Abstract
Although partial androgen insensitivity syndrome (PAIS) is caused by attenuated responsiveness to androgens, androgen receptor gene (AR) mutations on the coding regions and their splice sites have been identified only in <25% of patients with a diagnosis of PAIS. We performed extensive molecular studies including whole exome sequencing in a Japanese family with PAIS, identifying a deep intronic variant beyond the branch site at intron 6 of AR (NM_000044.4:c.2450−42 G > A). This variant created the splice acceptor motif that was accompanied by pyrimidine-rich sequence and two candidate branch sites. Consistent with this, reverse transcriptase (RT)-PCR experiments for cycloheximide-treated lymphoblastoid cell lines revealed a relatively large amount of aberrant mRNA produced by the newly created splice acceptor site and a relatively small amount of wildtype mRNA produced by the normal splice acceptor site. Furthermore, most of the aberrant mRNA was shown to undergo nonsense mediated decay (NMD) and, if a small amount of aberrant mRNA may have escaped NMD, such mRNA was predicted to generate a truncated AR protein missing some functional domains. These findings imply that the deep intronic mutation creating an alternative splice acceptor site resulted in the production of a relatively small amount of wildtype AR mRNA, leading to PAIS.
Similar content being viewed by others
Introduction
Partial androgen insensitivity syndrome (PAIS) is a rare endocrine disorder caused by attenuated responsiveness to androgens1. Affected 46,XY patients frequently exhibit undermasculinized genitalia since birth and gynecomastia with puberty, in the presence of age-appropriate serum androgen values1. Testis development is usually normal, while small testes have been identified in a substantial fraction of pubertal or adult patients with molecularly confirmed PAIS2.
Notably, androgen receptor gene (AR) mutations on the coding regions and their splice sites have been identified only in <25% of patients with a clinical diagnosis of PAIS, even after excluding other representative causes for PAIS-like phenotypes1,3. Furthermore, such AR mutations remain undetected in a certain fraction of patients with compromised endogenous AR activity4. These findings imply the possible presence of hidden mutations in non-coding regions of AR or in other genes involved in AR-signaling. Here, we report a deep intronic mutation creating an alternative splice acceptor site at intron 6 of AR.
Results
Clinical report
We encountered a Japanese family with an X-linked recessive or sex-limited autosomal dominant form of 46,XY disorder of sex development (DSD) (Fig. 1a). The proband (II-3) was referred to us because of undermasculinized genitalia at 18 years of age. Subsequently, two similarly affected boys (III-1 and III-2) were born to his elder sister (II-2). The two obligatory carrier females (I-2 and II-2) were clinically normal, as were their non-consanguineous husbands (I-1 and II-1).
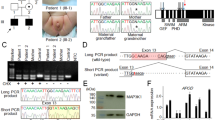
Clinical and genetic findings of this family. (a) The pedigree of this family, and external genital findings of the affected boys (III-1 and III-2). (b) CAG repeat length polymorphism at exon 1 of AR and c.245–42 G > A variant at intron 6 of AR. The “286 bp” peak and the “A” allele are shared by affected males II-3, III-1, and III-2, and obligatory carriers I-2 and II-2. The c.245–42 G > A variant creates the “AG” splice acceptor motif. WT: wildtype, and MT: mutant.
Clinical findings of the three affected males are summarized in Table 1. They exhibited undermasculinized genitalia including penoscrotal hypospadias and microphallus since birth (Fig. 1a). Patient II-3 also manifested gynecomastia with puberty, and small testes at 18 years of age. Müllerian derivatives were absent. Endocrine studies revealed sufficient androgen production capacities and hypergonadotropinism. Surgical treatment for undermasculinized genitalia and/or gynecomastia, and penile elongation therapy with testosterone enanthate and dihydrotestosterone cream, were performed or planned for the three affected males. Penile length responses to elongation therapy remained quite poor in patients II-3 and III-1. Thus, the three affected males were diagnosed as having PAIS.
