Abstract
The concentrations of tropospheric CO2 and O3 have been rising due to human activities. These rising concentrations may have strong impacts on soil functions as changes in plant physiology may lead to altered plant-soil interactions. Here, the effects of eCO2 and eO3 on the removal of polycyclic aromatic hydrocarbon (PAH) pollutants in grassland soil were studied. Both elevated CO2 and O3 concentrations decreased PAH removal with lowest removal rates at elevated CO2 and elevated O3 concentrations. This effect was linked to a shift in soil microbial community structure by structural equation modeling. Elevated CO2 and O3 concentrations reduced the abundance of gram-positive bacteria, which were tightly linked to soil enzyme production and PAH degradation. Although plant diversity did not buffer CO2 and O3 effects, certain soil microbial communities and functions were affected by plant communities, indicating the potential for longer-term phytoremediation approaches. Results of this study show that elevated CO2 and O3 concentrations may compromise the ability of soils to degrade organic pollutants. On the other hand, the present study also indicates that the targeted assembly of plant communities may be a promising tool to shape soil microbial communities for the degradation of organic pollutants in a changing world.
Similar content being viewed by others
Introduction
Global industrialization has led to an increase of tropospheric carbon dioxide (CO2) concentration from approximately 280 ppm in pre-industrial times to approximately 380 ppm nowadays, and it is expected to continue increasing in the future1,2. Alongside, the average surface ozone (O3) concentration has increased from an estimated pre-industrial value of 10 ppb to 20–45 ppb in the mid-latitudes of the northern hemisphere at a rate of 0.5–2% per year over the last decades2,3,4.
Industrialization has further led to a global pollution by organic pollutants, including polyaromatic hydrocarbons (PAHs)5. PAHs have mutagenic and carcinogenic properties, and show a high persistency in the environment6,7. Due to PAH contamination, huge areas are not suitable for agriculture or livestock anymore, and remediation of PAHs from soils is a priority goal to ensure food safety8,9. Among the proposed approaches, phytoremediation appears as an efficient and environment-friendly approach to remove PAHs from soils from large surfaces10. In most cases, phytoremediation of PAHs from soils was conducted by a single plant species11,12 and the mechanisms linking plants and PAH removal are still elusive. Moreover, it remains unclear whether and how global environmental change agents will affect phytoremediation of PAHs from soils.
In the present study, the potential effects of rising tropospheric O3 and CO2 concentrations on the natural ability of soils to degrade PAHs were investigated, and if such potential effects are altered by plant diversity. Soil microorganisms are important mediators of global change effects as several soil bacteria and fungi produce enzymes that break down PAHs, contributing to soil remediation. This effect is especially pronounced in microbial communities associated with plant roots13,14, where microbes are directly stimulated by the presence of labile plant-derived carbon. Since soil microbial communities and their activity are profoundly affected by plant community composition15,16,17,18, PAH removal may be influenced by changes in plant community composition and diversity. Plant diversity effects on soil biota may alter the functioning of soils, such as changes in the decomposition of organic matter19,20 and the sequestration of carbon in soil18. Increasing plant species richness may enhance the diversity of soil biota by combining different root morphology, root chemical composition, and temporal variability of resource inputs18,21. In addition, the functional composition of plant communities can drive belowground communities and processes22,23, e.g. through specific plant traits affecting nutrient availability24,25,26.
Both elevated CO2 (later: eCO2) and elevated O3 (later: eO3) have long been known to affect physiological and biochemical processes of plants and change rhizosphere conditions27,28. There is evidence that the composition and functioning of soil microbial communities change under eCO229,30 and eO331,32. A previous stuty showed that eCO2 increased soil bacterial abundance in soil contaminated with cadmium at various levels of concentration (0, 1.5, 3.0, 6.0 mg Cd kg−1soil) across the experimental period (2, 4, 6, 8 weeks)29, and eCO2 was reported to select for fungal communities that are more adapted to drought conditions30. Moreover, eO3 has been shown to eliminate the significant positive effect of eCO2 on cellobiohydrolase activity, but it did not alter the positive effect of eCO2 on N-acetylglucosaminidase activity31. Furthermore, eO3 substantially reduced the ectomycorrhiza colonization rate and ectomycorrhiza diversity in larch32. Tropospheric eCO2 has been shown to increase not only plant biomass, but also carbon inputs to soil, associated with higher soil microbial activity, and labile soil C representing elevated root exudation33. By contrast, eO3 decreases inputs of assimilates into the rhizosphere32, while both eCO2 and eO3 change the composition of root exudates released into the rhizosphere, thereby altering microbial biomass and activity in soils34. All these previous results suggest that plant diversity, eCO2, and eO3 may have strong interactive effects on ecosystem processes. As loss of plant diversity is likely to occur together with changes in tropospheric gas concentration, the present study for the first time tested their interactive effects on pollutant degradation, a central ecosystem function of soils15. Pollutant degradation is crucial as increasing anthropogenic activities pollute various ecosystems worldwide35,36.
Plant communities with different combinations of grasses, herbs, and legumes were set up in microcosms with PAHs-contaminated soil from a chemical plant in Nanjing, China. These microcosms were subjected to a full-factorial combination of ambient and predicted concentrations of CO2 and O3 in 20502. After ten weeks of cultivation, PAH residuals in soils, soil microbial biomass and composition, soil enzymes, and plant biomass were determined. We expected (1) plant functional group richness to increase plant productivity37 and soil microbial biomass and activity17,38, (2) eCO2 to increase plant productivity and soil microbial biomass and activity39,40, (3) eO3 to decrease plant biomass and soil microbial biomass and activity32, and (4) the three global change drivers to interactively influence plant biomass, soil microbial functions, and the degradation of PAHs, e.g., with plant diversity amplifying the effect of eCO241 or eCO2 buffering negative effects of eO342.
