Abstract
The sulfated polysaccharides from Enteromorpha prolifera (PE) are a potential source of anticoagulant agents. In this study, the PE was degraded by specific degradase and five hydrolysis products with different molecular weights were prepared. The product of 206 kDa is a kind of high rhamnose-containing polysaccharide with sulfate ester (34.29%). It could effectively prolong the activated partial thromboplastin time (APTT), which indicated inhibition of the intrinsic coagulation pathway. The artificial neural network (ANN) was built to realize the directional preparation of anticoagulant-active polysaccharides. Based on monitoring glucose concentration on-line, a visualization system of enzymatic hydrolysis was developed to simplify the operation of ANN. The model could be further applied to predict molecular weights of polysaccharides that possess diverse biological activities.
Similar content being viewed by others
Introduction
Enteromorpha prolifera, one of the most common green algae, is widely distributed along the intertidal zone of coastal areas1. It is regarded as an abundant source of carbohydrates, protein, minerals, crude fiber, and vitamins2,3. Polysaccharides from E. prolifera (PE) possess a wide range of pharmacological properties which can be evidenced by their antitumor, antioxidant, and anticoagulant activities4,5. Among these activities, the anticoagulant property is becoming a hot topic in natural products research.
The molecular weight and sulfate esters are the most important parameters for determining anticoagulant activities6. However, the high molecular weight and high viscosity of polysaccharides would restrict their pharmaceutical applications. Degradation of PE to low molecular weight polysaccharides might result in more exposure of their active moieties7. In recent years research on enzymatic hydrolysis has become popular because of its advantages such as high efficiency and environmental compatibility8. However, enzymatic hydrolysis will lead to the formation of complex mixture of molecules which makes the assessment of individual species unfeasible9. Consequently, it is essential to establish a method with less experimental effort to conveniently assess the process of the enzymatic hydrolysis reaction.
Artificial neural network (ANN), a non-parametric model, utilizes interconnected mathematical neurons to form a network that could model complex functional relationships10. ANN could discover behaviors and patterns from a finite set of data (called the “training set” or “training patterns”). If an adequate training set is provided, the ANN is able to generalize the knowledge acquired during the “learning” process, responding adequately to new values not contained in the “training set”11. The ANN is an effective and applicable method to predict the relationship between dependent and independent parameters while the mathematical formulation is unavailable12.
In this study, PE with high molecular weight was extracted from E. prolifera with hot water. The PE was degraded to five fragments with low molecular weight. The anticoagulant activities of the six sulfated polysaccharides were investigated. The ANN was developed to predict molecular weights of sulfated polysaccharides that exhibited potent anticoagulant activity.
Results and Discussion
Chemical characterizations
As shown in Table 1, PE consisted of four monosaccharides, including rhamnose (41.42%), glucose (32.53%), xylose (13.78%), and glucuronic acid (11.48%). Shi et al.13 reported a polysaccharide from E. prolifera (Zhejiang, China), which was composed of rhamnose (67.8%), glucose (18.6%), xylose (7.7%), galactose (4.0%), and mannose (1.4%). Kim et al.14 prepared three polysaccharides from E. prolifera (Wando, Korea), and they were composed of rhamnose (57.1–87.6%), glucose (3.6–39.1%), and xylose (2.4–8.8%). Altogether, rhamnose was the major component of the polysaccharides in E. prolifera.
As can be seen from Table 1, the sulfate ester content of PE was 23.96%. Li et al.15 reported that content of sulfate ester in the polysaccharide from E. prolifera was 23.17%. Zhang, Wang, Mo, and Qi7 reported that the polysaccharide from E. linza contained 22.4% sulfate ester. Qi et al.5 prepared the polysaccharide from E. clathrate in which the sulfate ester content was 31%. Consequently, polysaccharides from green alga Enteromorpha were rich in sulfate esters, compared with those from brown alga Eisenia bicyclis (13.2%)16 and red alga Furcellaria lumbricalis (15.2%)17. Qiao et al.18 reported that polysaccharide from E. prolifera exhibited well-behaved pseudoplastic and thixotropic property. As is shown in Table 1, the molecular weight of PE was 1012 kDa. Polysaccharides with high molecular weights and apparent viscosity are difficult to pass through organizational barriers, which might limit their biological activities19.