Comprehensive sequencing analyses of exons and their splice sites
We first performed Sanger direct sequencing for AR and for NR5A1, MAMLD1, and SRD5A2 whose mutations often cause PAIS-like phenotypes5 in the three affected males, identifying no pathogenic sequence variant on the coding exons and their splice sites. We next analyzed 25 hypospadias-related genes by target enrichment system6,7, revealing no pathogenic variant on the coding exons and their splice sites shared by the three affected males.
Thus, we carried out whole exome sequencing in all the seven subjects of this family. However, no pathogenic variant co-segregating with the PAIS phenotype was identified on the coding exons and their splice sites (~10 bp of intronic sequences) of all the previously known genes involved in hypospadias or DSD1,6,7,8. Furthermore, although a total of 20 autosomal, but not X-linked, gene variants (variants 1–20 in Supplementary Table S1) with minor allele frequencies (MAFs) of ≤0.01 in all the four public databases and in-house database were found to be present in the three affected males (II-3, III-1, and III-3) and the two carrier females (I-2 and II-2) and absent from the two non-affected males (I-1 and II-1), they were assessed as apparently irrelevant to the PAIS phenotype on the basis of the frequencies in the normal populations, the results of in silico pathogenic analyses, and/or the available information regarding the corresponding genes. Indeed, while missense variants 2, 4, and 15 were completely absent from the public and in-house databases, variants 4 and 15 were evaluated as non-pathogenic by ≥ 3 of the four in silico analyses. Similarly, while missense variants 1, 2, and 12 were evaluated as pathogenic by all the four in silico analyses, variants 1 and 12 were, though extremely rare, registered in the public databases, as were a nonsense variant 5 and a splice donor site variant 14. Furthermore, while variant 2 was completely absent from the public and in-house databases and was assessed as pathogenic by all the four in silico analyses, the tissue expression pattern of the corresponding gene (SOAT1) was apparently inconsistent with the PAIS phenotype.
Genomewide array comparative genomic hybridization (aCGH)
We also performed aCGH in the seven subjects in this family, to examine the presence or absence of a copy-number variant as an underlying factor for PAIS. However, aCGH identified no pathologic copy number variant co-segregating with the PAIS phenotype in this family.
Identification of a deep intronic variant in AR
Since no definitive variant was detected by the above studies, we suspected that a pathogenic variant might be hidden in non-coding regions of AR. In support of this, the analysis of CAG repeat length polymorphism on exon 1 of AR showed co-segregation of the same allele with the PAIS phenotype (Fig. 1b). Thus, we carefully searched AR non-coding regions covered by the whole exome sequencing for a rare variant (MAFs of ≤ 0.01 in the four public databases and in-house database shown in Supplementary Table S1) co-segregating with the PAIS phenotype. Consequently, only a single variant, i.e., a deep intronic variant beyond the branch site at intron 6 (NM_000044.4:c.2450−42 G > A) creating a new “AG” splice acceptor motif (https://www.ncbi.nlm.nih.gov/genbank/) was found to be present in the three affected males and the two carrier females and absent from the two non-affected males, as shown by Sanger sequencing (Fig. 1b). This variant was completely absent from the four public databases and in-house database.
In silico analyses for the deep intronic variant
Thus, we examined whether the new “AG” splice acceptor motif is accompanied by a pyrimidine (Y)-rich sequence and a branch site required for splicing9, as for the wildtype “AG” splice acceptor site at intron 6 (Fig. 2a). Human Splicing Finder (www.umd.be/HSF3/HSF.html) indicated the presence a pyrimidine-rich sequence and two candidate branch sites near the newly created “AG” splice acceptor motif (Fig. 2a). Furthermore, it was predicted that aberrant mRNA produced by the new “AG” motif undergoes nonsense-mediated decay (NMD)10 due to retention of a 40-bp intronic sequence and resultant occurrence of premature termination at the 842th codon on exon 7. In addition, if a small amount of aberrant mRNA could escape NMD, such mRNA was predicted to generate a truncated AR protein missing some functional domains (Fig. 2b).