Results
PAH residuals in soil
Both eCO2 and eO3 increased total PAH residuals significantly (Table 1, Figs 1 and 2), i.e. decelerated PAH degradation, and also altered the composition of remaining PAHs (increased PC1 of PAHs; Fig. 2). CO2 × O3 had a significant interactive effect on total PAH residuals as remaining PAHs were lowest at aCO2 and aO3, but substantially increased by eCO2 and eO3 and highest at both eCO2 and eO3 (+43% in comparison to ambient conditions). Plant functional group richness had no significant effects on PAH residuals (Table 1). The PLFAs i17:0, cy 17:0, and i16:0 were most strongly associated with PAH removal, and Benzo(k)fluoranthene and Indene(1,2,3-c,d)pyrene were the most recalcitrant PAHs (Fig. 3). Gram-negative bacteria were positively associated with total PAH residuals and PC1 of PAH residuals, while Gram-positive bacteria strongly reduced total PAHs and PC1 of PAH residuals (Fig. 2).
Total amount of polycyclic aromatic hydrocarbons (PAHs) after the experiment as affected by elevated CO2, elevated O3, and plant diversity (0, 1, 2, 3 plant functional groups). Means ± SE (n = 4). amb, eCO2, eO3, and eCO2 + eO3 means that microcosms were incubated in chambers with ambient air, with elevated CO2, with elevated O3, and with elevated CO2 and O3, respectively. Bars with different letters vary significantly (Tukey’s HSD test, a <0.05).
Structural equation model showing the effects of eCO2, eO3, and plant functional group richness on soil microorganisms and polycyclic aromatic hydrocarbon (PAHs) residuals in the soil. Red arrows: negative relationships, blue arrows: positive relationships, asterisks on numbers indicate significant relationships (see Table S1 for details).
Heat map illustrating the relationships between different phospholipid fatty acids (PLFAs) and different polycyclic aromatic hydrocarbons (PAHs). Phen, BaP, Fluor, DahA, Ant, BbF, BaA, BghiP, Chr, Pyr, BkF, and I123cdP represent Phenanthrene, Benzo(a)pyrene, Fluoranthene, Dibenzo(a,h)anthracene, Anthracene, Benzo(b)fluoranthene, Benzo(a)anthracene, Benzo(g,h,i)perylene, Chrysene, Pyrene, Benzo(k)fluoranthene, and Indene(1,2,3-c,d)pyrene, respectively. Red plots: negative correlations, blue plots: positive correlations, white plots: no correlations.
Although the fungal PLFA 18:2ω6, 9 was very abundant in the experimental soil (Fig. S3), it played a minor role in PAH degradation (Fig. 3). Furthermore, Gram-positive bacteria were the most important group of soil microbes in degrading PAHs (Figs 2 and 3).
Soil enzymes and microorganisms
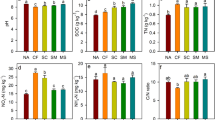
Both eCO2 and eO3 significantly reduced polyphenol oxidase activity, but plant functional group richness increased polyphenol oxidase activity (Table 1, Fig. S4b). However, enzyme activity depended on significant interactions between experimental factors (Table 1). CO2 × O3 had significant interactive effect on phenol oxidase and polyphenol oxidase activity. While the activity of phenol oxidase was lowest at aCO2 and aO3 and highest at eCO2 and aO3 (Fig. S4a), polyphenol oxidase activity was highest at aO3 and aCO2 or eCO2, but lowest at aCO2 and eO3 (Fig. S4b).
Furthermore, phenol oxidase activity was marginally affected by eO3 in the presence of 0 and 1 plant functional group, but increased phenol oxidase activity in the presence of 2 and 3 plant functional groups (significant O3 × plant functional group richness interaction). Phenol oxidase activity decreased with increasing plant functional group richness and was consistently higher at eCO2 and aO3, but did not vary with plant functional group richness and CO2 at eO3 (significant CO2 × O3 × plant functional group richness interaction). Moreover, the increase of polyphenol oxidase activity at eCO2 was most pronounced in the presence of two plant functional groups (significant CO2 × plant functional group richness interaction).
Elevated CO2 significantly reduced the biomass of Gram-positive bacteria (Table 1, Fig. S1a), and the composition of microbial communities changed as indicated by increased PC1 of microbial PLFAs (Table S1, Fig. 2). Furthermore, eO3 significantly reduced Gram-positive bacteria and Gram-negative bacteria (Tables 1, S1, Figs 2, S1a,b) and changed the composition of the soil microbial community (increased PC1 of microbial PLFAs; Table S1, Fig. 2). The biomass of Gram-positive bacteria, Gram-negative bacteria, and fungi increased significantly with increasing plant functional group richness (Table 1, Fig. S1a,b,c), which was also reflected by a significant change of soil microbial community composition (significantly reduced PC1 of microbial PLFAs; Table S1, Fig. 2). CO2 × O3 and CO2 × plant functional group richness had significant interactive effects on Gram-positive bacteria (Table 1). The biomass of Gram-positive bacteria was highest at aCO2 and aO3, lowest at aCO2 and eO3, and intermediate in the other treatments (Fig. S1a). Further, the biomass of Gram-positive bacteria increased with increasing plant functional group richness and was higher at aCO2, except in the treatment with two plant functional groups, where aCO2 and eCO2 had similar values.
