Abstract
Rpb9 is a non-essential subunit of RNA polymerase II that is involved in DNA transcription and repair. In budding yeast, deletion of RPB9 causes several phenotypes such as slow growth and temperature sensitivity. We found that simultaneous mutation of multiple N-terminal lysines within histone H3 was lethal in rpb9Δ cells. Our results indicate that hypoacetylation of H3 leads to inefficient repair of DNA double-strand breaks, while activation of the DNA damage checkpoint regulators γH2A and Rad53 is suppressed in Rpb9-deficient cells. Combination of H3 hypoacetylation with the loss of Rpb9 leads to genomic instability, aberrant segregation of chromosomes in mitosis, and eventually to cell death. These results indicate that H3 acetylation becomes essential for efficient DNA repair and cell survival if a DNA damage checkpoint is defective.
Similar content being viewed by others
Introduction
To protect genomic integrity, cells must continuously detect and repair DNA damage. Among different types of DNA lesions, double-strand DNA breaks (DSBs) are the most harmful, as they can lead to translocations and deletions of large fragments of chromosomes. To ensure efficient repair of DSBs, cells activate DNA damage checkpoints–mechanisms that halt progression of the cell cycle to provide extra time for DNA repair1. Numerous endo- and exogenous factors, including biochemical processes like cellular respiration or gene transcription may lead directly or indirectly to DNA damage. One example of such an endogenous trigger involves collisions between RNA and DNA polymerases that may occur in the S phase of the cell cycle and may in turn give rise to DSBs2. In such instances, efficient removal of RNA polymerase from DNA is essential for DSB repair and for continuation of DNA replication3.
Transcription of protein-coding genes is conducted by RNA polymerase II (RNAPII), which is comprised of 12 subunits encoded by genes RPB1 to RPB12 in yeast. Among these, two subunits–Rpb4 and Rpb9–are non-essential for cell viability and gene transcription. However, their deletion gives rise to several diverse phenotypes such as slow growth, sensitivity to high and low temperatures and to nucleotide-depleting drugs4,5,6,7,8,9,10,11. Rpb9 promotes ubiquitylation and degradation of stalled RNAPII in response to UV-induced DNA damage12 and is also involved in transcription-coupled repair through its role in regulation of transcription elongation13,14,15,16. At most RNAPII promoters, selection of the proper transcription initiation start site is altered in the rpb9 mutant cells17. Additionally, Rpb9 is important for maintaining transcriptional fidelity as evidenced by the fact that RNAPII lacking the Rpb9 subunit pauses at obstacles of transcription elongation at a much lower frequency than wild type RNAPII. However, once stopped, the rpb9Δ polymerase is inefficient at resuming transcription, as Rpb9 is needed for efficient recruitment of TFIIS–the factor required for activation of nascent transcript cleavage activity of RNAPII and reactivation of the stalled polymerase18,19,20,21. Although Rpb9 is not essential for cell viability, deletion of RPB9 is synthetically lethal with disruption of the SAGA complex - the main H3 acetyltransferase in yeast9,22, as well as with the Rad6-Bre1 complex23 that is required for monoubiquitylation of histone H2B24,25. Ubiquitylation of H2B has been implicated both in regulation of RNAPII-dependent transcription and in DNA damage response. It is needed for proper activation of the DNA damage checkpoint, timely initiation of DSB repair, and for recruitment of structure-specific endonucleases to the sites of DNA repair26,27,28. These genetic interactions suggest that chromatin modifications and careful regulation of the DNA damage response become essential for cell viability in the absence of Rpb9.
Acetylation of lysine residues within N-terminal tails of histone proteins is one of the most common chromatin modifications. It weakens histone-DNA and histone-histone interactions, and also serves as a signal for recruitment of several effector proteins. In higher eukaryotes, abnormal patterns of histone acetylation and deregulated expression of chromatin modifiers have been found in various cancers29,30,31. While elevated levels of histone acetylation lead to a more open chromatin in general, some acetylation sites on histone H3 (K14, 23, 56) and histone H4 (K5, 12, 91) have been shown to be important in regulation of DNA repair pathways in particular32,33,34,35. The precise roles of different histone modifications in this process remain the subject of debate. In fission yeast, acetylation of H3 K14 has been shown to be important for DNA damage checkpoint activation36. Specifically, it was found that this modification facilitates DNA repair by directly regulating the compaction of chromatin via recruitment of the chromatin remodelling complex RSC37. Another study has revealed that budding yeast strains lacking acetylatable lysines 14 and 23 on histone H3 are sensitive to the DNA-damaging agent methyl methanesulfonate (MMS) and defective in homologous recombination (HR) repair33.
To study the role of chromatin modifications in Rpb9-mediated processes, we examined the genetic interactions between Rpb9 and acetylation of histone H3. We found that deletion of Rpb9 was lethal in cells where three or more acetylatable lysine residues were mutated in the H3 N-terminal tail. Our results show that depletion of Rpb9 leads to elevated DNA recombination and impaired activation of the DNA damage checkpoint, while repair of DSBs is inefficient in H3 hypoacetylated cells. When H3 hypoacetylation is combined with depletion of Rpb9, defective DNA damage response and unrepaired DNA lesions lead to genomic instability, aberrant segregation of DNA in mitosis and eventually cell death.
