Abstract
Upon invasion, Plasmodium falciparum exports hundreds of proteins across its surrounding parasitophorous vacuole membrane (PVM) to remodel the infected erythrocyte. Although this phenomenon is crucial for the parasite growth and virulence, elucidation of precise steps in the export pathway is still required. A translocon protein complex, PTEX, is the only known pathway that mediates passage of exported proteins across the PVM. P. falciparum Parasitophorous Vacuolar protein 1 (PfPV1), a previously reported parasitophorous vacuole (PV) protein, is considered essential for parasite growth. In this study, we characterized PfPV1 as a novel merozoite dense granule protein. Structured illumination microscopy (SIM) analyses demonstrated that PfPV1 partially co-localized with EXP2, suggesting the protein could be a PTEX accessory molecule. Furthermore, PfPV1 and exported protein PTP5 co-immunoprecipitated with anti-PfPV1 antibody. Surface plasmon resonance (SPR) confirmed the proteins’ direct interaction. Additionally, we identified a PfPV1 High-affinity Region (PHR) at the C-terminal side of PTP5 where PfPV1 dominantly bound. SIM analysis demonstrated an export arrest of PTP5ΔPHR, a PTP5 mutant lacking PHR, suggesting PHR is essential for PTP5 export to the infected erythrocyte cytosol. The overall results suggest that PfPV1, a novel dense granule protein, plays an important role in protein export at PV.
Similar content being viewed by others
Introduction
Malaria, caused by protozoan parasite Plasmodium falciparum, is a leading cause of morbidity and mortality in endemic regions1. Upon invasion, the parasite exports hundreds of proteins through its surrounding parasitophorous vacuole membrane (PVM) into the cytosol of the host erythrocyte2. Through this process, the infected erythrocyte is modified with changes that are crucial to the parasite’s survival and virulence3. P. falciparum infected erythrocytes have increased rigidity and adhere to endothelial cells (cytoadherence) in the microcirculation of various organs. This phenomenon is believed to aid the parasite evade the host immune system, and contributes to malaria pathogenesis3.
In order to traverse the PVM, exported proteins must be initially unfolded4. A translocon protein complex, Plasmodium translocon of exported proteins (PTEX), mediates the unfolding and passage of the parasite proteins5,6,7. To date, five PTEX constituent proteins have been characterized including; EXP2, HSP101, PTEX150, PTEX88 and TRX25. EXP2, HSP101 and PTEX150 constitute the core PTEX, whereas PTEX88 and TRX2 are thought to play accessory roles8,9,10. P. falciparum Parasitophorous Vacuolar protein 1 (PfPV1; PF3D7_1129100), a previously reported bona fide PV protein is essential for the parasite growth11. A recent report indicates that PfPV1 co-precipitates with PTEX complex12,13, suggesting that PfPV1 constitutes PTEX accessory molecules13.
Cytoadherence of infected erythrocytes to microvasculature is mediated by members of the P. falciparum Erythrocyte Membrane Protein-1 (PfEMP1) family14. The PfEMP1 proteins are trafficked through cytosol to the surface of host erythrocytes15. A large-scale gene knockout study identified several exported proteins that play important roles in PfEMP1 trafficking16. PfEMP1-trafficking protein (PTP5) also known as PF70 (PlasmoDB PF3D7_1002100) is among the exported proteins essential for PfEMP1 trafficking16,17.
Most exported proteins, including PTP5, possess a Plasmodium export element (PEXEL) motif (RxLx/E/D/Q) at the N-terminus18,19. The exported proteins are proteolytically cleaved by Plasmepsin V (PMV) at the C-terminal side of the conserved leucine in the PEXEL motif followed by new N-terminal acetylation (Ac-xE/Q/D)20. There are approximately 500 proteins with PEXEL motif, representing ~10% of parasite genome, and are predicted to be exported proteins4,18,19,21. However, several exported proteins, named PEXEL-negative exported proteins (PNEPs), contain neither a PEXEL motif nor other conserved export sequences. PNEPs are a rather small group in P. falciparum and include proteins such as ring-exported proteins 1 and 2 (REX1, REX2)22,23, skeleton binding protein 1 (SBP1)24, and PfEMP121. Reported knockdown studies of PTEX150 and HSP101 suggest that PTEX plays a role in the export of both PEXEL and PNEPs proteins6,7.
PTEX core proteins, EXP2 and PTEX150, are stored in merozoite dense granules, with subsequent translocation to the PVM upon invasion8. Moreover, another protein, Ring-infected Erythrocyte Surface Antigen, (RESA), is stored in the dense granules but translocated to the sub-plasma membrane region of infected erythrocyte25. Therefore, it could be considered that proteins localized to the dense granules might be involved in protein export in infected erythrocytes.
In this study, we have characterized PfPV1 as a novel merozoite dense granule protein. We observed that PfPV1 partially co-localizes with EXP2, suggesting that the protein could be a PTEX accessory molecule. PfPV1 and the exported protein PTP5 co-immunoprecipitated with anti-PfPV1 antibody. Surface plasmon resonance (SPR) confirmed the proteins’ direct interaction. Moreover, we identified a PfPV1 High-affinity Region (PHR) at the C-terminal side of PTP5 where PfPV1 dominantly bound. We demonstrated the arrest of PTP5ΔPHR export in PV, suggesting that PHR is essential for PTP5 export. The overall results suggest that PfPV1 a novel dense granule protein that plays an important role in protein export at PV.