The composition of microbial PLFAs corresponded to the functional composition of plant communities (Fig. S3). Plant communities containing legumes were more strongly associated with the PLFAs cy17:0, a15:0, 16:1ω7, i17:0, 18:0, i15:0, and cy19:0, almost all of them indicators of bacteria (except 16:1ω7 and 18:0). Plant communities containing herbs were more strongly associated with a different set of PLFAs, namely i16:0, 16:0, 18:1ω9t, 18:1ω9c, 20:4ω6, 16:1ω5, and 18:2ω6,9, with almost none of them being indicators of bacteria (except i16:0). Plant communities with grasses were associated with both bacterial indicator PLFAs and non-bacterial indicator PLFAs. (Fig. S3).
Discussion
The present study shows that elevated CO2 and O3 concentrations may erode essential ecosystem services like the degradation of pollutants in soil by inducing significant shifts in soil microbial community structure and enzyme activity. These detrimental effects were consistent across plant communities differing in functional diversity. However, pronounced alterations of microbial community structure along the functional plant diversity gradient suggests that targeted and trait-based phytoremediation may help to counteract detrimental global change effects in long-term approaches.
In contrast to our hypothesis (1) stating that plant functional group richness to increase plant productivity37, plant biomass declined with increasing plant functional group diversity in the present study. These results highlight the context-dependency of biodiversity–ecosystem function relationships43 and the need to study biodiversity effects under stressful conditions44. Nevertheless and in line with our hypothesis17,38, soil microbial biomass and activity increased with plant functional group diversity, stressing the significance of plant diversity for soil functions. Diverse plant communities are expected to produce and release a higher quantity and diversity of organic compounds into their rhizosphere, which may sustain higher soil microbial biomass and activity45. Using long-term data from a grassland biodiversity experiment, Lange et al. found higher plant biodiversity to increase rhizosphere carbon inputs into the soil microbial community resulting in increased microbial diversity and activity18. These findings are consistent with the results of increased biomass of Gram-positive, Gram-negative bacteria and fungi in the present study. A recent meta-analysis reported that plant diversity effects on soil microbial biomass C were strong in long-term experiments and across various environmental contexts46. The present study extends those findings by showing that bacterial and fungal biomass increased with plant diversity, which also altered the activities of different soil enzymes. Similar with this study, Steinauer et al. found that soil microbial biomass and some enzyme activities increased with increasing plant diversity47.
In contrast to our hypotheses, no effect of plant functional group richness on PAH removal was observed. This is in line with a previous study that species richness had no significant effect on 14C-phenanthrene mineralization48. However, structural equation modeling (SEM) reveals a range of processes coupling plant functional group richness and PAH degradation. The SEM showed that plant diversity altered soil microbial community composition and favored both Gram-positive (accelerating PAH degradation) and Gram-negative bacteria (decelerating PAH degradation; Fig. 2). Plant community may therefore be an important driver of PAH degradation, even if lumping community composition into functional group richness doesn’t provide the adequate explanatory power. The present study suggests that it may be possible to assemble plant communities showing a high phytoremediation by steering soil microbial communities.
Elevated CO2 tended to increase plant productivity, although the results were only marginally significant. In addition, eCO2 had a negative impact on microbial processes linked to PAH degradation. Although eCO2 increased total soil microbial biomass and activity (Fig. 2, PC1 microbial PLFAs), it led to a decrease in Gram-positive bacteria, a microbial group linked to PAH degradation49 and the most important microbial group involved in the removal of PAHs from soil in the present study (Fig. 2). Furthermore, eCO2 altered the soil microbial community composition, which is also in line with previous studies38,50,51 and calls for more detailed investigations of shifts in soil microbial communities with sequencing techniques.
We propose that this effect of elevated tropospheric CO2 may be due to the higher plant carbon input in soil resulting from enhanced photosynthesis52. This may lead to higher soil microbial activity29,53, as the pool of labile soil C may be increased by elevated root exudation33,54,55. In the present study, eCO2 had non-significant effects on the biomass of fungi and Gram-negative bacteria, but decreased the biomass of Gram-positive bacteria (Fig. S1). Consistent with the present study, both Larson et al.31 and Grueter et al.56 found that eCO2 had no significant influence on microbial biomass and activity, while Manninen et al.57 found a negative effect of eCO2 on soil microbial biomass. These variable results indicate that eCO2 effects on soil microbial communities may depend on the environmental context, such as soil conditions and/or plant community composition38.
Elevated O3 decreased plant biomass, soil microbial biomass and activity, and PAH removal. Ozone is a toxic compound that can induce oxidative stress in plants, and high tropospheric O3 concentrations have been reported to decrease inputs and to change the composition of assimilates into the rhizosphere34, which in turns affects soil microbial communities. Results of the present study indicate that ozone-mediated changes in soil communities may have dramatic effects on soil self-cleaning potential. Consistent with past studies58,59,60, a strong decrease in the biomass of Gram-positive and Gram-negative bacteria and shifts in microbial community composition in response to eO3 was observed (Figs 2, S1).