Results
H3 acetylation is required for the viability of rpb9Δ cells
RNAPII is directly and indirectly involved in the regulation of DNA transcription, repair and recombination–all processes that require access to DNA in chromatin. Although Rpb9-deficient cells are viable, they display several phenotypes like slow growth and sensitivity to elevated temperatures and genotoxic agents. Genetic interactions have revealed that RPB9 deletion is synthetically lethal with deletions of the SAGA histone acetyl-transferase complex subunits9,22. Based on these observations, we hypothesized that rpb9Δ cells might be sensitive to H3 modifications that are crucial for chromatin regulation and genome maintenance. To investigate the role of H3 N-terminal acetylation in rpb9Δ cells, we systematically mutated H3 N-terminal lysine residues to arginines to see whether any combination of H3 mutations would affect cell viability. We found that in wild type strain background all H3 mutants were viable and showed no obvious growth defects (Fig. 1a). However, in the rpb9Δ strain, several H3 mutations confer lethality (Fig. 1b). While any combination of three or more H3 acetylation site mutations was lethal in the rpb9Δ background, some diversity in the phenotypes of H3 double lysine mutants was observed. Specifically, loss of K14 acetylation had the strongest effect on viability of rpb9Δ cells, as all non-viable double mutants contained the K14R mutation. However, intact K14 alone could not rescue lethality of rpb9Δ cells when three or more other lysine residues were mutated to arginines. This suggests that changes in the overall acetylation levels of the N-terminal tail of H3 may be the prime reason for synthetic lethality with the RPB9 deletion.
Analysis of genetic interactions between Rpb9 and H3 N-terminal mutations. Cells containing wild type (a) or rpb9Δ (b) RNAPII were transformed with plasmids encoding lysine-to-arginine mutations in histone H3 N-terminal tail. 10-fold serial dilutions of cells were spotted onto synthetic complete (SC) plates lacking histidine, or containing 5-FOA. Plates lacking histidine acted as a control, where strains expressed both wt and mutant versions of H3. On 5-FOA plates, only mutant versions of H3 were expressed. His plates were photographed 3 days and 5-FOA plates 5 days after incubation at 30 °C. Transformation with plasmid encoding wt histones was included on every plate as a control.
DNA damage checkpoint activation is impaired in Rpb9-depleted cells
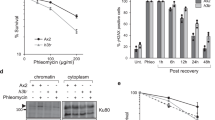
To investigate the mechanisms leading to lethality of the rpb9Δ strain in the H3 hypoacetylation background, we continued our study using the anchor-away method38 to remove Rpb9 from the H3 K9,14,23 R strain. This method allows pre-growing cells with intact RNAPII and subsequent removal of Rpb9 from the nucleus via addition of rapamycin to the growth medium, thereby phenocopying rpb9Δ cells. Since all combinations of three or more N-terminal lysine mutations of H3 were lethal in the rpb9Δ background, we continued our study using the H3 K9,14,23 R mutant as a representative example of H3 hypoacetylation. As RPB9 deletion causes slow growth in yeast, this phenotype can be used as an indicator of rapamycin-induced loss of Rpb9. When Rpb9 was removed from a strain carrying wt histone H3, cell growth rate decreased to levels comparable with the rpb9Δ strain, while depletion of Rpb9 from the H3 K9,14,23 R strain arrested cell growth entirely (Fig. 2a). These results confirmed that the anchor-away depletion of Rpb9 was efficient in our model system and was suitable for further studies of Rpb9-dependent survival of H3 K9,14,23 R cells. We additionally confirmed the efficiency of Rpb9 depletion by a spotting assay on rapamycin-containing media, where it was lethal in the H3 K9,14,23 R background (Supplementary Fig. S1).
S-phase checkpoint activation is defective in Rpb9-depleted cells. (a) Growth curves of Rpb9 anchor-away (AA) strains containing wt H3 or the K9,14,23 R mutant of H3. Rapamycin (−Rpb9) or DMSO (+Rpb9) was added to the cell growth medium at time-point 0, and culture density (cells/ml) was measured at indicated time-points. Wild type (wt H3) and rpb9Δ strains were used for reference. (b) 10-fold serial dilutions of cells with wt H3 or the H3 K9,14,23 R mutant were spotted onto SC plates containing DMSO (+Rpb9) or rapamycin (−Rpb9) with addition of indicated concentrations of MMS. (c) Western blot analysis of H2A and Rad53 phosphorylation in response to MMS treatment in Rpb9-depleted cells. Rpb9 anchor-away strains with wt or K9,14,23 R mutant H3 were incubated with DMSO (+Rpb9) or rapamycin (−Rpb9) for 6 hours before 0.01% MMS was added to the cells and samples were taken at indicated time-points. H3 is shown as a loading control. Full-size blots are shown in the Supplementary Fig. S3.
As Rpb9 is involved in DNA repair, we tested whether Rpb9-depleted, or H3 K9,14,23 R mutant cells can properly respond to DNA damage induced by MMS. While H3 K9,14,23 R mutation caused relatively mild MMS-sensitivity, the Rpb9-depleted cells were highly sensitive to long-term exposure to MMS (Fig. 2b). We confirmed that this result was not restricted to MMS treatment, as DSB induction with ionizing radiation or camptothecin caused identical phenotypes (Supplementary Fig. S2). Given that both Rpb9-depleted and H3 K9,14,23 R cells were sensitive to MMS, we hypothesized that these mutations may affect different steps in DNA repair pathway that can be tolerated separately, but become synthetically lethal in an Rpb9-deficient H3 K9,14,23 R strain.