Results
Production of recombinant PfPV1 protein
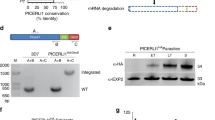
PfPV1 (PF3D7_1129100) is predicted as a 51.9 kDa protein (Fig. 1a). Signal peptide was predicted at the 1–22 amino acid residues (aa). The protein has neither PEXEL motif nor predicted functional domain. PfPV1 orthologs were identified in various plasmodium species including, P. vivax, P. knowlesi, P. chaubaudi and P. berghei11. Previous studies suggested that PfPV1 is essential for blood-stage parasite growth and localizes to PV11,26. In order to characterize the protein further, we utilized wheat germ cell-free system (WGCFS; CellFree Sciences, Matsuyama, Japan) to synthesize recombinant PfPV1. The expressed recombinant protein was Ni2+ affinity purified with an expected band at ~60 kDa (Fig. 1b). The purified recombinant PfPV1 protein was subsequently used to raise antibodies in rabbit. The antibodies specifically recognized blood-stage parasites native PfPV1 by Western blot, in both reducing and non-reducing conditions (Fig. 1c). Recognition of native PfPV1 under reducing conditions was at a slightly higher molecular weight.
Assessment of expressed recombinant PfPV1, and rabbit anti-PfPV1 antibody. (a) Schematic presentation of PfPV1. The thick black line indicates region of protein expressed using WGCFS; SP is the predicted signal peptide. (b) Purified recombinant PfPV1 resolved in SDS-PAGE under reduced conditions and stained with Coomassie brilliant blue (CBB), (c) Western blot analysis of PfPV1 in trophozoite and schizont-rich parasite lysate using rabbit anti-PfPV1 antibody. NR and R indicate results of non-reduced and reduced conditions respectively.
Subcellular localization of PfPV1 in comparison with EXP2
Using the rabbit anti-PfPV1 antibodies, we conducted subcellular localization of PfPV1 in schizont stage parasites by immunofluorescence assay (IFA) (Fig. 2a). We determined the co-localization intensity correlation by Pearson’s correlation coefficients (r) between PfPV1 and different organelle markers. PfPV1 showed tighter co-localization with RESA (r = 0.72), a dense granule protein, than with AMA1 (r = 0.04), RAP1 (r = 0.27) and RON2 (r = 0.02), which are microneme, rhoptry body and rhoptry neck proteins respectively. These results suggested that PfPV1 is a P. falciparum merozoite dense granule protein. Immunoelectronmicroscopy (IEM) examination confirmed the localization of PfPV1 to the dense granules (Fig. 2b). Moreover, consistent with a previous report indicating GFP tagged PfPV1 localizes to PV11, the IFA of ring and trophozoite stage parasites showed that PfPV1 co-localized with RAP1 a PV protein, but not with RESA or SBP1, which are erythrocyte membrane and Maurer’s clefts proteins respectively (Figure S1).
Subcellular localization of PfPV1 in schizont-stage parasites. (a) Results of IFA with Confocal Microscopy. Labeling of the utilized antibodies is on the upside of panels. PfPV1 (green) was counter stained with four different markers of subcellular localizations (red). The parasite nucleus was stained with DAPI (blue). PfPV1 co-localized with RESA a dense granule marker. DIC; Differential interference contrast microscope. Merge; the image created by merging image of IFA, and nuclear-staining. Pearson’s coefficient values ( = r) were calculated by using Zen 2010 software (Carl Zeiss MicroImaging, Thornwood, NY) and shown on the merged panels. Scale bar = 5 µm. (b) Results of IEM using rabbit anti-PfPV1 antibody. Gold particles can be observed at the Plasmodium falciparum merozoite dense granules at schizont stage. N; nuclear. Scale bar = 200 nm.
With an understanding that a previous report demonstrated that PTEX components localize to the merozoite dense granules8, we hypothesized that PfPV1 could play an important role in protein export to the erythrocyte cytosol after invasion. We therefore utilized structured illumination microscopy (SIM) to comparatively examine the sub-cellular localization of PfPV1 and EXP2. Consistent with previous reports27, EXP2 was observed in PV 1–12 h in early and mid trophozoite stages (Fig. 3a), while 18–24 h post invasion, it accumulated at a PV protrusion (Fig. 3a, arrowheads). Similar to other PTEX associated proteins27, PfPV1 localized proximal to EXP2, and remained partially co-localized in the time course, albeit at varying degree of co-localization (Fig. 3a). In addition, PfPV1 also accumulated at the base of the PV protrusion in partial co-localization with EXP2 18–24 h after invasion (Fig. 3a; yellow signals). IEM examination of trophozoite stage parasites with the rabbit anti-PfPV1 showed accumulated gold particles at the base of circular cleft or tubular structure (Fig. 3b, insets), hence confirming the SIM findings (Fig. 3a, 18–24 h).
Subcellular localization of PfPV1 in ring- and trophozoite-stage parasites. (a) Results of IFA with SIM. Samples were prepared shortly after merozoite invasion as indicated. PfPV1 (green) was counter stained with EXP2 (red). Parasite nucleus was stained with DAPI (blue). Arrowheads indicate observed region of PV protrusions with accumulated EXP2. The images were made from stacks of SIM optical sections (120 nm Z-step size). Scale bars = 2 µm. (b) Results of IEM using rabbit anti-PfPV1 antibody. Gold particle labeling was observed at the base of circular clefts. Scale bar = 2 µm.