The effects of eCO2, eO3, and plant diversity on PAH removal were mediated to some extent by alterations of soil enzymatic activity. Both elevated CO2 and O3 led to a decrease in polyphenol oxidase activity, while plant functional group richness increased polyphenol oxidase but decreased phenol oxidase. In line with the present study, eCO2 reduced the activity61 and abundance62 of polyphenol oxidase, suppressed phenol oxidase63, while enzymes including phenol oxidase were strongly affected by plant species richness64. These results indicate that simultaneous alteration of plant community composition and environmental conditions may have contrasting effects on enzyme activity involved in PAH removal. Notably, many enzymes are involved in the metabolism process of PAHs65,66, some of which were not measured here. Although the measured enzymes responded significantly to the treatments, this did not explain variation in PAH removal, which is why they were not considered in the structural equation model (Fig. 2).
Elevated CO2 and O3 concentrations and variations in plant diversity had significant interactive effects on plant biomass, soil microbial functions, and the degradation of PAHs. Plant diversity altered the effect of eCO2 on soil microbial biomass and activity, but the clear positive interaction effects as expected in hypothesis (4) were not detected. This highlights the importance of plant diversity and community composition in mediating soil microbial functions in a future world, but also calls for a better mechanistic understanding of interactive effects of plant diversity and global change drivers.
However, plant diversity did not alter eCO2 and eO3 effects on PAH removal in the present study. This is in line with a recent meta-analysis by Thakur et al.46 showing no interactive effects of plant diversity and global change factors in affecting soil microbial biomass in the short term. Potentially, plant diversity-induced differences in soil microbial community composition and subsequent effects on essential services like PAH degradation need a longer time than captured by the present experiment46. Moreover, we propose that lumping plant community composition into functional group richness may not provide the adequate explanatory level. Instead, we propose that future studies may use more targeted plant trait-based approaches67, e.g., by considering root/rhizosphere traits, to develop a better mechanistic understanding of the relationship between plant community composition and functioning of soil communities linked to pollutant removal.
Importantly and in contrast to our hypothesis (4), eCO2 amplified the inhibitory effect of eO3 on PAH removal. This effect was partly mediated by an enhancement of eO3 effects on most soil microbial groups at elevated CO2. It particularly amplified the negative effect of eO3 on Gram-positive bacteria, the most important microbial group driving the removal of PAHs from soil in this study. This result exemplifies how different global change drivers can have unexpected synergistic effects on soil functions and compromise important ecosystem services.
Conclusion
We highlight that global environmental change factors, such as human-induced alterations in tropospheric gas composition, may undermine the ability of ecosystems to degrade pollutants. Soil self-cleaning showed a high robustness to alterations in plant diversity and community composition, yet elevated CO2 and O3 concentrations may compromise efforts such as phytoremediation to restore polluted soils. On the other hand, the present study also indicates that the targeted assembly of plant communities applying a more comprehensive knowledge regarding plant effects on soil biota may be a promising tool to shape soil microbial communities for the degradation of organic pollutants.
Materials and Methods
Open top chambers
The open top chamber (OTC) system is located at Xianlin campus, Nanjing University, Nanjing, China (118°57′36.15″E, 31°7′23.99″N). Briefly, this system consists of four chambers with full control of atmospheric CO2 and O3 concentrations: one chamber with ambient CO2 (aCO2) and ambient O3 (aO3) levels, one with eCO2 and aO3 levels, one with aCO2 and eO3 levels, and one with both eCO2 and eO3 levels. The glass chambers are octagonal with 2 m in diameter and 2.8 m in height. CO2 was released from a tank (Q/JB-THB002, Beijing Tianhai Industry Co., Ltd.), and O3 was produced by an O3 generator (NPF10/W, Shandong Lvbang Ozone Co., Ltd.) from pure O2. CO2 and/or O3 were mixed with air from temperature-controlled rooms and conveyed by fans (SFG-2, Shanghai Jiabao Co., Ltd.) to the bottom of the chambers. Gases were released into the antra via tiny holes in the stainless steel plate between the bottom and the antrum, and then released into the air of the open top of chambers. The quantity of the CO2 and O3 release was controlled by a flowmeter (LZB-3WB, Changzhou Shuangbo Co., Ltd.), the concentration of CO2 was detected with a CO2 monitor (Li-7000, Li-Cor, USA), and the concentration of O3 was detected with an O3 monitor (Model 205, 2B Co., USA). The O3 fumigation was conducted between 9:00 a.m. and 5:00 p.m. until harvest, except during rain events, and the CO2 fumigation was all day long until harvest. The target CO2 concentration for the eCO2 treatment was 200 ppm higher than aCO2, and the target O3 concentration for the eO3 treatment was 50–60 ppb higher than aO3 in order to simulate the forecasted tropospheric CO2 and O3 levels in 20502.
Plant cultivation
Three species of grasses (Lolium perenne, Dactylis glomerata, Phleum pratense), herbs (Plantago lanceolata, Taraxacum officinale, Centaurea jacea), and legumes (Trifolium pratense, Trifolium repens, Medicago sativa) were germinated in trays filled with quartz sand in the lab. Ten days after germination, seedlings were transplanted into the microcosms (8 cm in diameter and 12 cm in height) with 250 g of PAHs contaminated soil collected from a chemical plant in Nanjing (118°44′51.87″E, 31°58′4.71″N). Plant communities consisting of nine individuals and differing in functional group richness (8 different communities) were set up: bare ground (no plants); functional group ‘monocultures’ of either three grass species, three herb species, or three legume species; mixtures of two functional groups (grasses plus herbs, grasses plus legumes, or herbs plus legumes); and the mixture containing all three plant functional groups (grasses plus herbs plus legumes), thereby yielding functional group richness levels of 0, 1, 2, and 3 and functionally dissimilar plant communities. Each plant community was replicated four times per CO2 × O3 treatment (32 microcosms per OTC, 128 microcosms in total). Plant communities were cultivated in the lab for one week, and dead seedlings were replaced before microcosms were transferred to OTCs.