In eukaryotic cells, genomic stability is maintained through careful coordination of DNA damage repair and cell cycle control. DNA damage checkpoints become activated to arrest the cell cycle, thereby allowing additional time for repair of DNA lesions. To test whether Rpb9-depleted cells can properly activate DNA damage checkpoints, we followed the kinetics of H2A and Rad53 phosphorylation in response to MMS treatment of cells. Phosphorylation of H2A Ser129 (γH2A) is one of the earliest checkpoint activating events that leads to Rad9-mediated recruitment and autophosphorylation of Rad53, and subsequent phosphorylation of multiple targets by Rad5339,40,41. We found that both wild type and H3 K9,14,23 R cells responded quickly to MMS, while DNA damage checkpoint activation was impaired in Rpb9-deficient cells (Fig. 2c). This indicates that activation of the γH2A-Rad9-Rad53 pathway is impaired in the absence of Rpb9 and that cells lacking this RNAPII subunit cannot adequately respond to DNA damage. Impaired activation of the DNA damage checkpoint in the Rpb9-depleted strain suggests that these cells may progress through the cell cycle with unrepaired DNA. Under normal growth conditions, this may merely lead to a decrease in the overall growth rate of the strain as only a subpopulation of cells acquires lethal amounts of DNA damage. However, if extensive DNA damage is induced throughout the entire cell population by treatment with genotoxic agents, none of the cells can survive without proper DNA damage checkpoint activation.
Depletion of Rpb9 leads to DNA repair by homologous recombination
Although DNA damage checkpoint activation was impaired in Rpb9-deficient cells, this did not explain the synthetic lethality of Rpb9 depletion and H3 hypoacetylation. With no exogenous DNA damage, the checkpoint functionality is not required for cell survival, as many checkpoint-deficient strains, such as mec1Δ and rad53Δ hypomorphs, and rad9Δ, are viable42,43,44. This suggests that in addition to aberrant checkpoint activation, Rpb9-deficiency actually induces genomic instability. To test whether depletion of Rpb9 leads to increased DNA damage, we determined the relative amount of cells with HR foci upon depletion of Rpb9 with and without exogenous induction of DNA damage by MMS. We used GFP-tagged Rad52 protein to reveal recombination sites in cells. During the S phase, Rad52 accumulates at nuclear foci that are indicative of active DNA repair by HR45. With no exogenous induction of DNA damage, Rad52 foci appeared in cells 6 hours after Rpb9 depletion (Fig. 3a). Importantly, Rpb9-depletion resulted in approximately the same amount of cells with Rad52 foci as did MMS treatment of wild type cells (Fig. 3c and d), highlighting the severity of DNA damage in the absence of Rpb9. When Rpb9-depleted strain was treated with MMS, the proportion of cells with Rad52 foci increased even further, reaching approximately 75% of the cell population (Fig. 3c and d). Interestingly, the elevated steady-state level of HR in the Rpb9-depleted strain coincides with delayed activation of the DNA damage checkpoint in these cells (Fig. 2c), suggesting that the checkpoint signalling might be saturated by the high background level of DNA repair. Moderate increases in numbers of Rad52 foci were also observed in H3 K9,14,23 R cells. When Rpb9 was depleted in this strain, Rad52 foci were detected in nearly 80% of cells (Fig. 3b and d). These results indicate that H3 K9,14,23 R and depletion of Rpb9 have a cumulative effect on induction of HR, suggesting that they act in different pathways of DNA repair.
Homologous recombination foci accumulate in Rpb9 depleted cells. Formation of Rad52-foci in response to 6-hour depletion of Rpb9 in cells with wt H3 (a), or H3 K9,14,23 R mutant (b). Additional DNA damage was induced in wt H3 cells with MMS (c). Scale bar 5 µm. (d) Quantification of S/G2 phase cells with Rad52-foci determined from three separate experiments.
H3 K9,14,23 R cells are ineffective in DSB repair and require DNA damage checkpoint activation for survival
Accumulation of DNA damage upon depletion of Rpb9 suggests that survival of these cells depends largely on the efficiency of DNA repair, and that any factor diminishing its effectiveness may become lethal. Given the sensitivity of the H3 K9,14,23 R strain to MMS (Fig. 2b), we next evaluated the efficiency of DSB repair in these cells. We transformed the strains with plasmid expressing the HO endonuclease under the control of a galactose-inducible promoter. The HO endonuclease introduces a single DSB at its recognition site in the MAT locus that is repaired primarily by HR using the silent HMR or HML loci as donor sequences46. Strains that are defective in repair of HO-induced DSB are not able to grow in the presence of continuously expressed HO endonuclease. Both wt H3 and H3 K9,14,23 R cells were able to grow on glucose-containing media, where expression of the nuclease was repressed. In contrast, when the HO nuclease was continuously activated on galactose-containing media, only cells with wt H3 were able to grow, indicating that repair of the HO-induced DSB was ineffective in the H3 K9,14,23 R strain (Fig. 4a). To estimate the efficiency of DSB repair in H3 K9,14,23 R cells, we followed the recovery of the MAT locus after shut-down of HO expression in wt H3 and H3 K9,14,23 R strains (Fig. 4b; detailed description of the assay is presented in the Supplementary Fig. S4). While the MAT locus was fully restored in cells with wt H3, it was repaired approximately in half of the H3 K9,14,23 R cells. Notably, depletion of Rpb9 did not influence the efficiency of DSB repair at the MAT locus (Fig. 4c). These results confirm that H3 acetylation is essential for efficient DSB repair and indicate that H3 hypoacetylation is lethal in the absence of Rpb9-mediated DNA damage checkpoint activation.