Anticoagulant activity assay
We prepared five low molecular weight fractions with the method of enzymatic hydrolysis. The anticoagulant activities of all the samples were determined with the classical coagulation APTT, TT, and PT assays in vitro that characterize different stages of the coagulation process. APTT is employed to evaluate the coagulation factors in the intrinsic blood coagulation pathway. PT is used to characterize the extrinsic coagulation factors while an increase in the TT suggests either thrombin inhibition or impaired fibrin20. In fact, there was a lack of PT and TT activities in all of the samples.
As seen in Fig. 1, the APTT were prolonged by PE, PE1, PE2, and PE3 in a concentration-dependent manner. The PE3 exhibited the most remarkable APTT activity and the clotting time was 60.56 s at 100 μg/mL. The prolongation of APTT suggests inhibition on the intrinsic coagulation pathway.
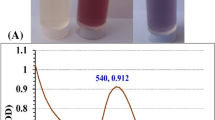
The HPGPC chromatogram of PE3 is illustrated in Fig. 2(A). The HPGPC chromatogram appeared as a single and symmetrical peak, which demonstrated PE3 was a kind of homogeneous polysaccharide. The FTIR spectrum of PE3 is shown in Fig. 2(C). The absorption bands at 3433 and 2927 cm−1 were generated by the stretching vibration of O-H and stretching vibration of C-H, respectively. The bands at 1635 and 1420 cm−1 suggested the stretching vibration of C=O and C-O of a carboxyl group, indicating the presence of uronic acid21. The bands at 1254 and 853 cm−1 were attributed to the stretching vibration of S=O and C-O-S, and indicated the existence of sulfate esters22.
The structural characteristics of the sulfated polysaccharides play an important role in the understanding of anticoagulant activity23. According to Kamide et al.24, anticoagulant activity of sulfated cellulose significantly increased with the sulfate ester in the C-2 and C-3 sites, but not in the C-6 site. Li et al.23 reported an anticoagulant-active polysaccharide from Monostroma angicava and sulfate esters substitution occurred at C-3 of (1 → 2)-linked-L-rhamnose residues. Wang et al.25 prepared three anticoagulant-active polysaccharides from E. linza, with sulfate esters residing at the C-3 of (1 → 4)-linked- L -rhamnose residues. Our research group reported that the backbone of the polysaccharides from E. prolifera consisted of D-GlcUAp-α-(1 → 4)-3-sulfate-L-Rhap-β-(1 → 4)-D-Xylp-β-(1 → 4)-3-sulfate-L-Rhap units, with sulphate ester linked on C-3 position of Rha26. On the other hand, Wang et al.25 reported sulfated rhamnose was possible the anticoagulant compound in green algae. The data above suggest that the anticoagulant activity of sulfated polysaccharide was related to sulfate ester position, monosaccharide composition, and glycosidic linkage.
Moreover, the content of sulfate esters also plays an important role in anticoagulant activities. It was reported that anticoagulant activity of the sulfated polysaccharides was partially caused by the strong interaction between the negatively charged sulfate esters and some positively charged peptidic sequences of proteins involved in coagulation process. It is known that Lysine and Arginine residues were present in the heparin-binding site27. Therefore, it is possible that the high level of sulfate ester (34.29%) increased the anticoagulant activity of PE3.
The molecular weight of sulfated polysaccharide is another important factor influencing its anticoagulant activity. It was reported that the sulfated polysaccharides from Monostroma latissimum required longer chains to achieve the inhibition of thrombin28. On the other hand, the spatial arrangement of sulfate ester was reported being important for anticoagulant activity29. Therefore, the spatial arrangement of PE3 might be the advantageous conformation and it could promote a better interaction with blood coagulation factors. As can be seen in Fig. 1, PE4 and PE5 with low molecular weights showed no prolongation of APTT. We assumed that some structural change, such as a shift in conformation, caused the extension of enzymatic hydrolysis time.
