Abstract
Harmonia axyridis is a voracious predator, a biological control agent, and one of the world most invasive insect species. The advent of next-generation sequencing platforms has propelled entomological research into the genomics and post-genomics era. Real-time quantitative PCR (RT-qPCR), a primary tool for gene expression analysis, is a core technique governs the genomic research. The selection of internal reference genes, however, can significantly impact the interpretation of RT-qPCR results. The overall goal of this study is to identify the reference genes in the highly invasive H. axyridis. Our central hypothesis is that the suitable reference genes for RT-qPCR analysis can be selected from housekeeping genes. To test this hypothesis, the stability of nine housekeeping genes, including 18S, 28S, ACTB, ATP1A1, GAPDH, HSP70, HSP90, RP49, and ATP6V1A, were investigated under both biotic (developmental time, tissue and sex), and abiotic (temperature, photoperiod, in vivo RNAi) conditions. Gene expression profiles were analyzed by geNorm, Normfinder, BestKeeper, and the ΔCt method. Our combined results recommend a specific set of reference genes for each experimental condition. With the recent influx of genomic information for H. axyridis, this study lays the foundation for an in-depth omics dissection of biological invasion in this emerging model.
Similar content being viewed by others
Introduction
The multicolored Asian lady beetle, Harmonia axyridis (Coleoptera: Coccinellidae), a generalist predator, preys on aphids and scale insects on crops and other plants1. Harmonia axyridis is native to central and eastern Asian. To exploit its ecosystem services, numerous releases were attempted in North America and Europe, as early as 19162,3. Due to its broad range of preys and incredible consumption rate, H. axyridis indeed has been used to control aphids4,5,6 and other sap-sucking arthropod pests7,8. However, the worldwide propagation of H. axyridis threatens the indigenous lady beetles and other non-target species9,10,11. Considered as “the most invasive ladybird on Earth”, the role of H. axyridis has shifted from a global biological control agent to an invasive alien species12. Multiple factors contribute to this transition. Predation of eggs and larvae of other lady beetle species is one of the reasons which leads to the decline of native species13,14. A higher level of resistance to infection is the other major reason to benefit its competition in the field15,16,17. The molecular basis of this resistance, however, is poorly understood.
Double-stranded RNA (dsRNA) can induce sequence-specific posttranscriptional gene silencing in many organisms, i.e., RNA interference (RNAi)18,19. RNAi can not only investigate gene functions in vivo or in vitro, but also offers a novel approach with a brand new mode-of-action to control arthropod pests20,21,22,23,24. With a recent influx of genomic information for H. axyridis, there is an increasing need for the development of genetic tools to functionally interpret the sequencing data20,24,25,26.
Real-time quantitative PCR (RT-qPCR) has been used primarily for gene expression quantification27,28,29. RT-qPCR analysis is highly sensitive, and its accuracy can be affected by RNA quantity, transcription efficiency, amplification efficiency and experimental procedures between samples. To avoid biases, normalization of gene expression is an essential step30. The most common practice is to compare a target gene expression with an internal reference gene31. Housekeeping genes, such as beta-actin (ACTB), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and translation elongation factor 1-alpha (EF1A)32,33 have been used extensively for RT-qPCR analysis. However, under any given experimental condition, the expression of these commonly used reference genes may vary substantially34,35,36,37. A systematic and customized study for each tested species is recommended for identifying appropriate reference genes38,39.
The overall goal of this study is to identify candidate reference genes in the highly invasive H. axyridis. Our objective is to determine the suitable reference genes for RT-qPCR analysis in H. axyridis from selected housekeeping genes, an array of constitutively expressed genes maintaining the basic cellular functions in an organism. We evaluated the stability of nine housekeeping genes under selected biotic and abiotic conditions, respectively. The candidate genes include 18S ribosomal RNA(18S), 28S ribosomal RNA (28S), Na+/K+-ATPase subunit alpha 1 (ATP1A1), heat shock protein 70 (HSP70), heat shock protein 90 (HSP90), ribosomal protein 49 (RP49), V-ATPase subunit A (ATP6V1A), ACTB and GAPDH from H. axyridis. All these housekeeping genes have been used empirically as the reference genes for RT-qPCR analyses in other organisms, especially in insects. The specific environmental conditions range from biotic (developmental stage, tissue type, and sex) to abiotic treatments (temperature, photoperiod, and in vivo RNAi). As a result, a specific set of reference genes is recommended for each given condition.