The microcosms were randomly placed in the OTCs, and each microcosm was watered with 10–20 ml of distilled water per day. After 10 weeks of cultivation in OTCs, plants and soils were sampled, survival of plants and plant community biomass was measured, and soils for PAHs determination were stored at −20 °C, whereas soils for the measurement of microbial parameters were stored at 4 °C.
Determination of soil enzymatic activity
A very important step of PAH metabolism by bacteria and fungi is the breaking of PAH rings by phenol oxidase or polyphenol oxidase65,66. Therefore, these enzymes were used as proxy for general microbial processes linked to PAH degradation. For enzyme measurements, 0.5 g fresh soil was mixed with 20 ml milli-Q-water in 50 ml falcon tubes, shaken at 250 rpm for 30 min, centrifuged at 3000 rpm for 10 min, supernatants mixed with substrates and buffer in 96-well plates (Corning 96 Flat Bottom Transparent Polystyrol), then determined on a plate reader (Infinite M200, Tecan, Germany). Phenol oxidase activity was measured according to a modified protocol68. Briefly, 20 μl soil supernatant was mixed with 100 μl 5 mM bicarbonate buffer and 100 μl 5 mM L-3,4-dihydroxyphenylalanine (L-DOPA) solution, incubated at 27 °C, and absorbance was measured at 460 nm for 1 h. ΔA460/min from the initial linear portion of the curve was calculated. Polyphenol oxidase activity was measured according to Montgomery and Sgarbieri69. Briefly, 20 μl soil supernatants was mixed with 100 μl 0.5 M potassium phosphate buffer and 100 μl 1 mM 3-(4-hydroxyphenyl) alanine (L-Tyrosine) solution, incubated at 25 °C, and absorbance was measured at 280 nm for 12 min. ΔA280/min from the initial linear portion of the curve was calculated.
PLFA analysis
PLFAs were extracted according to Bligh and Dyer70 modified by Kramer and Gleixner71. Briefly, soil lipids were extracted by a mixture of chloroform, methanol, and 0.05 M phosphate buffer (pH 7.4) and split up into phospholipids by eluting with chloroform, acetone, and methanol from a silica-filled solid phase extraction column. Subsequently, the phospholipids were hydrolyzed and methylated by a methanolic KOH solution, and the PLFA-methyl esters were identified and quantified by GC-ECD (PerkinElmer, Clarus 500, USA). PLFA 19:0 was used as internal standard. Separated phospholipid fatty acid methyl-esters were identified by chromatographic retention time and mass spectral comparison with a mixture of standard qualitative bacterial acid methyl-ester that ranged from C11 to C24 (Supelco). For each sample, the abundance of individual phospholipid fatty acid methyl-esters was expressed in nmol per g dry soil. The nomenclature for PLFAs followed that of Frostegård et al.72. The sum of PLFAs i14:0 i15:0, a15:0, i16:0, i17:0, and i18:0 represented the biomass of Gram-positive bacteria, that of PLFAs cy17:0 and cy19:0 represented the biomass of Gram-negative bacteria, and the amount of the fungal-specific fatty acid 18:2ω6,9 was used as an indicator of fungal biomass73,74.
Determination of PAHs in soils
Soil samples stored at −20 °C were freeze-dried (Labconco 12 L, Labconco Co., USA) for 96 h, ground by mortars, and passed through a 2 mm sieve. Samples (5 g) were extracted with 20 mL methanol: methylene dichloride (1:2, v-v), concentrated in a rotary evaporator, and dried under a fine stream of nitrogen. The residues were dissolved in 0.5 ml acetonitrile. Samples were analyzed by high-performance liquid chromatography on a SpuelcosilTM LC-PAH column (250 × 4.6 mm, 5 μm) (Supelco, Bellefonete, PA, USA) with UV detector at 254 nm (HPLC-UV, Hitachi L2000). The temperature of the column was kept constant at 30 °C to obtain reproducible retention times. The mobile phase consisted of water and acetonitrile in gradient mode at flow rate of 1 ml/min. The gradient solvent system started with 60% acetonitrile in water (v/v) during 10 min, then increasing linearly to 100% acetonitrile within 10 min, the 100% acetonitrile was maintained for 20 min, and finally returned to the initial conditions in 2 min.
Statistical analyses
Analyses of variance (ANOVAs) were performed to test effects of CO2 (ambient and elevated), O3 (ambient and elevated), plant functional group richness (1, 2, 3 functional groups present), and all interactions on total plant biomass, plant shoot biomass, plant root biomass, plant survival, biomass of Gram-positive bacteria, biomass of Gram-negative bacteria, biomass of fungi, phenol oxidase activity, polyphenol oxidase activity, and total PAH residuals (for the latter, treatments with 0, 1, 2, and 3 functional groups were considered). If significant treatment effects were detected, additional Tukey’s HSD tests were performed to test for differences among means. ANOVAs were performed using Statistica 7.1 (Statsoft). Furthermore, structural equation modeling (SEM) was used to shed light on the mechanisms of PAH degradation by accounting for multiple potentially correlated effect pathways to disentangle the direct and indirect effects75 of experimental treatments and soil microbial community properties. The initial model was based on previous knowledge with experimental treatments as exogenous variables and the endogenous variables “Gram-negative bacteria”, “Gram-positive bacteria”, “PC1 microbial PLFAs” (representing PLFA composition), “PC1 PAHs” (representing PAH composition), and “total PAHs”. The adequacy of the models was determined via chi²-tests, AIC, and RMSEA76. Model modification indices and stepwise removal of non-significant relationships were used to improve the models; however, only scientifically sound relationships were considered75. Structural equation modeling was performed using Amos 5 (Amos Development Corporation, Crawfordville, FL, USA).
