nature 526, 75, https://doi.org/10.1038/nature15394 (2015).
Huck, K. et al. Girls homozygous for an IL-2-inducible T cell kinase mutation that leads to protein deficiency develop fatal EBV-associated lymphoproliferation. Journal of Clinical Investigation 119, 1350–1358, https://doi.org/10.1172/jci37901 (2009).
Alkhairy, O. K. et al. Novel mutations in TNFRSF7/CD27: Clinical, immunologic, and genetic characterization of human CD27 deficiency. Journal of Allergy and Clinical Immunology 136, 703, https://doi.org/10.1016/j.jaci.2015.02.022 (2015).
Coffey, A. J. et al. Host response to EBV infection in X-linked lymphoproliferative disease results from mutations in an SH2-domain encoding gene. Nature Genetics 20, 129–135, https://doi.org/10.1038/2424 (1998).
Rigaud, S. et al. XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature 444, 110–114, https://doi.org/10.1038/nature05257 (2006).
Ochs, H. D. & Thrasher, A. J. The Wiskott-Aldrich syndrome. Journal of Allergy and Clinical Immunology 117, 725–738, https://doi.org/10.1016/j.jaci.2006.02.005 (2006).
Moshous, D. et al. Whole-exome sequencing identifies Coronin-1A deficiency in 3 siblings with immunodeficiency and EBV-associated B-cell lymphoproliferation. J Allergy Clin Immunol 131, 1594–1603, https://doi.org/10.1016/j.jaci.2013.01.042 (2013).
Stray-Pedersen, A. et al. Compound Heterozygous CORO1A Mutations in Siblings with a Mucocutaneous-Immunodeficiency Syndrome of Epidermodysplasia Verruciformis-HPV, Molluscum Contagiosum and Granulomatous Tuberculoid Leprosy. Journal of Clinical Immunology 34, 871–890, https://doi.org/10.1007/s10875-014-0074-8 (2014).
Nehme, N. T. et al. MST1 mutations in autosomal recessive primary immunodeficiency characterized by defective naive T-cell survival. Blood 119, 3458–3468, https://doi.org/10.1182/blood-2011-09-378364 (2012).
Li, F.-Y. et al. Second messenger role for Mg2+ revealed by human T-cell immunodeficiency. Nature 475, 471–U463, https://doi.org/10.1038/nature10246 (2011).
Gough, D. J., Messina, N. L., Clarke, C. J., Johnstone, R. W. & Levy, D. E. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity 36, 166–174, https://doi.org/10.1016/j.immuni.2012.01.011 (2012).
Karaghiosoff, M. et al. Partial impairment of cytokine responses in Tyk2-deficient mice. Immunity 13, 549–560 (2000).
Shimoda, K. et al. Tyk2 plays a restricted role in IFN alpha signaling, although it is required for IL-12-mediated T cell function. Immunity 13, 561–571 (2000).
Wallweber, H. J. A., Tam, C., Franke, Y., Starovasnik, M. A. & Lupardus, P. J. Structural basis of recognition of interferon-alpha receptor by tyrosine kinase 2. Nature Structural & Molecular Biology 21, 443–448, https://doi.org/10.1038/nsmb.2807 (2014).
Ragimbeau, J. et al. The receptor interaction region of Tyk2 contains a motif required for its nuclear localization. Journal of Biological Chemistry 276, 30812–30818, https://doi.org/10.1074/jbc.M103559200 (2001).
Ivashkiv, L. B. & Donlin, L. T. Regulation of type I interferon responses. Nat Rev Immunol 14, 36–49, https://doi.org/10.1038/nri3581 (2014).
Zhang, S. Y. et al. Inborn errors of interferon (IFN)-mediated immunity in humans: insights into the respective roles of IFN-alpha/beta, IFN-gamma, and IFN-lambda in host defense. Immunol Rev 226, 29–40, https://doi.org/10.1111/j.1600-065X.2008.00698.x (2008).
Zhang, Q. et al. Combined immunodeficiency associated with DOCK8 mutations. N Engl J Med 361, 2046–2055, https://doi.org/10.1056/NEJMoa0905506 (2009).
Martin, E. et al. CTP synthase 1 deficiency in humans reveals its central role in lymphocyte proliferation. Nature 510, 288–292, https://doi.org/10.1038/nature13386 (2014).