Results
RT-qPCR analysis
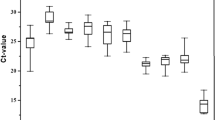
For each candidate reference gene, a single amplicon was produced, as detected by agarose gel electrophoresis analysis and the melting curve analysis. Nonspecific bands were not found, and a single peak was observed in the melting curve analysis. A standard curve was generated for each gene, using a five-fold serial dilution of the pooled cDNA. Efficiency of RT-qPCR ranged between 90 and 110% (Table 1), which is considered standard40. Ct values of the nine candidate reference genes ranged from 8 to 27, covering all the experimental conditions (Fig. 1). While the vast majority of Ct values were found between 17 and 26, 18S was the most abundant transcript. ATP1A1, VATP6V1A, and RP49 were the least abundant candidate reference genes.
Stability of candidate reference genes under biotic conditions
For different developmental stages, geNorm ranked the stability from high to low as 18S = HSP70, 28S, ATP6V1A, ATP1A1, ACTB, HSP90, GAPDH, and RP49. Normfinder provided a ranking as 18S, HSP70, ATP6V1A, 28S, ATP1A1, ACTB, HSP90, GAPDH, and RP49. Bestkeeper offered a list as follows: 18S, HSP70, 28S, ATP1A1, GAPDH, HSP90, ACTB, ATP6V1A, and RP49 (Table 2). The best set of reference genes was recommended in Table 2. Integrating the results from all four programs, RefFinder identified the consensus top three candidates, 18S, HSP70 and 28S, across different developmental stages. 18S was the most stable gene, while RP49 was the least stable candidate (Table 2, Fig. 2A).
For different tissues, the consensus top three candidates were 28S, 18S and RP49 according to RefFinder (Table 2, Fig. 2B). Specifically, 28S and ATP6V1A were the most and the least stable genes, respectively. For different sexes, the top three most stable candidates in both sexes were HSP90, RP40, and HSP70 according to RefFinder (Table 2, Fig. 2C). HSP90 and ATP1A1 were the most and the least stable genes, respectively. Based on the comprehensive ranking of RefFinder, the most to the least stable candidate reference genes under the biotic conditions was: 18S, 28S, ATP1A1, ACTB, HSP70, ATP6V1A, GAPDH, RP49, and HSP90 (Table 2; Fig. 2D).
Stability of candidate reference genes under abiotic conditions
According to RefFinder, the consensus top three candidate reference genes under different temperature regime were 18S, 28S and GAPDH (Table 3, Fig. 2E). Specifically, 18S and ATP6V1A was the most and least stable candidate, respectively. For different photoperiods, the top three candidates were 18S, 28S and HSP90 (Table 3, Fig. 2F), in which 18S and RP49 was the most and the least stable candidates, respectively. For in vivo RNAi experiments, the top three candidates were RP49, ATP1A1, and 28S (Table 3, Fig. 2G), in which RP49 and HSP90 was the most and the least stable candidates, respectively. Based on the comprehensive ranking of RefFinder, the most to the least stable candidate reference genes under the abiotic conditions was: 18S, 28S, GAPDH, HSP90, ATP6V1A, ACTB, ATP1A1, HSP70, and RP49 (Table 3; Fig. 2H).
Recommended reference genes
For repeatable and consistent results, multiple normalizers (≥2 reference genes) are required for RT-qPCR analysis. GeNorm analysis evaluated all pairwise variations under each experimental conditions (Fig. 3). According to Vandesompele et al.31, a Vn/Vn + 1 cutoff value of 0.15 means the addition of n + 1 reference gene is not necessary, i.e., the first n references genes are sufficient to normalize qRT-PCR results. The optimal number of reference genes was recommended in Tables 2 and 3, respectively, for biotic and abiotic conditions. Specifically, for different developmental stages, the recommended reference genes were 18S, HSP70, and 28S. For different tissues, the recommendation was 28S, 18S, and RP49. For different sexes, the recommendation was HSP90 and RP49. For different temperature treatments, the recommendation was 18S, 28S, and GAPDH. For different photoperiods, the recommendation was 18S, 28S, and HSP90. Finally, for in vivo RNAi, the best combination was RP49 and ATP1A1.
Optimal number of reference genes required for accurate normalization of gene expression. Based on geNorm analysis, average pairwise variations are calculated between the normalization factors NFn and NFn + 1. Values less than 0.15 indicate that n + 1 genes are not required for the normalization of gene expression.
Validation of selected reference genes
The expression of TPS, a target gene, was evaluated to validate the recommended reference genes under different temperature treatments. Using the most stable reference gene 18S (NF 1), the top two stable reference genes 18S and 28S (NF 1–2), and the top three stable reference genes, 18S, 28S, and GAPDH (NF 1–3) for normalization, TPS expression profiles were similar throughout all three temperature regimes (Fig. 4). In comparison, when ATP6V1A, the least stable candidate (NF 9), was used as the reference gene, TPS expression patterns were inconsistent across different temperature treatments. Specifically, TPS expression was numerically higher at 10 °C, and lower at 22 and 30 °C (Fig. 4).