Alternative splicing caused by the c.245–42 G > A variant creating the “AG” splice acceptor site. (a) Normal splicing identified in the affected boys (III-1 and III-2). Normal splicing has occurred using the wildtype “AG” splice acceptor motif, pyrimidine (Y)-rich sequence, and the branch site, generating wildtype mRNA, whereas aberrant splicing has taken place using the newly created “AG” splice acceptor motif that is associated with pyrimidine (Y)-rich sequence and two putative branch sites, producing mRNA with retention of a 40 bp intronic segment (highlighted with red) between exon 6 (highlighted with green) and exon 7 (highlighted with blue). Red asterisks indicate the highly conserved “A” nucleotide at branch sites. Exonic and transcribed intronic nucleotides are written with large letters, and intronic nucleotides with small letters. (b) Schematic representation of AR exons on the genome, and wildtype (WT) and mutant (MT) mRNA and protein. AF-1: activation function 1; NTD: N-terminal transactivation domain; DBD: DNA binding domain; LBD: ligand binding domain; and AF-2: activation function 2. The coding regions are shown with solid boxes, and the untranslated regions are depicted with open boxes; exon 6, exon 7, the transcribed intronic sequence, and the frameshifted sequence before the premature termination are shown in green, blue, red, and gray, respectively. It is predicted that the mutant mRNA primarily undergoes nonsense mediated decay (NMD), while it could escape NMD, producing a small amount of a truncated protein with a loss-of-function (LOF) activity. (c) RT-PCR analysis for mRNA samples extracted from the LCLs of the non-affected father (II-1), the affected boys (III-1 and III-2), and a healthy control adult male. The band intensity ratio has been obtained between the wildtype (WT)-specific and the mutant (MT)-specific bands produced by the P1/P2 primers in the affected boys. The WT mRNA expression ratio has been calculated with the data of quantitative PCR performed for CHX-treated LCLs using PW/Pc or PM/Pc; the black and the gray bars have been obtained using GAPDH and ACTB as internal controls, respectively.
Expression studies of the wildtype and aberrant AR mRNAs
We further studied whether an alternative splicing actually occurs using the newly created “AG” splice acceptor site. For this purpose, we performed reverse-transcriptase (RT)-PCR analysis, using Epstein-Barr virus (EBV)-transformed B-cell lymphoblastoid cell lines (LCLs) established from the affected boys (III-1 and III-2), their 30-year-old non-affected father (II-1), and a 28-year-old healthy adult male with proven fertility. The mRNA samples were obtained after 8-hour incubation with or without the NMD inhibitor cycloheximide (CHX).
RT-PCR analysis was performed with primers which could amplify both the wildtype mRNA and the predicted aberrant mRNA (P1 primer for exon 5; and P2 primer for exon 8) (Fig. 2a). Consequently, 387 bp and 427 bp products were identified in the affected boys (III-1 and III-2), whereas a 387 bp product alone was detected in the non-affected father (II-1) and the control male (Fig. 2c). There was no other RT-PCR product, such as that formed with skipping of exon 6 or exon 7. While the band intensity of the 387 bp product was grossly similar between CHX-untreated and CHX-treated LCLs of each subject, that of the 427 bp product was obviously stronger for CHX-treated LCLs than for CHX-untreated LCLs of the affected boys (III-1 and III-2), with the band intensity ratio between the 387 bp and 427 bp products obtained from CHX-treated LCLs being ~1:6. Sequencing of the two RT-PCR products showed that the 387 bp product was derived from the normally spliced wildtype mRNA, whereas the 427 bp product was derived from the aberrant mRNA with retention of a 40-bp intronic sequence, as predicted (Fig. 2a).