Data availability
All data generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Cramer, W. et al. Global response of terrestrial ecosystem structure and function to CO2 and climate change: results from six dynamic global vegetation models. Global Change Biol. 7, 357–373 (2001).
IPCC, 2007: Climate Change 2007: Synthesis Report. Contribution of Working Groups I, II and III to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change [Core Writing Team, Pachauri, R.K. & Reisinger, A. (Eds.)]. IPCC, Geneva, Switzerland, pp 104.
Vingarzan, R. A review of surface ozone background levels and trends. Atmos. Environ. 38, 3431–3442 (2004).
Morgan, P. B., Mies, T. A., Bollero, G. A., Nelson, R. L. & Long, S. P. Season-long elevation of ozone concentration to projected 2050 levels under fully open-air conditions substantially decreases the growth and production of soybean. New Phytol. 170, 333–343 (2006).
Bandowe, B. M., Srinivasan, P., Seelge, M., Sirocko, F. & Wilcke, W. A 2600-year record of past polycyclic aromatic hydrocarbons (PAHs) deposition at Holzmaar (Eifel, Germany). Palaeogeogr. Palaeocl. 401, 111–121 (2014).
Johnsen, A. R. & Karlson, U. PAH degradation capacity of soil microbial communities - Does it depend on PAH exposure? Microb. Ecol. 50, 488–495 (2005).
Augusto, S., Pereira, M. J., Máguas, C. & Branquinho, C. A step towards the use of biomonitors as estimators of atmospheric PAHs for regulatory purposes. Chemosphere. 92, 626–632 (2013).
Ferrarese, E., Andreottola, G. & Oprea, I. A. Remediation of PAH-contaminated sediments by chemical oxidation. J. Hazard. Mater. 152, 128–139 (2008).
Gan, S., Lau, E. V. & Ng, H. K. Remediation of soils contaminated with polycyclic aromatic hydrocarbons (PAHs). J. Hazard. Mater. 172, 532–549 (2009).
Khan, S., Wang, N., Reid, B. J., Freddo, A. & Cai, C. Reduced bioaccumulation of PAHs by Lactuca satuva L. grown in contaminated soil amended with sewage sludge and sewage sludge derived biochar. Environ. Pollut. 175, 64–68 (2013).
Liu, R., Xiao, N., Wei, S., Zhao, L. & An, J. Rhizosphere effects of PAH-contaminated soil phytoremediation using a special plant named Fire Phoenix. Sci. Total Environ. 473, 350–358 (2014).
Liao, C. et al. Effect of surfactant amendment to PAHs-contaminated soil for phytoremediation by maize (Zea mays L). Ecotox. Environ. Safe. 112, 1–6 (2015).
Wang, Y., Fang, L., Lin, L., Luan, T. & Tam, N. Effects of low molecular-weight organic acids and dehydrogenase activity in rhizosphere sediments of mangrove plants on phytoremediation of polycyclic aromatic hydrocarbons. Chemosphere. 99, 152–159 (2014).
Aranda, E. et al. Role of arbuscular mycorrhizal fungus Rhizophagus custos in the dissipation of PAHs under root-organ culture conditions. Environ. Pollut. 181, 182–189 (2013).
Wardle, D. A. et al. Ecological linkages between aboveground and belowground biota. Science. 304, 1629–1633 (2004).
Scherber, C. et al. Bottom-up effects of plant diversity on multitrophic interactions in a biodiversity experiment. Nature. 468, 553–556 (2010).
Eisenhauer, N. et al. Plant diversity effects on soil food webs are stronger than those of elevated CO2 and N deposition in a long-term grassland experiment. P. Natl. Acad. Sci. USA 110, 6889–6894 (2013).
Lange, M. et al. Plant diversity increases soil microbial activity and soil carbon storage. Nat. Commun. 6 (2015).
Hättenschwiler, S., Tiunov, A. V. & Scheu, S. Biodiversity and litter decomposition in terrestrial ecosystems. Annu. Rev. Ecol. Evol. S. 36, 191–218 (2005).
Orwin, K. H., Wardle, D. A. & Greenfield, L. G. Ecological consequences of carbon substrate identity and diversity in a laboratory study. Ecology. 87, 580–593 (2006).
Hooper, D. U. et al. Interactions between aboveground and belowground biodiversity in terrestrial ecosystems: Patterns, mechanisms, and feedbacks. Bioscience. 50, 1049–1061 (2000).
Eisenhauer, N. et al. Plant diversity effects on soil microorganisms support the singular hypothesis. Ecology. 91, 485–496 (2010).
Milcu, A. et al. Functionally and phylogenetically diverse plant communities key to soil biota. Ecology. 94, 1878–1885 (2013).
Spehn, E. M., Joshi, J., Schmid, B., Alphei, J. & Korner, C. Plant diversity effects on soil heterotrophic activity in experimental grassland ecosystems. Plant Soil. 224, 217–230 (2000).