Identification of PfPV1 associated proteins
To investigate the role of PfPV1 in protein export, we explored for possible interacting proteins. We conducted immunoprecipitation experiments with rabbit anti-PfPV1 antibody on Nonidet P-40 solubilized parasites samples. We then carried out Western blot analysis on the immunoprecipitate with a panel of mouse antibodies raised against 286 P. falciparum recombinant proteins (Table S2). PfEMP1-trafficking protein (PTP5, PF3D7_1002100)16,17 was co-immunoprecipitated with PfPV1 (Fig. 4b arrowhead, and S2a). The signal intensity derived from PTP5 was significantly higher than that of samples co-immunoprecipitated with anti-GST antibody (Fig. 4c). Inversely, rabbit anti-PTP5 antibody co-immunoprecipitated PV1 with PTP5 (Figure S2b). PTP5 consists of 631 amino acids (aa), with a theoretical MW of 69.9 kDa. The protein has a signal peptide (Fig. 4a, SP) and a PEXEL motif (RLLSE) starting at R57 (Fig. 4a, PEXEL). In addition, PF3D7_0801000, one of the proteins exported through PTEX5,8, was also co-immunoprecipitated with PfPV1 (Figure S3). EXP2 and HSP101 were also co-immunoprecipitated (Figure S4), in line with previous studies12,13. In contrast, RAP1, which localizes to PV space, was not co-immunoprecipiated with PV1 (Figure S2c).
Analysis of recombinant proteins; PTP5 and PfPV1 interaction. (a) Schematic presentation of PTP5 protein. SP (Red) is the predicted signal peptide, PEXEL (Orange) is the PEXEL motif position, the repetitive sequence is in Blue, Thick black lines indicate truncation design of PTP5 proteins expressed using WGCFS. (b) Results of immunoprecipitation experiment using rabbit anti-PfPV1 antibody. For Western blot analysis, we used mouse anti-PTP5 antibody to detect PTP5. Marker; protein molecular weight marker, Lysate; parasite lysate derived from 108 trophozoite-/schizont-rich parasite pellet, GST IP; sample immunoprecipitated with rabbit anti-GST antibody and included as Negative control, PV1 IP; sample immunoprecipitated with anti-PfPV1 antibody. The arrowhead indicates a signal that corresponds to native PTP5. Three independent experiments were carried out (Figure S2). (c) Relative intensities derived from PTP5 on the Western blot analysis are presented as a bar graph. The background signal was defined as the signal in the empty lane that corresponded to the molecular weight of native PTP5 (approximately 80 kDa). Error bars represent SD. Significantly higher signals from PTP5 were detected from the sample immunoprecipitated with anti-PV1 antibody (p < 0.05). (d) Purified GST-tagged recombinant PTP5 resolved in SDS-PAGE and stained with CBB. Lane 1; PTP5_Tr0, Lane 2; PTP5_Tr1, Lane 3; PTP5_Tr2. (e) SPR kinetics analyses sensorgrams. His-tagged purified recombinant PfPV1 was used for ligand. Analytes used for the analyses were indicated in sensorgrams. Black lines indicate fits used for the calculation of kinetics parameters. The PTP5_Tr0 concentrations used for the single-cycle kinetic analysis; 0.16, 0.8, 4, 20 and 100 nM, The PTP5_Tr1 concentrations used for the multi-cycle kinetic analysis; 10, 25, 50, 75, 100, 125, 150, 200, 250 and 500 nM, The PTP5_Tr2 concentrations used for the single-cycle kinetic analysis; 0.24, 1.2, 6, 30, and 150 nM. The GST concentrations used for the single-cycle kinetic analysis; 0.96, 4.8, 24, 120, 600 nM.
PfPV1 and PTP5 protein-protein interaction
We utilized surface plasmon resonance (SPR) to demonstrate and characterize direct protein-protein interaction between PfPV1 and PTP5. Kinetics analysis of the WGCFS expressed PfPV1 and PTP5_Tr0; 36 aa – 631 aa (Figs 1b and 4a,d) showed direct interaction with a KD value of 2.705 × 10−9 M in 1:1 stoichiometry (Fig. 4e). To identify the PTP5 region that is essential for interaction with PfPV1, we truncated PTP5 into PTP5_Tr1; 36 aa – 334 aa, and PTP5_Tr2; 537 aa – 631 aa (Fig. 4a,d). The interaction between PTP5_Tr2 and PfPV1 exhibited a KD value of 1.236 × 10−9 M, while as that of PTP5_Tr1 and PfPV1 showed a KD value of 9.381 × 10−7M (Fig. 4e). The PTP5_Tr1-PfPV1 interaction was 759 fold weaker than that of PTP5_Tr2-PfPV1, which prompted determination of interaction kinetics parameters through multi-cycle kinetic analyses (Fig. 4e). The results indicated that, at least, PTP5 region 335 aa − 631 aa is essential for PfPV1 high affinity interaction, and thus hereinafter referred to as PfPV1 High-affinity Region (PHR). Similarly, we also observed a direct interaction between PfPV1 and PF3D7_0801000 (Figure S3).
Subcellular localization of PTP5FL-HA and PTP5ΔPHR-HA
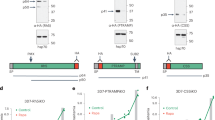
To analyze whether the PfPV1-PTP5 interaction contributes to PTP5 export, we generated transgenic parasite lines episomally expressing either PTP5FL (1 aa – 631 aa), PTP5ΔPHR (1 aa – 334 aa) or PTP5C (1 aa – 61 aa fused to 537 aa – 631 aa) followed by 3 × HA at the C-terminus (Fig. 5a). We confirmed expression of the proteins by Western blot and IFA using anti-HA antibodies. Notably, PTP5FL-HA, truncates PTP5ΔPHR-HA and PTP5C-HA were detected by immunoblot at the expected molecular weights (Fig. 5b). We however also observed an extra band with PTP5C-HA probably arising from a non-processed episomally expressed protein (Fig. 5b). Moreover, analysis with IFA demonstrated patchy signals of PTP5FL-HA in the cytosol of the infected erythrocyte existing in co-localization with SBP1; a Maurer’s cleft marker (Fig. 5c), suggesting that this PTP5 is exported to Maurer’s clefts. However, PTP5ΔPHR-HA and PTP5C-HA were not exported but arrested in the PV space and co-localized with PfPV1 (Fig. 5d,e). We therefore, conducted additional localization experiments using 3D-SIM (Fig. 6a,b and Movies S1, S2), to explore the loci of PTP5ΔPHR-HA accumulation in the PV space. We observed that only 1% of PTP5FL-HA co-localized with both PV1 and EXP2, while 30% of PTP5ΔPHR-HA had co-localization with both of the two PV proteins (Fig. 6c). Taken together, these results suggested that PTP5ΔPHR-HA translocation was arrested at an intermediate complex with PfPV1 and PTEX.