In conclusion, the potent anticoagulant activity of PE3 depended on the integration of many factors. Realization of accurate preparation of PE3 would enhance its application in pharmaceutical industries.
Artificial neural network for predicting molecular weight distributions
The ANN is a parallel processing network which determined the complex nonlinear relationships between independent and dependent variables30. The back-propagation (BP) network is one of the most widely used ANN for multilayered feed-forward networks31. The learning rule of BP network is to use the steepest descent method to continuously adjust the weights and thresholds32. In this work, a feed-forward neural network trained with an error BP algorithm was employed in constructing and training the ANN. It is noteworthy that the Levenberg-Marquardt algorithm was applied in the training process. This is a combination of the Grade method and the Gauss-Newton method. It was reported that when such an algorithm is used, time-consuming search can be significantly reduced33.
According to Kolmogorov theorem, a three-layer network is sufficient to complete any n-dimensional to m-dimensional nonlinear mapping, thereby setting only one hidden layer structure34. In the network, enzyme concentration, substrate concentration, enzymatic temperature, enzymatic time, and glucose concentration were assigned as five input layer nodes while molecular weight was output layer node (Fig. 3). The input layer provides the hidden layer with the sum of the weighted input parameters. These weights are adjusted automatically by the BP algorithm during training.
Different combinations of the transfer functions (logsig, tansig, and purelin) were studied to determine the best combination of two transfer functions that will yield accurate results (Table 2). The performance of these combinations was evaluated on the basis of mean absolute percentage error (MAPE). The MAPE of 0–10%, 10–20%, 20–50%, and >50% represents high prediction accuracy, good prediction, normal accuracy, and bad accuracy, respectively35. As can be seen in Table 2, the MAPE of all the combinations is in the range of 0–10%. The tansig and purelin were used as the transfer functions for input and output layer, respectively, because of the minimum MAPE (2.14%).
The mean square error (MSE) versus the number of repetitions for training is illustrated in Fig. 4(A). In this case, the MSE of training data is equal to 0.00016, after 173 epochs. As shown in Fig. 4(B), the R value is 0.99923. The model fits well with the actual data when R approaches to 136. Histogram of deviation margin for the optimal ANN is presented in Fig. 4(C). It can be seen that deviation distribution is concentrated around zero, which indicates that the ANN has a remarkable accuracy. A comparison between experimental data and outputs of ANN model is shown in Fig. 4(D). Overall, there is a good fitting between the estimated values and the actual values.
The 10% of the testing sets (Table 3) were used for testing the precision of the ANN model. The mean absolute percentage error (MAPE) of molecular weights for testing sets is 1.86% (Table 3). According to Khoshnevisan et al.37, a MAPE value of less than 10% indicates that the best prediction has been achieved. Therefore, this network could be used with high reliability to estimate the molecular weights. The testing performance and regression of ANN (Fig. 5). As can be seen in Fig. 5(A), testing was stopped after 150 epochs with MSE value of approximately 0.0034. As shown in Fig. 5(B), the R value is 0.99558, which implies good fits between predicted values and the actual values. Therefore, this network could be used with high reliability to estimate the molecular weights.
When enzymatic hydrolysis condition is changed, the users need to run MATLAB software and modify parameters in the procedure. However, this will make the manipulation complicated. In order to solve the problem, the visual prediction system was developed to create a convenient user interface (Fig. 6). According to Li et al.38, glucose was one of the hydrolysates. Glucose concentration could reflect the enzymatic hydrolysis process to a certain extent. Consequently, the ANN model could predict molecular weight of the sulfated polysaccharide efficiently by monitoring glucose concentration on-line.
Conclusions
In the present work, enzymatic products PE3 exhibited the most potent anticoagulant activity. Results indicate that sulfate ester, monosaccharide composition, and molecular weight are important factors for determining the anticoagulant activity in polysaccharides from E. prolifera. The ANN model was built to simulate the distributions of molecular weight during the complex enzymatic reaction. The developed vision-based ANN model could be an applicable and convenient tool to obtain the desirable polysaccharides with prominent anticoagulant activity. Further efforts along this direction may provide a new dimension for the treatment of thrombosis.