DSB repair is impaired in H3 K9,14,23 R cells. (a) Strains with wt H3 or H3 K9,14,23 R were transformed with GAL-HO-pRS412 plasmid and 10-fold serial dilutions of cells were spotted onto SC media lacking adenine and containing either glucose (−Ade, Glc) or galactose (−Ade, Gal). Plates were photographed after 3 days of growth at 30 °C. (b) Outline of the DSB repair assay. Schematic representation of the mating type (MAT) locus and silent mating loci (HMLα and HMRa) in chromosome III is shown on the left. The site of HO cutting is indicated by red arrow, the centromere of the ChrIII is indicated as a blue circle and the approximate locations of PCR products shown on panel C are indicated as black thin lines under the MAT loci. The experimental scheme is shown on the right. The HO endonuclease was expressed under the control of a galactose-inducible promoter. Cells were pre-grown in raffinose-containing media where the expression of HO is off. HO was induced by galactose for 3 hours and repressed by shifting cells to glucose-containing media. The intactness of the MAT locus was assayed by PCR that spanned the HO cut site and generated a 1.1 kb product in MATα cells or 1.0 kb product in MATa cells. For more detailed description of the assay see Supplementary Fig. S4. (c) Top: Agarose gel electrophoresis analysis of HO cutting and repair of the MAT locus. PCR products were obtained from cells before HO induction (Raf), 3 hours after HO induction (Gal) and 3, 6 and 20 hours after repression of HO (Glc). Note that during the repair, either MATa or MATα genes can be copied into MAT locus. PCR product of the VPS13 locus was used as an internal control. Bottom: quantification of MAT locus repair from at least three independent replicates, the error bars indicate standard error. MATa/α signals were quantified, normalized to the VPS13 signal and plotted relative to raffinose time points. Full-size blots are shown in the Supplementary Fig. S5. (d) 10-fold serial dilutions of rad53Δ cells combined with wt H3 or H3 K9,14,23 R were spotted onto MMS-containing plates.
These results also suggest that H3 acetylation may become critical for the survival if cells fail to properly activate DNA damage checkpoint. To test this hypothesis, we introduced H3 K9,14,23 R mutation into the checkpoint-deficient rad53Δ background. This strain was viable, although it grew slower than rad53Δ with wt H3 and was very sensitive to DNA damaging agents (Fig. 4d and Supplementary Fig. S2), confirming the importance of H3 acetylation in cells with a compromised DNA damage checkpoint. However, the H3 K9,14,23 R mutation causes a more severe phenotype in rpb9Δ cells (Fig. 2b) than in those lacking Rad53, suggesting that the elevated level of DNA lesions in rpb9Δ strain becomes detrimental to cell survival.
Rpb9-depleted H3 K9,14,23 R cells exhibit abnormal morphology and go through lethal mitosis
To further examine the reasons behind the lethality in Rpb9-deficient H3 K9,14,23 R cells, we analysed cell cycle distribution and DNA content in the H3 K9,14,23 R strain upon depletion of Rpb9. Remarkably, soon after removal of Rpb9, DNA content in this strain becomes heterogeneous, with some cells containing less DNA than in the normal G1 cells and others with abnormally high DNA content indicative of irregular DNA ploidy (Fig. 5a). This suggests that in the absence of Rpb9, H3 K9,14,23 R cells go through mitosis with unrepaired DNA, leading to unequal distribution of the genome between the daughter cells. To confirm this, we studied cell morphology and DNA distribution in H3 K9,14,23 R cells upon Rpb9 depletion. Compared to wild type cells, most of the Rpb9-depleted H3 K9,14,23 R cells displayed a swollen cell phenotype. Additionally, there was a large increase in the number of cells with aberrantly elongated bud morphologies and defects in chromosome segregation (Fig. 5b). Thus, we conclude that collectively inefficient DNA repair and impaired checkpoint signalling lead to unequal mitotic distribution of genomic DNA in Rpb9-deficient H3 K9,14,23 R cells, resulting in aneuploidy and ultimately cell death.
Cell cycle and morphology analysis of Rpb9-depleted H3 K9,14,23 R cells. (a) Cell cycle analysis of Rpb9 anchor-away strains with wt or K9,14,23 R mutant H3. Rapamycin was added to growth media of Rpb9 anchor-away strains at time-point 0. Samples were collected at indicated time-points and DNA content of cells was analysed by flow cytometry. (b) Rpb9 was depleted from H3 K9,14,23 R cells for 4 hours, after which the cells were fixed and stained with DAPI. Red arrows show aberrations in DNA segregation and black arrows point to abnormally oblong and elongated buds. Scale bar 5 µm, BF–bright field.