RT-PCR analysis was also carried out with primers (PW/Pc) designed to amplify the wildtype mRNA alone and those (PM/Pc) designed to amplify the mutant mRNA alone (PW: a primer specific to the wildtype splice junction between exon 6 and exon 7; PM: a primer specific to the mutant splice junction between exon 6 and retained intron 6; and PC: a primer for the splice junction between exon 7 and exon 8 commonly utilized for the wildtype and mutant mRNA amplifications) (Fig. 2a). The band intensity for the wildtype mRNA was grossly similar between CHX-untreated and CHX-treated LCLs of the four subjects examined, whereas that for the aberrant mRNA was weak for the CHX-untreated LCLs and strong for the CHX-treated LCLs of the affected boys (III-1 and III-2) (Fig. 2c). Furthermore, quantitative RT-PCR was performed with the PW/Pc and PM/Pc primers, using GAPDH or ACTB as an internal control, indicating that the wildtype mRNA expression ratio (the ratio between the wildtype mRNA expression dosage and the sum of the wildtype and aberrant mRNA expression dosages obtained from CHX-treated LCLs) was 25–30% in the affected boys and ~100% in the non-affected male and the control male (Fig. 2c). However, precise comparisons of the wildtype or aberrant mRNA expression dosage among different subjects was impossible, because of variable expression dosages among subjects. For example, the sum of the wildtype and aberrant mRNA expression dosage obtained from CHX-treated LCLs was apparently large in the affected boy (III-1) and small in the non-affected male (II-1).
Discussion
We identified a deep intronic mutation (c.2450−42 G > A) creating an alternative splice acceptor site at intron 6 of AR in a family with PAIS. This new acceptor site was accompanied by a pyrimidine-rich sequence and two candidate branch sites required for splicing and, consistent with this, RT-PCR experiments for CHX-treated LCLs revealed a relatively large amount of aberrant mRNA produced by the newly created alternative splice acceptor site and a relatively small amount of wildtype mRNA yielded by the original normal splice acceptor site. Furthermore, most of the aberrant mRNA was shown to undergo NMD and, if a small amount of aberrant mRNA may have escaped NMD in vivo, as suggested by the weak but discernible bands for the aberrant mRNA obtained from CHX-untreated LCLs of the affected boys (III-1 and III-2), such mRNA was predicted to generate a truncated AR protein missing some functional domains. These findings imply that the deep intronic mutation has resulted in the production of a relatively small amount of wildtype AR mRNA, leading to PAIS.
To our knowledge, three intronic aberrations have been identified in AR, except for splice site mutations at the conserved “GT-AG” motif: [1] a > 6 kb deletion at intron 2 involving a putative branch site, which produced ~90% of aberrant mRNA encoding non-functional AR protein due to in-frame skipping of exon 3 and ~10% of wildtype mRNA produced by a cryptic branch site, in a family with typical PAIS11; [2] c.1769−11 T > A at the pyrimidine-rich region at intron 2, which produced two types of aberrant mRNA encoding non-functional AR proteins due to in-frame retention of 69-bp intronic sequence transcribed by a cryptic splice acceptor site and in-frame skipping of exon 3, as well as a very low level of wildtype mRNA, in a family with apparently complete AIS (CAIS)12 (though described as PAIS in the original paper, the combination of normal female external genitalia and Wolffian development is frequently observed in CAIS with a residual AR activity)1; and [3] c.2450−118 A > G at intron 6, which produced two types of mRNA subject to NMD due to retention of 85 bp and 202 bp sequences of intron 6 respectively, in siblings with CAIS13. These findings, together with the present data, indicate that several types of intronic mutations other than splice site mutations at the conserved “GT-AG” motif do exist in AR, although they remain extremely rare, and that PAIS takes place when a relatively small amount of wildtype AR mRNA is produced. In this regard, it would be worth pointing out that the c.2450−42 G > A identified in this study is the first PAIS-causing deep intronic mutation beyond the branch site.
Two points should be made with regard to the present study. First, although we performed extensive molecular studies, possible relevance of other variant(s) has not been excluded formally. Indeed, a true pathogenic variant for 46,XY DSD might have been overlooked by the whole exome sequencing, if it has been registered in the public and/or in-house databases because of its detection in normal females or in the general population due to incomplete penetrance, or if its pathogenic assessment remains more or less equivocal. In addition, such a variant might be hidden in non-examined sequences of AR or other genes.