Materials and Methods
Materials
E. prolifera was collected from the coasts of Qingdao, China. Dialysis membrane was purchased from Lvniao (Yantai, China) and molecular weight was cut off at 3500 Da. Dextran standards (50, 80, 150, 270, 410, and 670 kDa), D-glucose, L-rhamnose, D-xylose, and D-glucuronic acid were purchased from Sigma Co. (USA). Activated partial thromboplastin time (APTT), thrombin time (TT), and prothrombin time (PT) reagent kits were purchased from Nanjing Jiancheng Bioengineering Institute. Coagulation analyzer SYSMEX CA 6000 was purchased from TOA Medical Electronics Co. (Kobe, Japan). All other chemicals and reagents were analytical grade.
Extraction and quantification of PE
The PE was extracted with the improved method as described by Li et al.38. The milled alga (60 g) was dipped into 40 volumes of tap water, homogenized and extracted at 100 °C for 1 h. The water extraction solution was centrifuged (4000 × g, 10 min), and the supernatant was dipped in 2 times volume of 95% ethanol to remove pigment. After centrifugation, the sediment was collected, and dissolved in distilled water.
Preparation for degradase for PE (DPE) and Enzyme assay
Alteromonas sp. A321 was cultivated and its extracellular supernatant was extracted according to Li et al.38. Then, the supernatant was brought to 60% (W/V) saturation with (NH4)2SO4. The precipitate collected was dissolved in 20 mM phosphate buffer saline (pH 7.5), yielding DPE. The degradase activity was assayed by the method described by Fenice, Selbmann, Zucconi, and Onofri39.
Preparation of polysaccharides with different molecular weights
The PE [200 mL; 1% (w/v)] was hydrolyzed by the DPE (0.81 U/mL, 20 mL) for 8 h under the conditions of 35 °C, pH 6.5. The 10 mL enzymatic hydrolysate was taken out after two hours and quenched by heating at 100 °C for 5 min. Under the condition of continuous stirring the solution was precipitated by adding 2 times the volume of 95% ethanol. After centrifugation, the sediment was collected, dissolved in distilled water, and dialyzed to give one degraded product named after PE1. The PE2, PE3, and PE4 were gained in the same manner as PE1 except that the hydrolysate was drawn out after 4 h, 6 h, and 8 h, respectively. During the process of preparing the PE4, the supernatant was collected after centrifugation. The supernatant was concentrated, dialyzed, and referred as PE5.
Characterization
The molecular weights of PE, PE1, PE2, PE3, PE4, and PE5 were determined by using an Agilent 1260 HPLC system (Wilmington, USA) equipped with PL Aquagel-OH 50 column (0.75 cm × 30 cm) and a refractive index detector (RID). The column was eluted with 0.2 M NaNO3 and 0.01 M NaH2PO4 at a flow rate of 0.8 mL/min. The molecular weights of all samples were estimated by referencing the calibration curve made from dextran standards.
The total sugar was determined by phenol-sulfuric acid method using glucose as standard40. Sulfate ester content was estimated according to the method described by Therho and Hartiala41. The monosaccharide composition was analyzed by using reversed-phase HPLC after pre-column derivatization42. The vibrational spectra of different atomic and polar bonds were studied by Fourier transform infrared spectroscopy (FTIR). The FTIR spectra was recorded in the wavelength range of 4000-400 cm−1 at a resolution of 4 cm−1 using MAGNA-IR 560 E.S.P (Nicolet, USA).
Anticoagulant activity assays
Activated partial thromboplastin time (APTT), thrombin time (TT), and prothrombin time (PT) assays were carried out according to the method described by Pawlaczyk et al.43 Saline and the unfractionated heparin were used as negative and positive controls, respectively. All clotting assays were performed with three individual replicates to estimate the mean clotting time. The anticoagulant activity was expressed as the clotting time.