Discussion
The role of Rpb9 is not unambiguously clear as the cells lacking this protein display a variety of defects, including impaired efficiency of RNAPII elongation and transcription-coupled repair6,14,15,18,19, transcription start site selection17, impaired degradation of RNAPII in response to UV-induced DNA damage12, proteotoxic stress and shortened life span47. In order to identify the histone modifications that are critical for viability under stress conditions, we screened a panel of H3 N-terminal lysine-to-arginine mutants in yeast cells lacking the Rpb9 protein, a non-essential subunit of RNAPII. While mutations of acetylatable H3 N-terminal lysines had no detectable effect on survival or growth in cells with wt RNAPII, simultaneous mutation of several N-terminal H3 lysines was lethal in the rpb9Δ background (Fig. 1). In particular, loss of K14 acetylation had the strongest effect, as this was the single common site in all lethal double-lysine mutants. However, any combination of three or more mutated lysines in H3 became lethal in the rpb9Δ background, suggesting that the overall hypoacetylation of the H3 N-terminal tail was the primary cause for inviability of the cells. Synthetic lethality of hypoacetylated H3 together with loss of Rpb9 is in concordance with a previous finding that RPB9 deletion is not tolerated in strains lacking the main H3 acetyltransferase Gcn5 or other components of SAGA complex22. In this respect, mutation of K14 is indeed expected to have the strongest influence on the acetylation state of H3, as this residue is the preferred target of the SAGA48,49. Moreover, acetylated K14 interacts with the bromodomain of Gcn5, and this interaction strongly influences acetylation of other H3 N-terminal lysine residues by the enzyme50,51.
We found that removal of Rpb9 induced appearance of active homologous recombination centres. This induction was very prominent, comparable with treatment of the cells with the genotoxic agent MMS. Rad52 foci characteristic of HR emerged in response to Rpb9-depletion, indicating that DNA damage was detected and actively repaired in these cells (Fig. 3). The initial origin of DNA damage is unclear, but it is reasonable to assume that collisions between RNA and DNA polymerases and concurrent collapse of replication fork may induce DSBs and elevated DNA recombination activity in these sites2. Several mechanisms have evolved to minimize these collisions52, but in the rare cases they do occur, replication can resume only after the RNA polymerase is removed from the site. In this respect, it is important to note that degradation of stalled RNAPII is inefficient in rpb9Δ cells12, suggesting that proper resolution of transcription-replication collisions may be hampered in the absence of Rpb9, which in turn may lead to elevated levels of DSBs. This is also supported by the fact that high levels of DNA recombination and impairment of replication fork progression were observed in the rpb9Δ strain3.
We also found that cellular response to DNA damage was compromised in the absence of Rpb9. The principal checkpoint signals of DSBs–phosphorylations of H2A and Rad53–were delayed in Rpb9-deficient cells (Fig. 2c). This finding was unexpected, particularly in light of the fact that DNA damage was actively repaired by HR in the absence of Rpb9. We propose that the excessive DNA damage in these cells exhausts the checkpoint signalling machinery, depleting some essential component(s) of the pathway. For example, the ssDNA binding protein RPA may become limiting in conditions of replication stress, and this may hamper the efficiency of DNA damage checkpoint activation53,54. Nevertheless, Rpb9-deficient cells are viable with wt H3, indicating that defective DNA damage checkpoint activation can be compensated by H3 acetylation. This suggests that in the presence of extensive DNA damage, acetylation of histones is essential, as even a minor decline in DNA repair efficiency can be detrimental to cell viability. Indeed, our results confirm that repair of DSBs is inefficient in the H3 K9,14,23 R strain (Fig. 4c), indicating that H3 acetylation may become critical for cell survival in the presence of elevated levels of DSBs. Alternatively, H3 acetylation can be required for viability of DNA damage checkpoint-deficient cells in general. To test that, we introduced the H3 K9,14,23 R mutation into the checkpoint-defective rad53Δ strain. These cells remained viable; however, they were very sensitive to DNA damage, confirming that H3 acetylation becomes crucial for cell survival when DNA damage checkpoint is not functional (Fig. 4d and Supplementary Fig. S2).
We found that removal of Rpb9 form H3 K9,14,23 R cells resulted in unequal distribution of DNA between daughter cells as well as large variations in cell morphology (Fig. 5). Analysis of the cell cycle profile revealed aberrant DNA content in most cells and no clearly visible G1 and G2 subpopulations. Collectively these results suggest that Rpb9-deficient H3 K9,14,23 R cells enter mitosis with unrepaired DNA, leading to mitotic catastrophe and cell death. In conclusion, our findings demonstrate that despite accumulation of active HR centres in Rpb9-deficient cells, DNA damage checkpoint is not properly activated in the absence of Rpb9. In these conditions, cell survival depends on efficient DNA repair, which is supported by H3 acetylation.