Zak, D., Holmes, W. E., White, D. C., Peacock, A. D. & Tilman, D. Plant diversity, soil microbial communities, and ecosystem function: are there any links? Ecology. 84, 2042–2050 (2003).
Milcu, A., Partsch, S., Scherber, C., Weisser, W. W. & Scheu, S. Earthworms and legumes control litter decomposition in a plant diversity gradient. Ecology. 89, 1872–1882 (2008).
Chakraborty, S., Pangga, I. B. & Roper, M. M. Climate change and multitrophic interactions in soil: the primacy of plants and functional domains. Global Change Biol. 18, 2111–2125 (2012).
Rajkumar, M., Prasad, M. N. V., Swaminathan, S. & Freitas, H. Climate change driven plant-metal-microbe interactions. Environ. Int. 53, 74–86 (2013).
Chen, Y., Liu, Q., Liu, Y., Jia, F. & He, X. Responses of soil microbial activity to cadmium pollution and elevated CO2. Sci. Rep. 4, 2045–2322 (2014).
Curlevski, N., Drigo, B., Cairney, J. & Anderson, I. C. Influence of elevated atmospheric CO2 and water availability on soil fungal communities under Eucalyptus saligna. Soil Biol. Biochem. 70, 263–271 (2014).
Larson, J. L., Zak, D. R. & Sinsabaugh, R. L. Extracellular enzyme activity beneath temperate trees growing under elevated carbon dioxide and ozone. Soil Sci. Soc. Am. J. 66, 1848–1856 (2002).
Wang, X. et al. Ectomycorrhizal colonization and growth of the hybrid larch F-1 under elevated CO2 and O3. Environ. Pollut. 197, 116–126 (2015).
Eisenhauer, N., Cesarz, S., Koller, R., Worm, K. & Reich, P. B. Global change belowground: impacts of elevated CO2, nitrogen, and summer drought on soil food webs and biodiversity. Global Change Biol. 18, 435–447 (2012).
Formánek, P., Rejsek, K. & Vranova, V. Effect of Elevated CO2, O3, and UV Radiation on Soils. The Scientific World Journal. article ID 730149, 8 pages (2014).
Samanta, S. K., Singh, O. V. & Jain, R. K. Polycyclic aromatic hydrocarbons: environmental pollution and bioremediation. Trends Biotechnol. 20, 243–248 (2002).
Bostrom, C. E. et al. Cancer risk assessment, indicators, and guidelines for polycyclic aromatic hydrocarbons in the ambient air. Environ. Health Persp. 110, 451–488 (2002).
Reich, P. B. et al. Impacts of biodiversity loss escalate through yime as redundancy fades. Science. 336, 589–592 (2012).
Chung, H., Zak, D. R., Reich, P. B. & Ellsworth, D. S. Plant species richness, elevated CO2, and atmospheric nitrogen deposition alter soil microbial community composition and function. Global Change Biol. 13, 980–989 (2007).
Reich, P. B. et al. Nitrogen limitation constrains sustainability of ecosystem response to CO2. Nature. 440, 922–925 (2006).
Blankinship, J. C., Niklaus, P. A. & Hungate, B. A. A meta-analysis of responses of soil biota to global change. Oecologia. 165, 553–565 (2011).
Reich, P. B. et al. Plant diversity enhances ecosystem responses to elevated CO2 and nitrogen deposition. Nature. 410, 809–812 (2001).
Chung, H. & Zak, D. R. Elevated atmospheric CO2 and O3 alter soil fungal community composition and function. Ecological Society of America Annual Meeting Abstracts. 88, 65–65 (2003).
Jousset, A., Schmid, B., Scheu, S. & Eisenhauer, N. Genotypic richness and dissimilarity opposingly affect ecosystem functioning. Ecol. Lett. 14, 537–545 (2011).
Steudel, B. et al. Biodiversity effects on ecosystem functioning change along environmental stress gradients. Ecol. Lett. 15, 1397–1405 (2012).
Eisenhauer, N. et al. Root biomass and exudates link plant diversity with soil bacterial and fungal biomass. Sci. Rep. 7, 44641 (2017).
Thakur, M. P. et al. Plant diversity drives soil microbial biomass carbon in grasslands irrespective of global environmental change factors. Global change boil. 21, 4076–4085 (2015).
Steinauer, K. et al. Plant diversity effects on soil microbial functions and enzymes are stronger than warming in a grassland experiment. Ecology. 96, 99–112 (2015).
Oyelami, A. O. et al. Effects of plant species identity, diversity and soil fertility on biodegradation of phenanthrene in soil. Environ. Pollut. 173, 231–237 (2013).
Krivobok, S. et al. Identification of pyrene-induced proteins in Mycobacterium sp. Strain 6PY1: evidence for two ring-hydroxylating dioxygenases. J. Bacteriol. 185, 3828–3841 (2003).
Lee., S. H. & Kang, H. Elevated CO2 causes a change in microbial communities of rhizosphere and bulk soil of salt marsh system. Appl. Soil Ecol. 108, 307–314 (2016).
Du, W. et al. Elevated CO2 levels modify TiO2 nanoparticle effects on rice and soil microbial communities. Sci. Total Environ. 578, 408–416 (2016).
Phillips, R. P., Finzi, A. C. & Bernhardt, E. S. Enhanced root exudation induces microbial feedbacks to N cycling in a pine forest under long-term CO2 fumigation. Ecol. let. 14, 187–194 (2011).
Inubushi, K. et al. Microbial biomass carbon and methane oxidation influenced by rice cultivars and elevated CO2 in a Japanese paddy soil. Eur. J. Soil Sci. 62, 69–73 (2011).