Experimental design
Enzymatic temperature (25, 30, and 35 °C), enzymatic time (1, 2, 3, 4, 5, 6, 7, 8, 9, 10, and 11 h), substrate concentration (4, 6, 8, 10, and 12 mg/mL), and enzyme dose (8.10, 9.72, and 12.15 U) were considered according to the situation of enzymolysis. In order to construct and train the ANN model, two hundred and thirty-one experimental groups were designed referring to the uniform orthogonal design method.
ANN modeling
The training of ANN was accomplished by adapting the strengths or weights of the connections among the input, intermediate and output neurons, which were capable of storing memory and information. By achieving the learning ability, ANN produced the desired responses according to the given decision variables44.
In this study, the ANN model was developed using the neural network toolbox in MATLAB version 8.1. The 231 experimental data points were employed to build the ANN model. They were divided into training and testing sets with a ratio of 90% and 10%, respectively. Flowchart of the optimal neural network is presented in Fig. 7. The equation below was used for data normalization:
where Xi, X, Xmin, and Xmax represented the normalized value, actual value, minimum value, and maximum value, respectively.
The BP network and Levenberg-Marquardt optimization algorithm was applied in the training process. The MSE between the output values and targeted values are calculated and returned back to the hidden layer. Weights between inputs and hidden layer are also calculated, which are updated between the hidden layer and the output layer.
Statistical analysis
All experiments were repeated in triplicate, and the data were reported as mean ± standard deviation and evaluated by one-way ANOVA. Statistical analysis was performed using MATLAB mathematical software (version 8.1).
References
Zhao, J. et al. Genetic variation of Ulva (Enteromorpha) prolifera (Ulvales, Chlorophyta)-the causative species of the green tides in the Yellow Sea, China. J. Appl. Phycol. 23, 227–233 (2011).
Aguilera-Morales, M., Casas-Valdez, M., Carrillo-Dominguez, S., Gonzalez-Acosta, B. & Perez-Gil, F. Chemical composition and microbiological assays of marine algae Enteromorpha spp. as a potential food source. J. Food Compos. Anal. 18, 79–88 (2005).
Mamatha, B. S., Namitha, K. K., Senthil, A., Smitha, J. & Ravishankar, G. A. Studies on use of Enteromorpha in snack food. Food Chem. 101, 1707–1713 (2007).
Cho, M. L., Lee, H. S., Kang, I. J., Won, M. H. & You, S. G. Antioxidant properties of extract and fractions from Enteromorpha prolifera, a type of green seaweed. Food Chem. 127, 999–1006 (2011).
Qi, X. H. et al. Chemical characteristic of an anticoagulant-active sulfated polysaccharide from Enteromorpha clathrate. Carbohyd. Polym. 90, 1804–1810 (2012).
Mulloy, B. The specificity of interactions between proteins and sulfated polysaccharides. An. Acad. Bras. Cienc. 77, 651–664 (2005).
Zhang, Z., Wang, X., Mo, X. & Qi, H. Degradation and the antioxidant activity of polysaccharide from Enteromorpha linza. Carbohyd. Polym. 92, 2084–2087 (2013).
Tzu-Wen, L., Jia-Lin, H. & San-Lang, W. Production and purification of a protease, a chitosanase, and chitin oligosaccharides by Bacillus cereus TKU022 fermentation. Carbohyd. Res. 362, 38–46 (2012).
Pinto, G. A., Giordano, R. L. C. & Giordano, R. C. Neural Network Inference of Molar Mass Distributions of Peptides during Tailor-Made Enzymatic Hydrolysis of Cheese Whey: Effects of pH and Temperature. Appl. Biochem. Biotech. 143, 142–152 (2007).
Sha, W. & Edwards, K. L. The use of artificial neural networks in materials science based research. Mater. Design. 28, 1747–1752 (2007).
Salgado, C. M., Brandão, L. E. B., Conti, C. C. & Salgado, W. L. Density prediction for petroleum and derivatives by gamma-ray attenuation and artificial neural networks. Appl. Radiat. Isotope. 116, 143–149 (2016).