Works, M. G. et al. Inhibition of TYK2 and JAK1 Ameliorates Imiquimod-Induced Psoriasis-like Dermatitis by Inhibiting IL-22 and the IL-23/IL-17 Axis. Journal of Immunology 193, 3278–3287, https://doi.org/10.4049/jimmunol.1400205 (2014).
Schattner, A., Meshorer, A. & Wallach, D. Involvement of interferon in virus-induced lymphopenia. Cellular Immunology 79, 11–25, https://doi.org/10.1016/0008-8749(83)90046-1 (1983).
Prchal-Murphy, M. et al. In vivo tumor surveillance by NK cells requires TYK2 but not TYK2 kinase activity. Oncoimmunology 4, https://doi.org/10.1080/2162402x.2015.1047579 (2015).
Stoiber, D. et al. TYK2 is a key regulator of the surveillance of B lymphoid tumors. Journal of Clinical Investigation 114, 1650–1658, https://doi.org/10.1172/jci22315 (2004).
Velusamy, T. et al. A novel recurrent NPM1-TYK2 gene fusion in cutaneous CD30-positive lymphoproliferative disorders. Blood 124, 3768–3771, https://doi.org/10.1182/blood-2014-07-588434 (2014).
Sanda, T. et al. TYK2-STAT1-BCL2 Pathway Dependence in T-cell Acute Lymphoblastic Leukemia. Cancer Discovery 3, 564–577, https://doi.org/10.1158/2159-8290.cd-12-0504 (2013).
Tsurusawa, M. et al. Improved Treatment Results of Children With B-Cell Non-Hodgkin Lymphoma: A Report From the Japanese Pediatric Leukemia/Lymphoma Study Group B-NHL03 Study. Pediatric Blood & Cancer 61, 1215–1221, https://doi.org/10.1002/pbc.24968 (2014).
Patte, C. et al. Results of the randomized international FAB/LMB96 trial for intermediate risk B-cell non-Hodgkin lymphoma in children and adolescents: it is possible to reduce treatment for the early responding patients. Blood 109, 2773–2780, https://doi.org/10.1182/blood-2006-07-036673 (2007).
Ye, K., Schulz, M. H., Long, Q., Apweiler, R. & Ning, Z. M. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25, 2865–2871, https://doi.org/10.1093/bioinformatics/btp394 (2009).
Rausch, T. et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28, I333–I339, https://doi.org/10.1093/bioinformatics/bts378 (2012).
Imahashi, M. et al. Lack of Association between Intact/Deletion Polymorphisms of the APOBEC3B Gene and HIV-1 Risk. Plos One 9, https://doi.org/10.1371/journal.pone.0092861 (2014).
Kodama, Y. et al. The DDBJ Japanese Genotype-phenotype Archive for genetic and phenotypic human data. Nucleic Acids Research 43, D18–D22, https://doi.org/10.1093/nar/gku1120 (2015).
Shearer, W. T. et al. Lymphocyte subsets in healthy children from birth through 18 years of age: The pediatric AIDS clinical trials group P1009 study. Journal of Allergy and Clinical Immunology 112, 973–980, https://doi.org/10.1067/mai.2003.1778 (2003).
Schatorje, E. J. H. et al. Paediatric Reference Values for the Peripheral T cell Compartment. Scandinavian Journal of Immunology 75, 436–444, https://doi.org/10.1111/j.1365-3083.2012.02671.x (2012).
Duchamp, M. et al. B-cell subpopulations in children: National reference values. Immunity, Inflammation and Disease 2, 131–140 (2014).
Acknowledgements
We all appreciate the family for their continuous cooperation. The authors also thank Dr. Kazuhiro Matsuoka for helpful discussions and Dr. Jun Komano for establishing the EBV-BCLs. This work was partly supported by research funding from Grant-in-Aid for Young Scientists (B) from Japan Society for the Promotion of Science to M.N.
Author information
Authors and Affiliations
Contributions
M.N., H.H. and Y.I. designed the research; M.N., H.M., S.M. and T.K. performed the experiments; H.H., N.M. and K.H. treated the patients; M.N., H.H., N.M., N.A., H.M., S. M., T.K., M.M., H.O., A.H., W.S., Y.Y., K.H. and Y.I. analyzed the data; M.N., H.H. and Y.I. wrote the paper.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nemoto, M., Hattori, H., Maeda, N. et al. Compound heterozygous TYK2 mutations underlie primary immunodeficiency with T-cell lymphopenia. Sci Rep 8, 6956 (2018). https://doi.org/10.1038/s41598-018-25260-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-25260-8
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.