Characterization of episomally expressed PTP5, PTP5ΔPHR and PTP5C. (a) Schematic presentation of PTP5 protein episomally expressed in P. falciparum 3D7 strain. The color code is same as Fig. 4a. (b) Western blot analysis of parasite lysate expressing PTP5FL-HA, PTP5ΔPHR-HA and PTP5C-HA. Rat anti-HA antibody was used for the primary antibody. M; All Blue prestained protein molecular weight marker, 1; 3D7 parasite lysate (negative control, prepared from 1 × 106 infected RBC), 2; PTP5FL-HA parasite lysate (prepared from 1 × 106 infected RBC), 3; PTP5ΔPHR-HA parasite lysate (prepared from 1 × 106 infected RBC), 4; 3D7 parasite lysate (negative control, prepared from 1 × 107 infected RBC), 5; PTP5C parasite lysate (prepared from 1 × 107 infected RBC). (c,d,e) Subcellular localization of PTP5FL-HA, PTP5ΔPHR-HA and PTP5C-HA. The samples were stained with anti-HA (green) co-stained with anti-PfPV1 or anti-SBP1 (red). Parasite nucleus was stained with DAPI (blue). Scale bars = 5 µm.
3D-SIM observations of episomally expressed PTP5 and PTP5ΔPHR. (a) Subcellular localization of PTP5FL-HA (green) and (b) PTP5ΔPHR-HA (green) observed with SIM. EXP2 (red) and PfPV1 (cyan) were used as a counter-staining. The results of 3D-SIM for the identical erythrocytes are available in Movies S1 and S2. (c) Numerical analysis of protein localizations; vertical axis shows percentages of HA signals detected in 3D-SIM. HA; HA signals did not co-localized with other proteins, HA + PV1; HA signals co-localized with PV1 signals, HA + EXP2; HA signals co-localized with EXP2 signals, HA + PV1 + EXP2; HA signals co-localized with the other 2 proteins. The HA signal percentages are average values of 3 PTP5FL-HA, and 5 PTP5ΔPHR-HA observed parasite cells. (d) Schematic presentation of PTP5FL-HA (upper panel) and PTP5ΔPHR-HA (lower panel). PEXEL motif, PV1 high-affinity region (PHR), and HA tag of episomally expressed PTP5 are represented in orange, gray, and green colors respectively. The present study identified PHR as essential for PTP5 export, suggesting that PV1 plays an important role in protein translocation mediated by the PTEX complex. The PTP5FL-HA that interacted with PV1 successfully translocated to the cytosol of infected erythrocyte. PTP5ΔPHR-HA mutant weakly interacts PV1 and the export of this protein was arrested at PV.
In order to confirm the protein-protein interaction between PV1 and PTP5FL-HA or PTP5ΔPHR-HA in the parasite, we performed immunoprecipitation of the transfected parasites using anti-PfPV1 antibody. Consistent with the SPR findings, both PTP5FL-HA and PTP5ΔPHR-HA were co-immunoprecipitated by anti-PV1 antibody (Figure S5a). In addition, PV1 was co-immunoprecipitated with PTP5FL-HA or PTP5ΔPHR-HA by anti-HA antibody (Figure S5b).
Discussion
Plasmodium falciparum merozoite dense granules release their contents into PV space soon after erythrocyte invasion28. For instance, RESA, a protein stored in dense granules, is exported to the infected erythrocyte cytosol within 12 min post invasion27. Thus, it is hypothesized that proteins localized to the dense granules play a role in protein export into infected erythrocytes for hastened construction of export machinery. Therefore, there is need to screen and characterize novel dense granule proteins that could be involved in protein export. In this study, using IEM we demonstrated PfPV1 as a novel P. falciparum merozoite dense granule protein, and a bona fide PV protein as previously suggested11.
It’s recently reported that PfPV1 co-precipitated with PTEX complex12,13 suggesting this protein constitutes PTEX accessory molecules13. Consistently, our results here showed that PfPV1 is partially co-localized with EXP2 (Figs 3a and 6). We demonstrated that PV1 directly interacted with PTP5, thus an intermediate complex as PTEX-PTP5-PfPV1 is conjectured (Fig. 6d). The observed high affinity of PfPV1 and PTP5, suggests that the PfPV1-PTP5 is a “strong transient complex”29. Thus, it can be postulated that a structural change of the PfPV1-PTP5 complex caused by an unknown protein, and/or PTP5 unfolding mediated by PTEX would be required to dissociate PTP5 for export.