Parthiban, T., Ravi, R. & Kalaiselvi, N. Exploration of artificial neural network [ANN] to predict the electrochemical characteristics of lithium-ion cells. Electrochim. Acta. 53, 1877–1882 (2007).
Shi, M. J. et al. Carboxymethylated degraded polysaccharides from Enteromorpha prolifera: Preparation and in vitro antioxidant activity. Food Chem. 215, 76–83 (2017).
Kim, J. K., Cho, M. L., Karnjanapratum, S., Shin, I. S. & You, S. G. In vitro and in vivo immunomodulatory activity of sulfated polysaccharides from Enteromorpha prolifera. Int. J. Biol. Macromol. 49, 1051–1058 (2011).
Li, B. et al. Degradation of sulfated polysaccharides from Enteromorpha prolifera and their antioxidant activities. Carbohyd. Polym. 92, 1991–1996 (2013).
Ermakova, S. et al. Water-soluble polysaccharides from the brown alga Eisenia bicyclis: Structural characteristics and antitumor activity. Algal Res. 2, 51–58 (2013).
Yang, B. et al. Structural characterisation and bioactivities of hybrid carrageenan-like sulphated galactan from red alga Furcellaria lumbricalis. Food Chem. 124, 50–57 (2011).
Qiao, L. K. et al. Rheological properties, gelling behavior and texture characteristics of polysaccharide from Enteromorpha prolifera. Carbohyd. Polym. 136, 1307–1314 (2016).
Zhang, Y. et al. Modification, characterization and structure-anticoagulant activity relationships of persimmon polysaccharides. Carbohyd. Polym. 82, 515–520 (2010).
Ye, L., Xu, L. & Li, J. Preparation and anticoagulant activity of a fucosylated polysaccharide sulfate from a sea cucumber Acaudina molpadioidea. Carbohyd. Polym. 87, 2052–2057 (2012).
Zhao, X. et al. Preparation of low-molecular-weight polyguluronate sulfate and its anticoagulant and anti-inflammatory activities. Carbohyd. Polym. 69, 272–279 (2007).
Mao, W. J. et al. Heparinoid-active two sulfated polysaccharides isolated from marine green alage Monostroma nitidum. Carbohyd. Polym. 74, 834–839 (2008).
Li, N. et al. Structure and anticoagulant property of a sulfated polysaccharide isolated from the green seaweed. Monostroma angicava. Carbohyd. Polym. 159, 195–206 (2017).
Kamide, K., Okajima, K., Matsui, T., Ohnishi, M. & Kobayashi, H. Roles of Molecular Characteristics in Blood Anticoagulant Activity and Acute Toxicity of Sodium Cellulose Sulfate. Polym. J. 15, 309–321 (1983).
Wang, X., Zhang, Z., Yao, Z., Zhao, M. & Qi, H. Sulfation, anticoagulant and antioxidant activities of polysaccharide from green algae Enteromorpha linza. Int. J. Biol. Macromol. 58, 225–230 (2013).
Yu, Y., Li, Y., Du, C., Mou, H. & Wang, P. Compositional and structural characteristics of sulfated polysaccharide from Enteromorpha prolifera. Carbohyd. Polym. 165, 221–228 (2017).
Bourin, M. C. & Lindahl, U. Glycosaminoglycans and the regulation of blood coagulation. Biochem. J. 289, 313–330 (1993).
Zhang, H. J. et al. Chemical characteristics and anticoagulant activities of a sulfated polysaccharide and its fragments from Monostroma latissimum. Carbohyd. Polym. 71, 428–434 (2008).
Huynh, R., Chaubet, F. & Jozefonvicz, J. Anticoagulant properties of dextranmethylcarboxylate benzylamide sulfate (DMCBSu); a new generation of bioactive functionalized dextran. Carbohyd. Res. 332, 75–83 (2001).