Second, precise comparison of expression dosage between different subjects was impossible even by the quantitative RT-PCR, because of variable AR mRNA expressions among subjects. Thus, it remains to be demonstrated whether the wildtype mRNA expression dosage is actually reduced in the affected males as compared with that in the non-affected males, although the present study revealed markedly reduced relative wildtype mRNA expression ratios in the affected males. This would primarily be due to expression studies being performed for polyclonal EBV-transformed LCLs that could express multiple genes to highly variable degrees, probably depending on the character of clones examined14. For example, it is possible that the LCLs of the affected boy III-1, which showed a high level of AR expression, were primarily composed of clones that efficiently express AR. In addition, since we utilized LCLs of non-affected adult father (II-1) and a healthy adult male for controls primarily because of an ethical issue, the age difference between the affected boys and the control adults may also be relevant to the inter-individual difference in the AR expression dosage.
Despite such caveats, the present data strongly support that the deep intronic mutation of AR (c.2450−42 G > A) is the pathogenic variant for PAIS. Further studies will permit to clarify the relevance of AR intronic mutations to the development of PAIS and CAIS.
Methods
Ethical approval
This study was approved by the Institutional Review Board Committee at Hamamatsu University School of Medicine, and performed after obtaining written informed consent to participate in this study and to publish molecular and clinical data including external genital findings/images from the adult subjects or from the parents of the affected children. All experiments were performed in accordance with the relevant guidelines and regulations.
Reference sequence
Of plural AR isoforms, transcript variant 1 (NM_000044.4) consisting of eight exons and encoding 920 amino acids was utilized as a reference (GenBank, https://www.ncbi.nlm.nih.gov/genbank/), as suggested previously15.
Primers
Primers utilized in this study are shown in Supplementary Table S2.
Molecular studies using leukocyte genomic DNA samples
Genomic DNA was obtained from peripheral blood leukocytes using FlexiGene DNA Kit (QIAGEN).
Sanger direct sequencing for AR, NR5A1, MAMLD1, and SRD5A2 was performed from both directions on the ABI 3130xl Genetic Analyzer (Thermo Fisher Scientific).
Sequence analysis for 25 causative or candidate genes for hypospadias, i.e., AR, ATF3, BMP4, BMP7, BNC2, CTGF, CYP1A1, CYR61, DGKK, EGF, ESR1, ESR2, FGF8, FGFR2, GSTM1, GSTT1, HOXA4, HOXB6, HSD3B2, HSD17B3, MAMLD1, MID1, NR5A1, SRD5A2, and WT16,7, was performed using the Haloplex Target Enrichment System (Design ID 10380-1414463301) (Agilent Technologies). After amplification, the coding regions of the 25 genes were sequenced as 150 bp paired-end reads on a MiSeq sequencer (Illumina). Subsequently, nucleotide alterations in the samples were called by the Genome Analysis Toolkit (Broad Institute) and SAMtools 0.1.17 software (http://samtools.sourceforge.net)16.
Whole exome sequencing was carried out, using SureSelect Human All Exon V6 (Agilent Technologies). Captured libraries were sequenced by NextSeq. 500 (Illumina) with 150 bp paired-end reads. Reads were aligned to the reference genome (Human GRCh37/hg19) (UCSC Genome Browser; http://genome.ucsc.edu/) using BWA-MEM (Version 0.7.12) with default parameters. Duplicated reads are removed by Picard (Version 1.106), and local realignment and base quality recalibration were performed by GATK Version 3.5. Variants were identified with the GATK HaplotypeCaller, and variants with MAFs > 1% in at least one of four public databases and in-house database shown in Supplementary Table S1 were excluded. Final variants were annotated with Annovar17.