This study reports, for the first time, the biochemical properties of protein-protein interactions between PV1, PTP5 and PF3D7_0801000. To the best of our knowledge, this is also the first study reporting direct interaction between PTEX accessory proteins and exported proteins. PTP5_Tr0 and PTP5_Tr2 bound PfPV1 with comparable KD values of 2.705 × 10−9 M, and 1.236 × 10−9 M, respectively. We therefore, considered PTP5_Tr2 as a principal PTP5 region attributable to PfPV1 binding. We also demonstrated that PfPV1 and PF3D7_0801000 interact directly. However, we could not identify any shared amino acid sequence/motif(s) between PTP5_Tr2, and PF3D7_0801000 that would be responsible for the PfPV1 binding. These findings suggest two possibilities regarding the interaction of these molecules. Either, the interaction is probably dependent on multiple structural motifs or, the binding motif lacks conserved sequence, but is dependent on amino acid hydrophobicity and sequence length are contributory to binding, as observed in signal peptides30. Further biochemical analysis, in future, would elucidate the mechanism of interaction and identify the specific PfPV1 binding motifs.
The importance of PfPV1-PTP5 interaction to PTP5 export was explored through a comparative subcellular localization of PTP5FL-HA, PTP5ΔPHR-HA and PTP5C-HA. We demonstrated that PTP5ΔPHR-HA export was arrested in PV space (Figs 5d and 6b), suggesting a critical role of PHR in the PTP5 export. This observation highlights, for the first time, the essential role of a protein’s C-terminal region in its export. The PTP5ΔPHR-HA arrested in export has an intact N-terminal sequence consisting of native PEXEL and PEXEL-proximal downstream residues. Previous reports indicate that the less conserved residues downstream of matured N-terminus are considered responsible for protein export31,32. Furthermore and consistent with this observation, PTP5C-HA export was similarly arrested in the PV space (Fig. 5e). Hence, it was proposed that both the downstream PEXEL and PHR are essential for PTP5 export.
The exported proteins are proteolytically cleaved by PMV on the C-terminal side of the conserved leucine in the PEXEL sequence RxLxE/D/Q and is N-acetylated (Ac-xE/Q/D)20. After PMV cleavage, further steps in export such as the mechanism of protein recognition at PV space, remains unclear. Our findings suggest that PTEX auxiliary proteins such as PfPV1 play an important role in export protein recognition in the PV space. However, further in-depth biochemical analysis is required to elucidate PfPV1 – protein interactions required for its role in the export protein recognition.
Recently, while this paper was being reviewed, Steven, B. et al. reported on the findings of PV1-HA interacting proteins mass spectrometry as well as its knock down phenotype analyses33. Consistent with our results, it was reported that PV1-HA co-precipitated some exported proteins and PTEX components, as well as co-localized with dense granule protein when observed by IFA. Notably, knockdown of PV1 resulted in altered knob morphology, reduced cell rigidity, and significant decrease in expression of export proteins such as PfEMP1, KAHRP, and PfEMP3. This is also in line with our hypothesis that PV1 is important for protein export. Further study is needed to clarify the exact role of interaction between PV1 and PTP5 for subsequent PTP5 export.
In conclusion, PfPV1 was characterized as a novel merozoite dense granule protein that interacts with proteins exported to the cytosol of an infected erythrocyte. Our biochemical analyses demonstrated that PfPV1 interacts directly with PTP5 via PTP5 PHR; a region identified in this study. Moreover, the arrest of PTP5ΔPHR-HA export at the PV strongly suggests that PHR is essential for PTP5 export. Put together, the PfPV1 data presented here strengthens the need for further studies aimed at unraveling mechanisms of protein export crucial for parasite growth and virulence, which could offer novel approaches to malaria intervention.
Materials and Methods
Construction of plasmids and preparation of purified recombinant proteins
All the sequences of primers for the construction of plasmids used in this study are summarized in Table S1. Pfpv1 and ptp5_tr0 were amplified from cDNA derived from P. falciparum 3D7 strain using primers; PV1F and PV1R, PTP5F and PTP5R respectively. Ptp5_tr1 and ptp5_tr2 were amplified from plasmids harboring pfptp5_tr0 using primers; PTP5F and PTP5Tr1R, PTP5Tr2F and PTP5R respectively. N-terminal truncate of Pf3D7_0801000 (Pf3D7_0801000N) spanning 93 aa – 494 aa was amplified from cDNA derived from P. falciparum 3D7 strain using primers; 0801NF and 0801NR. The amplified DNA was restricted with XhoI and NotI, and ligated into WGCFS vector, pEU, for N-terminal glutathione S-transferase (GST) or C-terminal hexa-histidine (His) fusion protein expression (CellFree Sciences, Matsuyama, Japan). Ptp5fl was PCR amplified from P. falciparum 3D7 strain cDNA using primers PTP5FL-F and PTP5FL-R. Ptp5Δphr was amplified from plasmids harboring ptp5fl using primers PTP5FL-F and PTP5ΔPHR-R. Ptp5c gene was commercially synthesized by GenScript (Tokyo, Japan). The resulting DNA fragments were restricted with NotI and PstI, and ligated into pD3HA plasmids for episomal expression of PTP5FL, PTP5ΔPHR and PTP5C34.
Production of mouse and rabbit antisera
As previously described35, we generated rabbit polyclonal antisera against PfPV1 (V23 to S452 3D7 sequence; PF3D7_1129100) and mouse antisera against PTP5 (T36 to K631; PF3D7_ PF3D7_1002100), AMA1 (Q25 to K546; PF3D7_1133400), RAP1 (M1 to D782; PF3D7_1410400), EXP2 (D25 to E287; PF3D7_1471100), SBP1 (Q258 to T337; PF3D7_0501300) and RON2 (K84 to Q968; PF3D7_1452000). All antigens were synthesized by WGCFS with N or C-terminal His- or GST-tag fusion protein expression. All animal antisera were purchased from KITAYAMA LABES Co., LTD. (Ina, Japan). Mouse monoclonal antibody against RESA (PF3D7_0102200) was a kind gift from Robin F. Anders36.