Shakeri, S., Ghassemi, A., Hassani, M. & Hajian, A. Investigation of material removal rate and surface roughness in wire electrical discharge machining process for cementation alloy steel using artificial neural network. Int. J. Adv. Manuf. Tech. 82, 549–557 (2016).
Agatonovic-Kustrin, S. & Loescher, C. M. Qualitative and quantitative high performance thin layer chromatography analysis of Calendula officinalis using high resolution plate imaging and artificial neural network data modelling. Anal. Chim. Acta. 798, 103–108 (2013).
Li, W. X. et al. Analysis on correlation between general efficacy and chemical constituents of danggui-chuanxiong herb pair based on artificial neural network. China Journal of Chinese Materia Medica 37, 2935–2942 (2012).
Wang, Z., Wang, F. & Su, S. Solar irradiance short-term prediction model based on BP neural network. Energy Procedia 12, 488–494 (2011).
Liu, F., Liu, W. & Tian, S. Artificial neural network optimization of Althaea rosea seeds polysaccharides and its antioxidant activity. Int. J. Biol. Macromol 70, 100–107 (2014).
Lewis, C. D. Industrial and business forecasting methods: a practical guide to exponential smoothing and curve fitting. Butterworth Scientific (1982).
Sina, H. N., Yusof, S., Hamid, N. S. A. & Rahman, R. A. Optimization of enzymatic clarification of sapodilla juice using response surface methodology. J. Food Eng. 73, 313–319 (2006).
Khoshnevisan, B., Rafiee, S., Omid, M., Mousazadeh, H. & Sefeedpari, P. Prognostication of environmental indices in potato production using artificial neural networks. J. Clean Prod. 52, 402–409 (2013).
Li, Y. P. et al. Production of enzymes by Alteromonas sp. A321 to degrade polysaccharides from Enteromorpha prolifera. Carbohyd. Polym. 98, 988–994 (2013).
Fenice, M., Selbmann, L., Zucconi, L. & Onofri, S. Production of extracellular enzymes by Antarctic fungal strain. Polar Biol. 17, 275–280 (1997).
DuBois, M., Gilles, K. A., Hamilton, J. K., Rebers, P. A. & Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28, 350–356 (1956).
Terho, T. T. & Hartiala, K. Method for determination of the sulfate content of glycosaminoglycans. Anal. Biochem. 41, 471–476 (1971).
Sun, H. H. et al. Isolation, chemical characteristics and antioxidant properties of the polysaccharides from marine fungus Penicillium sp. F23-2. Carbohyd. Polym. 78, 117–124 (2009).
Pawlaczyk, I. et al. Anticoagulant and anti-platelet activity of polyphenolic-polysaccharide preparation isolated from the medicinal plant Erigeron canadensis L. Thromb. Res. 127, 328–340 (2011).
Miaa, M. & Dhar, N. R. Prediction of surface roughness in hard turning under high pressure coolant using Artificial NeuralNetwork. Measurement 92, 464–474 (2016).
Acknowledgements
This work was supported by the project of Qingdao marine biomedical science and technology innovation center (2017-CXZX01-3-13), the Innovation and Development of Oceanic Economy Demonstration City Program during the 13th Five-Year Plan Period, and Shandong support plan for Mount Tai Scholar team of blue industry (2015 No. 260).
Author information
Authors and Affiliations
Contributions
J.F.C. and P.W. designed the research. J.F.C., Y.P.L., S.X.W. and Y.Z.C. performed the research. J.F.C. and Y.P.L., analyzed the data. J.F.C. wrote the manuscript. J.F.C., Y.P.L., H.H. and P.W. revised the manuscript. All of the authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cui, J., Li, Y., Wang, S. et al. Directional preparation of anticoagulant-active sulfated polysaccharides from Enteromorpha prolifera using artificial neural networks. Sci Rep 8, 3062 (2018). https://doi.org/10.1038/s41598-018-21556-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-21556-x
This article is cited by
-
Enzymatic degradation of polysaccharide from Enteromorpha prolifera: an efficient way to enhance its antioxidant and tyrosinase inhibitory activities
Journal of Food Measurement and Characterization (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.