Huang, S., Jia, X., Zhao, Y., Bai, B. & Chang, Y. Elevated CO2 benefits the soil microenvironment in the rhizosphere of Robinia pseudoacacia L. seedlings in Cd- and Pb-contaminated soils. Chemosphere. 168, 606–616 (2017).
Wu, K. et al. CO2-induced alterations in plant nitrate utilization and root exudation stimulate N2O emissions. Soil Biol. Biochem. 106, 9–17 (2017).
Grueter, D., Schmid, B. & Brandl, H. Influence of plant diversity and elevated atmospheric carbon dioxide levels on belowground bacterial diversity. Bmc Microbiol. 6, 68 (2006).
Manninen, S., Aaltonen, H., Kanerva, T., Rämö, K. & Palojärvi, A. Plant and soil microbial biomasses in Agrostis capillaris and Lathyrus pratensis monocultures exposed to elevated O3 and CO2 for three growing seasons. Soil Biol. Biochem. 42, 1967–1975 (2010).
Chen, Z., Wang, X., Yao, F., Zheng, F. & Feng, Z. Elevated ozone changed soil microbial community in a rice paddy. Soil Sci. Soc. Am. J. 74, 829–837 (2010).
Chen, Z., Wang, X. & Shang, H. Structure and function of rhizosphere and non-rhizosphere soil microbial community respond differently to elevated ozone in field-planted wheat. J. Environ. Sci.-China. 32, 126–134 (2015).
Feng, Y. et al. The contrasting responses of soil microorganisms in two rice cultivars to elevated ground-level ozone. Environ. Pollut. 197, 195–202 (2015).
Sunoj, V. S. J., Kumar, S. N. & Muralikrishna, K. S. Effect of elevated CO2 and temperature on oxidative stress and antioxidant enzymes activity in coconut (Cocosnucifera L.) seedlings. Indian J. Plant Physi. 19, 382–387 (2014).
Casteel, C. L. et al. Transcriptional profiling reveals elevated CO2 and elevated O3 alter resistance of soybean (Glycine max) to Japanese beetles (Popillia japonica). Plant Cell Environ. 31, 419–434 (2008).
Fenner, N. et al. Interactions between elevated CO2 and warming could amplify DOC exports from peatland catchments. Environ. Sci. Technol. 41, 3146–3152 (2007).
Zhang, C. B. et al. Effects of plant diversity on nutrient retention and enzyme activities in a full-scale constructed wetland. Bioresource Technol. 101, 1686–1692 (2010).
Peng, R. H. et al. Mcirobial biodegradation of polyaromatic hydrocarbons. FEMS Microbio. Rev. 32, 927–955 (2008).
Bamforth, S. M. & Singleton, I. Bioremediation of polycyclic aromatic hydrocarbons: current knowledge and future directions. J. Chem. Technol. Biot. 80, 723–736 (2005).
Laliberté, E. Below-ground frontiers in trait-based plant ecology. New Phytol. 213, 1597–1603 (2017).
Gallo, M., Amonette, R., Lauber, C., Sinsabaugh, R. L. & Zak, D. R. Microbial community structure and oxidative enzyme activity in nitrogen-amended north temperate forest soils. Microbial Ecol. 48, 218–229 (2004).
Montgomery, M. W. & Sgarbieri, V. C. Isoenzynmes of banana polyphenol oxidase. Phytochemistry. 14, 1245–1249 (1975).
Bligh, E. G. & Dyer, W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Phys. 37, 911–917 (1959).
Kramer, C. & Gleixner, G. Variable use of plant- and soil-derived carbon by microorganisms in agricultural soils. Soil Biol. Biochem. 38, 3267–3278 (2006).
Frostegård, A., Baath, E. & Tunlid, A. Shifts in the structure of soil microbial communities in limed forests as revealed by phospholipid fatty-acid analysis. Soil Biol. Biochem. 25, 723–730 (1993).
Zelles, L. Phospholipid fatty acid profiles in selected members of soil microbial communities. Chemosphere. 35, 275–294 (1997).
Zelles, L. Fatty acid patterns of phospholipids and lipopolysaccharides in the characterisation of microbial communities in soil: a review. Biol. Fert. Soils. 29, 111–129 (1999).
Grace, J. B. & Kelley, J. E. A structural equation model analysis of postfire plant diversity in California shrublands. Ecol. Appl. 16, 503–514 (2006).
Eisenhauer, N., Bowker, M. A., Grace, J. B. & Powell, J. R. From patterns to causal understanding: Structural equation modeling (SEM) in soil ecology. Pedobiologia. 58, 65–72 (2015).
Acknowledgements
We gratefully acknowledge the Program of New Century Excellent Talents in University (NCET-12-0266) and the National Natural Science Foundation of China (grant no. 21177058). NE acknowledges funding by the German Research Foundation in the frame of the Emmy Noether research group (Ei 862/2) and the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, funded by the German Research Foundation (FZT 118).
Author information
Authors and Affiliations
Contributions
N.E., R.J., H.Y.G. and O.B. conceived and designed the experiment. F.X.A. performed the experiments. F.X.A., A.J., N.E. and O.B. analyzed the data. F.X.A. and N.E. wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ai, F., Eisenhauer, N., Jousset, A. et al. Elevated tropospheric CO2 and O3 concentrations impair organic pollutant removal from grassland soil. Sci Rep 8, 5519 (2018). https://doi.org/10.1038/s41598-018-23522-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-23522-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.