Western blot analysis
Western blot was conducted as previously described37. Purified schizont-rich parasite pellets from P. falciparum 3D7 strain were directly lysed in reducing or non-reducing SDS-PAGE sample buffer. The lysate was boiled at 95 °C for 5 min, centrifuged at 10,000 × g for 10 min at 4 °C, supernatant collected, and resolved by SDS-PAGE on a 12.5% e-PAGEL (ATTO). Following electroblotting, a polyvinylidene fluoride membrane was incubated with primary antibodies diluted at the following concentrations: rabbit anti-PfPV1 antibody, 1:1000; mouse anti-PTP5 antibody, 1:1000; rat anti-HA monoclonal antibody (Roche Diagnostics, Indianapolis, IN, USA), 1:100; mouse anti-PF3D7_0801000 antibody, 1:1000. After washing, the membrane was incubated with secondary antibodies diluted at the following concentrations: ECL peroxidase labeled anti-mouse antibody (GE Healthcare), 1:10000; ECL peroxidase labeled anti-rabbit antibody (GE Healthcare), 1:10,000; ECL peroxidase labeled anti-rat antibody (GE Healthcare), 1:1000.
Immunofluorescence Assay
IFA was conducted as previously described35. Briefly, samples were incubated with primary antibodies diluted at the following concentrations in blocking solution at 37 °C for 1 h. Rabbit anti-PfPV1 antibody, 1:1000; mouse anti-AMA1 antibody, 1:100; mouse anti-RAP1 antibody, 1:1,000; mouse anti-RON2 antibody, 1:100; mouse anti-RESA monoclonal antibody, 1:100; mouse anti-EXP2 antibody, 1:1000; mouse anti-SBP1 antibody, 1:100; rat anti-HA antibody, 1:100. Secondary antibodies, Alexa Fluor 488-conjugated goat anti-rabbit IgG, Alexa Fluor 488-conjugated goat anti-rat IgG, Alexa Fluor 568-conjugated goat anti-rabbit IgG, Alexa Fluor 568-conjugated goat anti-mouse IgG and Alexa Fluor 647-conjugated goat anti-mouse IgG (Invitrogen), were used at a 1:1000 dilution in blocking solution at 37 °C for 30 min. DAPI (4′, 6-diamidino-2-phenylindole) at 2 µg/ml was also added to the secondary-antibody solution to stain the nuclei. Slides were mounted in ProLong Gold Antifade reagent (Invitrogen) and observed under a 63 × oil immersion lens. Image capture and processing were performed with a confocal scanning laser microscope (LSM710; Carl Zeiss MicroImaging, Thornwood, NY). Images were processed in Adobe Photoshop (Adobe Systems Inc., San Jose, CA).
Structured illumination microscopy (SIM) imaging and image processing
The samples were prepared is same manner as for IFA. Nikon N-SIM system (Nikon, Tokyo, Japan) equipped with a 100×/1.49 TIRF oil immersion objective lens (Nikon), the iXon + electron multiplying charged-coupled device camera (Andor) and an excitation laser unit of 405 nm, 488 nm, 561 nm and 640 nm (Coherent) was used for a super-resolution optical imaging. Z-stacks of SIM optical sections were acquired with a 120 nm Z-step size. Image processing, including 3-dimensional reconstruction and co-localization analysis, were carried out using the NIS-Element Advanced Research software (Nikon).
Immunoprecipitation and detection
Immunoprecipitation was carried out as described38. Briefly, proteins were extracted from late trophozoites-/schizont-rich parasite pellets in PBS with 0.5% Nonidet P-40 containing cOmpleteTM protease inhibitor (Roche). Seventy-five μl of the supernatants were preincubated at 4 °C in suspension of protein G-conjugated beads (protein G-Sepharose 4 Fast Flow; GE Healthcare) in TNE buffer (50 mM Tris-HCl, 0.2 M NaCl, 5 mM EDTA). Aliquots of recovered supernatants were incubated with 3 μl of rabbit anti-PfPV1 or anti-GST antibodies. The recovered supernatant aliquots were again mixed with the suspension of protein G-conjugated bead. After 2 h incubation at 4 °C with continuous rotation, the beads were washed three times with TNE buffer. Finally, proteins were extracted from the protein G-conjugated beads by incubation with 1 × SDS-PAGE reducing loading buffer at 95 °C for 5 min. Final supernatants were used for Western blot analysis using panel of mouse antisera against 286 antigens synthesized by WGCFS (Table S2) at the same dilution (1/1000). When the immunoprecipitated samples were screened, we applied the following criteria for the selection; 1) single major band in the anti-PV1 imunoprecipitated sample should be detected; 2) the band should not be detected in negative control sample (immunoprecipitation sample prepared with anti-GST antibody). Only the two proteins among the 286 passed the criteria. The positive signal of the co-immunprecipitated PTP5 (n = 3) was quantified by Image-J (https://imagej.nih.gov/ij/). The relative density was determined in relation to the background in empty lane (Fig. 4c).
Surface Plasmon Resonance
All SPR experiments were performed using a Biacore X100 instrument (GE Healthcare) according to manufacturer’s instructions. The Biacore X100 evaluation software was used for single-cycle or multi-cycle kinetic analysis. Sensor CM5, amine coupling reagents, and buffers were purchased from GE Healthcare. Fresh HBS-EP + (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.05% (v/v) surfactant P20) was used as running buffer for all SPR experiments. Blank flow cells were used to subtract buffer effects on sensorgrams. After subtraction of the contribution of bulk refractive index, and nonspecific interactions with the CM5 chip surface, individual association (ka) and dissociation (kd) rate constants were obtained by global fitting of data. Measurement conditions were optimized so that the contribution of mass transport to the observed values of KD was negligible.