Genomewide array comparative genomic hybridization was performed with a catalog human array (1 × 1 M format, ID G4447A) (Agilent Technologies), using leukocyte genomic DNA from sex-matched normal subjects as controls. For autosomes and female X chromosomes, log2 signal ratios of <−0.8 and >+ 0.4 were regarded as indicative of heterozygous deletions and duplications, respectively. For male sex chromosomes that appear in a heterogametic condition, log2 signal ratios of −∞ and around +1.0 were interpreted as hemizygous deletions and duplications, respectively. When ≥three consecutive probes showed abnormal log2 ratios, the corresponding region was regarded as copy number variants (CNVs). The genomic position was based on human GRCh37/hg19. CNVs were regarded as normal variants if they have been registered in the public databases such as Database of Genomic Variants (http://dgv.tcag.ca/dgv/app/home) and ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/).
CAG repeat length on exon 1 of AR was determined for PCR products obtained with a fluorescently labeled forward primer and an unlabeled reverse primer on the ABI 3130xl Genetic Analyzer, using GeneMapper Software 5.
Molecular studies using lymphoblastoid cell line-derived mRNA
EBV transformed LCLs were established from the non-affected male (II-1), the affected boys (III-1 and III-2), and an adult healthy control male with proven fertility by a standard method18 with minor modifications, after obtaining written informed consent from the parents of the affected boys and from the non-affected father and the healthy adult male. Because of the ethical issue, we could not establish an LCL from age-matched healthy boy or utilize LCLs established from boys with congenital disorders other than 46,XY DSD.
Total RNA samples were isolated from immortalized LCLs cultured in media containing dimethyl sulfoxide or cycloheximide (Sigma) for 8 hours, using RNeasy Mini Kit (QIAGEN). RT-PCR with P1/P2 primers was performed with one μg of total RNA, using ReverTra Ace qPCR RT Kit (TOYOBO). Then, RT-PCR products were subjected to electrophoresis, and the band intensity was measured with ImageJ version 1.49 software (http://www.imagej.com-about.com/). The RT-PCR products were also subjected to direct sequencing. RT-PCR with PW/Pc or PM/Pc primers was similarly carried out with one μg of total RNA. Quantitative RT-PCR with PW/Pc or PM/Pc primers was also performed for mRNA samples extracted from CHX-treated LCLs by the SYBR Green methods on StepOnePlus system with Software v2.2.2 (Thermo Fisher Scientific), using GAPDH and ACTB as internal controls. We calculated the relative mRNA expression of each sample based on its threshold cycle (Ct) in comparison to the Ct of GAPDH or ACTB. Then, we calculated the ratio of the wildtype AR mRNA expression to the total (the sum of wildtype and mutant mRNAs) AR mRNA expression obtained from CHX-treated LCLs.
References
Achermann, J. C. & Hughes, I. A. Pediatric disorders of sex development. In: Melmed S, Polonsky KS, Larsen PR, Kronenberg HM, eds. Williams textbook of endocrinology. 13th ed. Philadelphia: Elsevier 893–963 (2016).
Hellmann, P. et al. Male patients with partial androgen insensitivity syndrome: a longitudinal follow-up of growth, reproductive hormones and the development of gynaecomastia. Arch. Dis. Child. 97, 403–409 (2012).
Deeb, A., Mason, C., Lee, Y. S. & Hughes, I. A. Correlation between genotype, phenotype and sex of rearing in 111 patients with partial androgen insensitivity syndrome. Clin. Endocrinol. 63, 56–62 (2005).
Hornig, N. C. et al. Identification of an AR mutation-negative class of androgen insensitivity by determining endogenous AR activity. J. Clin. Endocrinol. Metab. 101, 4468–4477 (2016).
Gaspari, L. et al. ‘Idiopathic’ partial androgen insensitivity syndrome in 28 newborn and infant males: impact of prenatal exposure to environmental endocrine disruptor chemicals? Eur. J. Endocrinol. 165, 579–587 (2011).
van der Zanden, L. F. et al. Aetiology of hypospadias: a systematic review of genes and environment. Hum. Reprod. Update 18, 260–283 (2012).
Kon, M. et al. Molecular basis of non-syndromic hypospadias: systematic mutation screening and genome-wide copy-number analysis of 62 patients. Hum. Reprod. 30, 499–506 (2015).
Eggers, S. et al. Disorders of sex development: insights from targeted gene sequencing of a large international patient cohort. Genome Biol. 17, 243 (2016).