Methods
Yeast strains, plasmids and antibodies
All Saccharomyces cerevisiae strains were derived from W303 background and are listed in Supplementary Table S1. Strains AKY796 (wt RNAPII) and AKY1037 (rpb9Δ) were used in plasmid shuffling assays. These strains express wild type copies of HHT2 and HHF2 from a URA3-based plasmid (YCp50:HHT2-HHF2) as a sole source for histones H3 and H4. Histone H3 point mutations were made in HIS3-based plasmids (pRS413-H3H4-3F12), transformed into AKY796 and AKY1037, and counter-selected on 5-FOA plates to obtain strains with H3 mutations. All histone expression plasmids are listed in Supplementary Table S2. To generate Rpb9 anchor-away strains, the RPB9 locus was replaced with rpb9-FRB-hphMX expression cassette in strain HHY168 (Euroscarf)38. In Rpb9 anchor-away strains AKY1162 and AKY1190, HIS3-based plasmid with wild type H3 or H3 K9,14,23 R mutant was introduced as a sole source for H3 and H4 genes. The rad53Δ strain was constructed by first replacing the SML1 gene with kanMX6 (AKY1438) and then RAD53 was replaced with URA3 to obtain AKY1459 (RAD53 deletion is viable only in sml1Δ background). yEGFP was fused to C-terminus of Rad52 in its native locus in AKY1551 strain. Plasmid with galactose inducible HO endonuclease pGALHO-pRS41255 was a gift from Dr. Jeff Thompson. Strain LS50 with GAL-HO integrated into the genome was a gift from Dr. Lena Ström. This strain was used to create AKY1390. Phosphorylated H2A was detected by anti γ-H2A antibody (ab15083, Abcam). Histone H3 was C-terminally tagged with E2-tag and detected with 5E11 antibody (Icosagen), Rad53 was tagged with Flag-tag and detected with M2 antibody (Sigma-Aldrich). For western blot, cell extracts were prepared as described56 and protein samples were separated on SDS-polyacrylamide gel.
Yeast growth assays and flow cytometry
For growth curve analysis, exponentially growing yeast cultures were inoculated into 10 ml fresh YPD media at density 5 × 106 cells per ml. Cells were grown further in a shaker at 30 °C and samples were collected at indicated time-points during the growth. Culture density was measured with Z2 Cell and Particle Counter (Beckman Coulter). For spot test assays, 10-fold serial dilutions of cell suspensions were made and 5 µl of each dilution was spotted onto plates with synthetic complete (SC) selective medium. For plasmid shuffling, 1 mg/ml 5-fluoroorotic acid (5-FOA) and indicated concentrations of MMS in SC plates were used to test viability of cells. In experiments with Rpb9 anchor-away strains, 1 µg/ml rapamycin (Cayman Europe) in 0.1% DMSO as a final concentration was added to the cultures (0.1% DMSO was used for controls). Plates were incubated at least 2 days at 30 °C. For flow cytometry analysis of cell cycle, 0.5 ml of yeast culture was fixed in 10 ml of ice-cold 70% ethanol for at least 15 min and washed once with 50 mM citric acid. RNA was degraded with RNase A (10 μg/ml) in 50 mM citric acid overnight at 37 °C. DNA was stained with 10× SYBR Green I (Invitrogen) in 50 mM citric acid for 30 min. Cells were analysed with FACS Aria, cell cycle distribution was analysed with Cyflogic software.
Fluorescence microscopy
For cell morphology analysis, cells were fixed with 70% ethanol and stained with 4′,6-Diamidino-2-Phenylindole (DAPI). Cells were imaged using Olympus BX61 microscope at 100× magnification. Rad52-GFP foci were detected in vivo from live S-G2 cells. For quantification at least 100 cells from three independent experiments were counted. For MMS-induction of Rad52 foci, Rpb9 was depleted from cells for 6 hours and treated with 0.1% MMS for 1 hour. All images were collected with cellSens software and analysed in ImageJ.
DSB repair analysis
For detection of DSB repair in MAT locus, cells were grown overnight in rich medium containing 2% raffinose and HO expression was induced for 1.5 hours in the media containing 2% galactose. Then rapamycin was added to the media for depletion of Rpb9 from nucleus and cells were grown for further 1.5 hours in galactose-containing media. After that, HO expression was repressed in 2% glucose-containing media. Cells were collected after each step and at 3, 6 and 20 hour time-points of HO repression in glucose-containing media. Cells were lysed with glass beads and genomic DNA was purified from the cell lysate by phenol-chloroform extraction, followed by chloroform extraction. PCR primers (HOcut-F 5′CTCTGGTAACTTAGGTAAATTACAGC and HOcut-R 5′GAATGATGCTAAGAATTGATTGTTTGC) flanking the HO cut site in MAT locus in chromosome III were used to determine the degree of MAT locus cutting and repair. VPS13 locus was amplified with primers VPSq2.6 F (5′ACGTTAATTACTCTTCTGGTTCCG) and VPSq3.5 R (5′AGGTGCCTTGATTACTATGTCCATT) as an internal control. PCR products were resolved in 2% agarose gel electrophoresis, stained with ethidium bromide and quantified with Image Studio Lite.
Data availability
All data generated or analysed during this study are included in this published article and its Supplementary Information files.
References
Hartwell, L. H. & Weinert, T. A. Checkpoints: controls that ensure the order of cell cycle events. Science 246, 629–634 (1989).
Gaillard, H., Herrera-Moyano, E. & Aguilera, A. Transcription-associated genome instability. Chemical reviews 113, 8638–8661 (2013).
Felipe-Abrio, I., Lafuente-Barquero, J., Garcia-Rubio, M. L. & Aguilera, A. RNA polymerase II contributes to preventing transcription-mediated replication fork stalls. The EMBO journal 34, 236–250, https://doi.org/10.15252/embj.201488544 (2015).