Immunoelectron microscopy
Parasites were fixed, embedded in LR White resin (Polysciences, Warrington, PA), and ultrathin sections were immunostained as previously described37. Rabbit anti-PfPV1 antibody was used at a dilution of 1:200. Samples were examined with transmission electron microscope (JEM-1230; JEOL, Tokyo, Japan).
P. falciparum culture and transfection
P. falciparum 3D7 strain was a kind gift from the National Institute of Allergy and Infectious Diseases (NIAID), and cultured as previously described39. Briefly, parasites were maintained in continuous culture of 2% hematocrit O+ human erythrocytes, incubated at 37 °C in RPMI-1640 complete medium. RPMI-1640 complete medium was supplemented with 5% heat-inactivated pooled type AB+ human serum and 0.25% Albumax II (Invitrogen, Carlsbad, CA), 200 mM hypoxanthine (Sigma, St. Louis, MO) and 10 µg/ml gentamicin (Invitrogen). The human erythrocytes and plasma were obtained from the Japanese Red Cross Society. For transfection, parasitized erythrocytes were resuspended in 200 µl of cytomix (120 mM KCl, 0.15 mM CaCl2, 2 mM EGTA, 5 mM MgCl2, 10 mM K2HPO4/KH2PO4, 25 mM HEPES, pH 7.4) containing 100 µg of plasmid DNA. Electroporation was performed in 2 mm cuvette using a Gene Pulser Xcell Electroporation System (Bio-Rad, Hercules, CA) at the following conditions; 0.31 kV, 950 µF and ∞ Ω. After electroporation, the erythrocytes were immediately resuspended in complete medium. WR99210 was added to 10 nM in the culture medium 1 day after electroporation, and culture was maintained under same drug pressure until drug-resistant parasites appeared.
Ethical approval
The use of human erythrocytes and human plasma for parasite culture was approved by the ethical review committee of Ehime University Hospital (Aidaiibyourin 1301005). The experiment was conducted in accordance with approved protocols and regulations.
References
WHO. World Malria Report2016 (2016).
Spillman, N. J., Beck, J. R. & Goldberg, D. E. Protein export into malaria parasite-infected erythrocytes: mechanisms and functional consequences. Annu Rev Biochem 84, 813–841, https://doi.org/10.1146/annurev-biochem-060614-034157 (2015).
Raventos-Suarez, C., Kaul, D. K., Macaluso, F. & Nagel, R. L. Membrane knobs are required for the microcirculatory obstruction induced by Plasmodium falciparum-infected erythrocytes. Proceedings of the National Academy of Sciences 82, 3829–3833 (1985).
Heiber, A. et al. Identification of new PNEPs indicates a substantial non-PEXEL exportome and underpins common features in Plasmodium falciparum protein export. PLoS Pathog 9, e1003546 (2013).
de Koning-Ward, T. F. et al. A newly discovered protein export machine in malaria parasites. Nature 459, 945–949 (2009).
Beck, J. R., Muralidharan, V., Oksman, A. & Goldberg, D. E. PTEX component HSP101 mediates export of diverse malaria effectors into host erythrocytes. Nature 511, 592–595 (2014).
Elsworth, B. et al. PTEX is an essential nexus for protein export in malaria parasites. Nature 511, 587–591 (2014).
Bullen, H. E. et al. Biosynthesis, localization, and macromolecular arrangement of the Plasmodium falciparum translocon of exported proteins (PTEX). Journal of Biological Chemistry 287, 7871–7884 (2012).
Matthews, K. et al. The Plasmodium translocon of exported proteins (PTEX) component thioredoxin‐2 is important for maintaining normal blood‐stage growth. Molecular microbiology 89, 1167–1186 (2013).
Matz, J. M., Matuschewski, K. & Kooij, T. W. Two putative protein export regulators promote Plasmodium blood stage development in vivo. Molecular and biochemical parasitology 191, 44–52 (2013).
Chu, T., Lingelbach, K. & Przyborski, J. M. Genetic evidence strongly support an essential role for PfPV1 in intra-erythrocytic growth of P. falciparum. PloS one 6, e18396 (2011).
Mesén-Ramírez, P. et al. Stable Translocation Intermediates Jam Global Protein Export in Plasmodium falciparum Parasites and Link the PTEX Component EXP2 with Translocation Activity. PLoS Pathog 12, e1005618 (2016).
Elsworth, B. et al. Proteomic analysis reveals novel proteins associated with the Plasmodium protein exporter PTEX and a loss of complex stability upon truncation of the core PTEX component, PTEX150. Cellular microbiology 18, 1551–1569 (2016).
Leech, J. H., Barnwell, J. W., Miller, L. H. & Howard, R. J. Identification of a strain-specific malarial antigen exposed on the surface of Plasmodium falciparum-infected erythrocytes. The Journal of experimental medicine 159, 1567–1575 (1984).
Su, X. Z. et al. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82, 89–100 (1995).
Maier, A. G. et al. Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell 134, 48–61 (2008).
Tshefu, K. & James, M. Relationship of antibodies to soluble Plasmodium falciparum antigen (Pf70) and protection against malaria in a human population living under intense transmission in Kinshasa, Zaire. Tropical medicine and parasitology: official organ of Deutsche Tropenmedizinische Gesellschaft and of Deutsche Gesellschaft fur Technische Zusammenarbeit (GTZ) 46, 72–76 (1995).
Hiller, N. L. et al. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science 306, 1934–1937 (2004).
Marti, M., Good, R. T., Rug, M., Knuepfer, E. & Cowman, A. F. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science 306, 1930–1933 (2004).