Stracham, T. & Read, A. Nucleic acid structure and gene expression. In: Stracham, T. & Read, A. eds. Human molecular genetics. 4th ed. New York: Garland Science 1–27 (2011).
Kuzmiak, H. A. & Maquat, L. E. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol. Med. 12, 306–316 (2006).
Ris-Stalpers, C. et al. Differential splicing of human androgen receptor pre-mRNA in X-linked Reifenstein syndrome, because of a deletion involving a putative branch site. Am. J. Hum. Genet. 54, 609–617 (1994).
Brüggenwirth, H. T. et al. Molecular analysis of the androgen-receptor gene in a family with receptor-positive partial androgen insensitivity: an unusual type of intronic mutation. Am. J. Hum. Genet. 61, 1067–1077 (1997).
Känsäkoski, J. et al. Complete androgen insensitivity syndrome caused by a deep intronic pseudoexon-activating mutation in the androgen receptor gene. Sci. Rep. 6, 32819 (2016).
Marinov, G. K. et al. From single-cell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome Res. 24, 496–510 (2014).
Gottlieb, B., Beitel, L. K., Nadarajah, A., Paliouras, M. & Trifiro, M. The androgen receptor gene mutations database: 2012 update. Hum. Mutat. 33, 887–894 (2012).
Li, H. et al. The sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from next-generation sequencing data. Nucleic Acids Res. 38, e164 (2010).
Neitzel, H. A routine method for the establishment of permanent growing lymphoblastoid cell lines. Hum. Genet. 73, 320–326 (1986).
Ishii, T., Matsuo, N., Inokuchi, M. & Hasegawa, T. A cross-sectional growth reference and chart of stretched penile length for Japanese boys aged 0–7 years. Horm. Res. Paediatr. 82, 388–93 (2014).
Matsuo, N. Skeletal and sexual maturation in Japanese children. Clin. Pediatr. Endocrinol. 2(suppl), 1–4 (1993).
Matsuo, N., Anzo, M., Sato, S., Ogata, T. & Kamimaki, T. Testicular volume in Japanese boys up to the age of 15 years. Eur. J. Pediatr. 159, 843–845 (2000).
Inada, H., Imamura, T. & Nakajima, R. Manual of endocrine examination for children. Osaka: Medical Review, (in Japanese) (2002).
Ishii, T., Hayashi, M., Suwanai, A., Amano, N. & Hasegawa, T. The effect of intramuscular testosterone enanthate treatment on stretched penile length in prepubertal boys with hypospadias. Urology 76, 97–100 (2010).
Acknowledgements
We thank Ms. Fumiko Kato and Ms. Maki Igarashi for their technical support. This study was supported by Grants-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology (17H06428), Grants from the Japan Agency for Medical Research and Development (16ek0109049h0003, 16ek0109166h0002, 17ek0109151h0003, and17ek0109266h0001), and Grant for Research on Intractable Diseases from the Ministry of Health, Labor and Welfare (H27-025).
Author information
Authors and Affiliations
Contributions
H.O., Y.F., and T.O. designed this study; R.H., Y.O., and T.O. collected clinical data; H.O., H.S., S.N., K.Y., K.N., and M.F. performed molecular and in silico analyses; and M.F., Y.F., and T.O. wrote the paper.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ono, H., Saitsu, H., Horikawa, R. et al. Partial androgen insensitivity syndrome caused by a deep intronic mutation creating an alternative splice acceptor site of the AR gene. Sci Rep 8, 2287 (2018). https://doi.org/10.1038/s41598-018-20691-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-20691-9
This article is cited by
-
Clinical characteristics, AR gene variants, and functional domains in 64 patients with androgen insensitivity syndrome
Journal of Endocrinological Investigation (2022)
-
A novel de novo androgen receptor nonsense mutation in a sex-reversed 46,XY infant
Human Genome Variation (2021)
-
Clinical, hormonal and genetic characteristics of androgen insensitivity syndrome in 39 Chinese patients
Reproductive Biology and Endocrinology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.