Choder, M. & Young, R. A. A portion of RNA polymerase II molecules has a component essential for stress responses and stress survival. Molecular and cellular biology 13, 6984–6991 (1993).
Desmoucelles, C., Pinson, B., Saint-Marc, C. & Daignan-Fornier, B. Screening the yeast "disruptome" for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. The Journal of biological chemistry 277, 27036–27044 (2002).
Hemming, S. A. et al. RNA polymerase II subunit Rpb9 regulates transcription elongation in vivo. The Journal of biological chemistry 275, 35506–35511 (2000).
Pillai, B., Sampath, V., Sharma, N. & Sadhale, P. Rpb4, a non-essential subunit of core RNA polymerase II of Saccharomyces cerevisiae is important for activated transcription of a subset of genes. The Journal of biological chemistry 276, 30641–30647 (2001).
Rosenheck, S. & Choder, M. Rpb4, a subunit of RNA polymerase II, enables the enzyme to transcribe at temperature extremes in vitro. Journal of bacteriology 180, 6187–6192 (1998).
Wery, M. et al. Members of the SAGA and Mediator complexes are partners of the transcription elongation factor TFIIS. The EMBO journal 23, 4232–4242 (2004).
Woychik, N. A., Lane, W. S. & Young, R. A. Yeast RNA polymerase II subunit RPB9 is essential for growth at temperature extremes. The Journal of biological chemistry 266, 19053–19055 (1991).
Woychik, N. A. & Young, R. A. RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Molecular and cellular biology 9, 2854–2859 (1989).
Chen, X., Ruggiero, C. & Li, S. Yeast Rpb9 plays an important role in ubiquitylation and degradation of Rpb1 in response to UV-induced DNA damage. Molecular and cellular biology 27, 4617–4625, https://doi.org/10.1128/MCB.00404-07 (2007).
Li, S. Transcription coupled nucleotide excision repair in the yeast Saccharomyces cerevisiae: The ambiguous role of Rad26. DNA Repair 36, 43–48, https://doi.org/10.1016/j.dnarep.2015.09.006 (2015).
Li, S., Ding, B., Chen, R., Ruggiero, C. & Chen, X. Evidence that the transcription elongation function of Rpb9 is involved in transcription-coupled DNA repair in Saccharomyces cerevisiae. Molecular and cellular biology 26, 9430–9441, https://doi.org/10.1128/MCB.01656-06 (2006).
Li, S. & Smerdon, M. J. Rpb4 and Rpb9 mediate subpathways of transcription-coupled DNA repair in Saccharomyces cerevisiae. The EMBO journal 21, 5921–5929 (2002).
Li, S. & Smerdon, M. J. Dissecting transcription-coupled and global genomic repair in the chromatin of yeast GAL1-10 genes. The Journal of biological chemistry 279, 14418–14426, https://doi.org/10.1074/jbc.M312004200 (2004).
Hull, M. W., McKune, K. & Woychik, N. A. RNA polymerase II subunit RPB9 is required for accurate start site selection. Genes Dev 9, 481–490 (1995).
Awrey, D. E. et al. Transcription elongation through DNA arrest sites. A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. The Journal of biological chemistry 272, 14747–14754 (1997).
Kaster, B. C., Knippa, K. C., Kaplan, C. D. & Peterson, D. O. RNA Polymerase II Trigger Loop Mobility: Indirect Effects of Rpb9. The Journal of biological chemistry 291, 14883–14895, https://doi.org/10.1074/jbc.M116.714394 (2016).
Knippa, K. & Peterson, D. O. Fidelity of RNA polymerase II transcription: Role of Rpb9 in error detection and proofreading. Biochemistry 52, 7807–7817, https://doi.org/10.1021/bi4009566 (2013).
Nesser, N. K., Peterson, D. O. & Hawley, D. K. RNA polymerase II subunit Rpb9 is important for transcriptional fidelity in vivo. Proceedings of the National Academy of Sciences of the United States of America 103, 3268–3273, https://doi.org/10.1073/pnas.0511330103 (2006).
Van Mullem, V., Wery, M., Werner, M., Vandenhaute, J. & Thuriaux, P. The Rpb9 subunit of RNA polymerase II binds transcription factor TFIIE and interferes with the SAGA and elongator histone acetyltransferases. The Journal of biological chemistry 277, 10220–10225 (2002).
Xiao, T. et al. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Molecular and cellular biology 25, 637–651 (2005).
Hwang, W. W. et al. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Molecular cell 11, 261–266 (2003).
Wood, A. et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Molecular cell 11, 267–274 (2003).
Giannattasio, M., Lazzaro, F., Plevani, P. & Muzi-Falconi, M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. The Journal of biological chemistry 280, 9879–9886 (2005).
Moyal, L. et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Molecular cell 41, 529–542 (2011).
Saugar, I., Jimenez-Martin, A. & Tercero, J. A. Subnuclear Relocalization of Structure-Specific Endonucleases in Response to DNA Damage. Cell reports 20, 1553–1562 (2017).
Di Cerbo, V. & Schneider, R. Cancers with wrong HATs: the impact of acetylation. Briefings in functional genomics 12, 231–243 (2013).