Boddey, J. A., Moritz, R. L., Simpson, R. J. & Cowman, A. F. Role of the Plasmodium export element in trafficking parasite proteins to the infected erythrocyte. Traffic 10, 285–299 (2009).
Boddey, J. A. et al. Role of plasmepsin V in export of diverse protein families from the Plasmodium falciparum exportome. Traffic 14, 532–550 (2013).
Dixon, M. W. et al. Targeting of the Ring Exported Protein 1 to the Maurer’s Clefts is Mediated by a Two‐Phase Process. Traffic 9, 1316–1326 (2008).
Haase, S. et al. Sequence requirements for the export of the Plasmodium falciparum Maurer’s clefts protein REX2. Molecular microbiology 71, 1003–1017 (2009).
Blisnick, T. et al. Pfsbp1, a Maurer’s cleft Plasmodium falciparum protein, is associated with the erythrocyte skeleton. Molecular and biochemical parasitology 111, 107–121 (2000).
Aikawa, M. et al. Pf155/RESA antigen is localized in dense granules of Plasmodium falciparum merozoites. Experimental parasitology 71, 326–329 (1990).
Nyalwidhe, J. & Lingelbach, K. Proteases and chaperones are the most abundant proteins in the parasitophorous vacuole of Plasmodium falciparum‐infected erythrocytes. Proteomics 6, 1563–1573 (2006).
Riglar, D. T. et al. Spatial association with PTEX complexes defines regions for effector export into Plasmodium falciparum-infected erythrocytes. Nature communications 4, 1415 (2013).
Torii, M., Adams, J. H., Miller, L. H. & Aikawa, M. Release of merozoite dense granules during erythrocyte invasion by Plasmodium knowlesi. Infection and immunity 57, 3230–3233 (1989).
Perkins, J. R., Diboun, I., Dessailly, B. H., Lees, J. G. & Orengo, C. Transient protein-protein interactions: structural, functional, and network properties. Structure 18, 1233–1243 (2010).
Chou, M. M. & Kendall, D. A. Polymeric sequences reveal a functional interrelationship between hydrophobicity and length of signal peptides. Journal of Biological Chemistry 265, 2873–2880 (1990).
Boddey, J. A. et al. Export of malaria proteins requires co-translational processing of the PEXEL motif independent of phosphatidylinositol-3-phosphate binding. Nature communications 7, 10470 (2016).
Grüring, C. et al. Uncovering common principles in protein export of malaria parasites. Cell host & microbe 12, 717–729 (2012).
Steven, B. et al. An exported protein-interacting complex involved in the trafficking of virulence determinants in Plasmodium-infected erythrocytes. Nature communications 8, 16044 (2017).
Riglar, D. T. et al. Super-resolution dissection of coordinated events during malaria parasite invasion of the human erythrocyte. Cell host & microbe 9, 9–20 (2011).
Ito, D. et al. RALP1 is a rhoptry neck erythrocyte-binding protein of Plasmodium falciparum merozoites and a potential blood-stage vaccine candidate antigen. Infect Immun 81, 4290–4298, https://doi.org/10.1128/IAI.00690-13 (2013).
Culvenor, J. G., Day, K. P. & Anders, R. F. Plasmodium falciparum ring-infected erythrocyte surface antigen is released from merozoite dense granules after erythrocyte invasion. Infect Immun 59, 1183–1187 (1991).
Arumugam, T. U. et al. Discovery of GAMA, a Plasmodium falciparum merozoite micronemal protein, as a novel blood-stage vaccine candidate antigen. Infect Immun 79, 4523–4532, https://doi.org/10.1128/IAI.05412-11 (2011).
Ito, D. et al. Plasmodial ortholog of Toxoplasma gondii rhoptry neck protein 3 is localized to the rhoptry body. Parasitology international 60, 132–138 (2011).
Trager, W. & Jensen, J. B. Human malaria parasites in continuous culture. Science 193, 673–675 (1976).
Acknowledgements
We thank the Japanese Red Cross Society for providing human erythrocytes and human plasma. We appreciate Masachika Shudo at the Division of Analytical Bio-Medicine, the Advanced Research Support Center (ADRES), Ehime University for their technical assistance. This work was supported in part by MEXT KAKENHI (JP23117008) and JSPS KAKENHI (JP25460517, JP26253026, JP26670202, JP26860279, JP15H05276, JP16K15266) in Japan. This work was also partly supported by IMSUT Joint Research Project (2016–2019). We also thank Jacobus Pharmaceuticals for providing the Plasmodium falciparum dihydrofolate reductase inhibitor, WR99120.
Author information
Authors and Affiliations
Contributions
M.M., H.N., B.N.K., D.I., T.N., J.W.L., K.T., T.I. and M.T. conducted experiments. E.T. and T.T. designed the study. E.T., E.H.N., B.N.K. and T.T. wrote the paper.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Morita, M., Nagaoka, H., Ntege, E.H. et al. PV1, a novel Plasmodium falciparum merozoite dense granule protein, interacts with exported protein in infected erythrocytes. Sci Rep 8, 3696 (2018). https://doi.org/10.1038/s41598-018-22026-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-22026-0
This article is cited by
-
A single-cell atlas of Plasmodium falciparum transmission through the mosquito
Nature Communications (2021)
-
Antibodies against a short region of PfRipr inhibit Plasmodium falciparum merozoite invasion and PfRipr interaction with Rh5 and SEMA7A
Scientific Reports (2020)
-
Cell-Free Protein Synthesis: A Promising Option for Future Drug Development
BioDrugs (2020)
-
Characterization of a novel secretory spherical body protein in Babesia orientalis and Babesia orientalis-infected erythrocytes
Parasites & Vectors (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.