Discussion
RT-qPCR has been used extensively for quantification of mRNA expression and is a primary tool for genetic research. Although multiple factors, such as RNA extraction, storage, cDNA synthesis, and handling of materials and reagents, can affect the RT-qPCR analysis, a reliable reference gene (set) to overcome confounding variations in an empirical dataset is of particular importance. Normalization by internal controls is an integral part of the quantification process. A single or multiple stably expressed reference genes are required for the normalization process to achieve accurate and reliable results. Each candidate reference gene should be evaluated under specific experimental conditions to ensure a constant level of expression35. Following the “Minimum Information for Publication of Quantitative Real-Time PCR Experiments” (MIQE) guideline41, reference gene selection study has been carried out for many insect species34,42,43, and has become a routine practice to standardize RT-qPCR analysis.
Due to different algorithms, stability ranking derived from the four analytical tools can vary. For example, when H. axyridis was injected with dsRNAs (in vivo RNAi), 28S was rated as the best reference gene by BestKeeper, RP49 was considered as the most stable by Normfinder as well as ΔCT method, whereas ATP1A1 and GAPDH were the top choice by geNorm. Despite some discrepancies in individual rankings, RP49 and ATP1A1 were consistently exhibited a higher level of stability than the rest of the candidates projected by all four algorithms (Table 3), suggesting the importance of (1) using a comprehensive analysis to interpret the dataset and (2) adopting the multiple instead of a single normalizer for RT-qPCR analysis.
In recent years, researchers have been more receptive to use multiple reference genes to replace a single normalizer in RT-qPCR analysis44. The optimal number of reference genes is typically determined by geNorm. In this study, three reference genes for recommended for different developmental stages (18S, HSP70, and 28S), tissues (28S, 18S, and RP49), temperatures (18S, 28S and GAPDH), and photoperiods (18S, 28S and HSP90), while two reference genes were required for the reliable normalization in different sexes (HSP90 and RP49), and in vivo RNAi (RP49 and ATP1A1). Our combined results are, in part, consistent with previous studies of other Coccinellidae predatory species (Table 4), especially for ribosome RNAs (rRNAs).
Not surprisingly, rRNAs (e.g., 18S and 28S) were consistently stably expressed throughout the vast majority of biotic and abiotic conditions among the four Coccinellidae species, including H. axyridis, Hippodamia convergens45, Coleomegilla maculate46, and Coccinella septempunctata47. The over-representation of rRNAs in the total RNA pool (>80%), however, can potentially mask the subtle changes of the target gene expression48. Therefore, customized reference gene study is still a prerequisite for standardized RT-qPCR analysis in predatory lady beetles. A large body of works has demonstrated that there are no “universal” reference genes applicable for all cell and tissue types and various experimental conditions49. As a major structural protein, Actin has been used extensively as the internal control without any validation. In this study, however, Actin was one of the least stable candidates under both biotic and abiotic conditions, except the temperature treatment, which is consistent with the other three Coccinellidae species45,46,47.
This study not only provides a standardized procedure for the quantification of gene expression, but also lays a foundation for the genomics and functional genomics dissection of H. axyridis, an emerging model in invasion biology50.
Materials and Methods
Insects
Harmonia axyridis was originally collected from the University of Kentucky North Farm (38°07′N, 84°30′W). Harmonia axyridis colony was maintained at 23 ± 1 °C, 12 L:12D photoperiod, 50% relative humidity, and provisioned with pea aphids and sugar water for more than two months. Pea aphid clones were a gift from Dr. John Obrycki (University of Kentucky) and maintained on seedlings of fava beans in a glasshouse.
Experimental conditions
Biotic conditions
The developmental stages include eggs (N = 15), four larval instars (N = 5 for each instar, respectively), pupae (N = 1), and adults (one male and one female). Sex of adult beetles was determined by the presence or absence of the male genitalia. Tissues, including head, midgut, and carcass, were dissected from the fourth instar larvae (N = 5).
Abiotic conditions
To examine the effects of temperature, third instars were exposed to 10, 22, and 30 °C for 3 hours. For photoperiod, third-instar larvae were treated with a series of light and dark regime of 16 L:8D, 12 L:12D, and 8 L:16D for two days. For in vivo RNAi, H. axyridis ATP6V1A was the intended molecular target. Specifically, 280 ng of dsRNAs (56 nl, 5 μg/μl), derived from H. axyridis ATP6V1A (HA-dsRNA) and a plant gene, β-glucuronidase (GUS-dsRNA), were injected into the abdomen of third instars (N = 5). GUS-dsRNA is an exogenous control for the unintended silencing effects, and H2O is the vehicle control for the delivery agent of dsRNAs. Samples were collected on day-3 for RT-qPCR analysis.