Gong, F., Chiu, L. Y. & Miller, K. M. Acetylation Reader Proteins: Linking Acetylation Signaling to Genome Maintenance and Cancer. PLoS genetics 12, e1006272 (2016).
Miremadi, A., Oestergaard, M. Z., Pharoah, P. D. & Caldas, C. Cancer genetics of epigenetic genes. Human molecular genetics 16, R28–49 (2007). Spec No 1.
Ge, Z. et al. Sites of acetylation on newly synthesized histone H4 are required for chromatin assembly and DNA damage response signaling. Molecular and cellular biology 33, 3286–3298, https://doi.org/10.1128/MCB.00460-13 (2013).
Qin, S. & Parthun, M. R. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Molecular and cellular biology 22, 8353–8365 (2002).
Tamburini, B. A. & Tyler, J. K. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Molecular and cellular biology 25, 4903–4913, https://doi.org/10.1128/MCB.25.12.4903-4913.2005 (2005).
Wurtele, H. et al. Histone H3 lysine 56 acetylation and the response to DNA replication fork damage. Molecular and cellular biology 32, 154–172, https://doi.org/10.1128/MCB.05415-11 (2012).
Wang, Y. et al. Histone H3 lysine 14 acetylation is required for activation of a DNA damage checkpoint in fission yeast. The Journal of biological chemistry 287, 4386–4393 (2012).
Duan, M. R. & Smerdon, M. J. Histone H3 lysine 14 (H3K14) acetylation facilitates DNA repair in a positioned nucleosome by stabilizing the binding of the chromatin Remodeler RSC (Remodels Structure of Chromatin). The Journal of biological chemistry 289, 8353–8363 (2014).
Haruki, H., Nishikawa, J. & Laemmli, U. K. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Molecular cell 31, 925–932 (2008).
Downs, J. A. et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Molecular cell 16, 979–990 (2004).
Downs, J. A., Lowndes, N. F. & Jackson, S. P. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408, 1001–1004 (2000).
Hammet, A., Magill, C., Heierhorst, J. & Jackson, S. P. Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO reports 8, 851–857 (2007).
Weinert, T. A. & Hartwell, L. H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241, 317–322 (1988).
Weinert, T. A. & Hartwell, L. H. Characterization of RAD9 of Saccharomyces cerevisiae and evidence that its function acts posttranslationally in cell cycle arrest after DNA damage. Molecular and cellular biology 10, 6554–6564 (1990).
Zhao, X., Muller, E. G. & Rothstein, R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Molecular cell 2, 329–340 (1998).
Lisby, M., Rothstein, R. & Mortensen, U. H. Rad52 forms DNA repair and recombination centers during S phase. Proceedings of the National Academy of Sciences of the United States of America 98, 8276–8282 (2001).
Haber, J. E. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191, 33–64, https://doi.org/10.1534/genetics.111.134577 (2012).
Vermulst, M. et al. Transcription errors induce proteotoxic stress and shorten cellular lifespan. Nat Commun 6, 8065, https://doi.org/10.1038/ncomms9065 (2015).
Cieniewicz, A. M. et al. The bromodomain of Gcn5 regulates site specificity of lysine acetylation on histone H3. Mol Cell Proteomics 13, 2896–2910 (2014).
Kuo, Y. M. & Andrews, A. J. Quantitating the specificity and selectivity of Gcn5-mediated acetylation of histone H3. PloS one 8, e54896 (2013).
Dhalluin, C. et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399, 491–496 (1999).
Syntichaki, P., Topalidou, I. & Thireos, G. The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature 404, 414–417 (2000).
Garcia-Muse, T. & Aguilera, A. Transcription-replication conflicts: how they occur and how they are resolved. Nature reviews 17, 553–563 (2016).
Toledo, L., Neelsen, K. J. & Lukas, J. Replication Catastrophe: When a Checkpoint Fails because of Exhaustion. Molecular cell 66, 735–749 (2017).
Toledo, L. I. et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103 (2013).
Herskowitz, I. & Jensen, R. E. Putting the HO gene to work: practical uses for mating-type switching. Methods in enzymology 194, 132–146 (1991).
Zhang, T. et al. An improved method for whole protein extraction from yeast Saccharomyces cerevisiae. Yeast 28, 795–798 (2011).
Acknowledgements
We thank Drs Jeff Thompson and Lena Ström for GAL-HO expression plasmid and yeast strains, Dr. Nikita Avvakumov for helpful discussion and critical reading of the manuscript, and Dmitri Lubenets for technical assistance. This work was supported by the Estonian Research Council (grants ETF9188 and IUT20-23 to AK).
Author information
Authors and Affiliations
Contributions
H.S., K.R., K.P. and K.K. made the yeast strains and performed the experiments, H.S., K.R. and K.P. analysed the data, H.S., K.R., K.P., S.V. and A.K. contributed to study design, H.S., K.P. and A.K. wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sein, H., Reinmets, K., Peil, K. et al. Rpb9-deficient cells are defective in DNA damage response and require histone H3 acetylation for survival. Sci Rep 8, 2949 (2018). https://doi.org/10.1038/s41598-018-21110-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-21110-9
This article is cited by
-
Taf14 is required for the stabilization of transcription pre-initiation complex in Saccharomyces cerevisiae
Epigenetics & Chromatin (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.