Total RNA extraction and reverse transcription
Total RNA was extracted separately from each developmental stage, including eggs (N = 15), pupa (N = 1), and adult (N = 1) for each sex. For other experiments involving larvae, five individuals were pooled as one sample. Each experiment was repeated three times independently. Samples were preserved in 1.5 ml centrifuge tubes and snap frozen immediately in liquid nitrogen before storage at −80 °C. Total RNA was extracted using TRIzol® (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. Each sample of 2.0 μg RNA was reverse transcribed with random primers using the M-MLV reverse transcription kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s recommendations.
Primer design and cloning of candidate reference genes
Primers for 18S, 28S, ATP1A1, HSP70, HSP90, and RP49 (Table 1) were designed based on their respective sequences from NCBI (http://www.ncbi.nlm.nih.gov/). Degenerate primers for ACTB, GAPDH, ATP6V1A were designed using CODEHOP (http://blocks.fhcrc.org/codehop.html). PCR amplifications were performed in 50 μl reactions containing 10 μl 5 × PCR Buffer (Mg2+ Plus), 1 μl dNTP mix (10 mM of each nucleotide), 5 μl of each primer (10 μM each), 0.25 μl of Go Taq (5 u/μl) (Promega, Madison, WI) and 25 ng first-strand cDNA. The PCR parameters were as follows: one cycle of 94 °C for 3 min; 35 cycles of 94 °C for 30s, 55 °C for 1 min and 72 °C for 1 min; a final cycle of 72 °C for 10 min. PCR products were purified and cloned into the pCR™4-TOPO® vector (Invitrogen, Carlsbad, CA) for sequencing confirmation. The primers for the target gene, TPS, were obtained from a previous work51.
Quantitative real-time PCR (RT-qPCR)
Gene-specific primers (Table 1) were used in PCR reactions (20 μl) containing 7.0 μl water, 10.0 μl 2 × SYBR Green MasterMix (BioRad, Hercules, CA), 1.0 μl each specific primer (10 μM), and 10 ng first-strand cDNA. The RT-qPCR program included an initial denaturation for 3 min at 95 °C followed by 40 cycles of denaturation at 95 °C for 10 s, annealing for 30 s at 55 °C, and extension for 30 s at 72 °C. For melting curve analysis, a dissociation step cycle (55 °C for 10 s, and then 0.5 °C for 10 s until 95 °C) was added. Three technical replicates were analyzed for each biological replicate.
Reactions were performed in a MyiQ Single Color Real-Time PCR Detection System (BioRad). The existence of one peak in melting curve analysis was used to confirm gene-specific amplification and to rule out non-specific amplification and primer-dimer generation. The RT-qPCR was determined for each gene using slope analysis with a linear regression model. Relative standard curves for the transcripts were generated with a serial dilution of cDNA. The corresponding RT-qPCR efficiencies (E) was calculated according to the equation:
Stability of gene expression
The stability of the nine candidate reference genes were evaluated using RefFinder (http://www.leonxie.com/referencegene.php), a web-based analysis tool which integrates all four major computational programs, including geNorm31, NormFinder52, BestKeeper53, and the comparative ΔCt method54. geNorm calculates an expression stability value (M) for each gene and a pair-wise comparison. NormFinder ranks the set of candidate genes based on their expression stability in the given sample set. BestKeeper considers the Ct values of all candidate reference genes, to calculate standard deviation and coefficient of variation. ΔCt approach directly compares relative expression of ‘pairs of genes’ within each sample. Then, RefFinder assigned an appropriate weight of the four methods to an individual gene and calculated the geometric mean of their weights for the overall final ranking.
Validation of selected reference genes
Trehalose-6-phosphate synthase (TPS), the intermediate of trehalose, is a key component in insect energy metabolism and resilience25,51,55. The stability of candidate reference genes was evaluated using TPS as the target gene. TPS expression levels under different temperature treatments were calculated based on selected sets of candidate reference genes. Two separate normalization factors (NFs) have been computed based on (1) the geometric mean of the genes with the lowest Geomean values (as determined by RefFinder), and (2) a single normalizer with the lowest or highest Geomean value. Relative expression of TPS in different samples was calculated using the 2−ΔΔCt method56.
References
Brown, P. M. J. et al. Intraguild predation by Harmonia axyridis (Coleoptera: Coccinellidae) on native insects in Europe: molecular detection from field samples. Entomol Sci 18, 130–133, https://doi.org/10.1111/ens.12092 (2015).
Gordon, R. D. The Coccinellidae (Coleoptera) of America North of Mexico. J the New York Entomol Society 93, 1–912 (1985).
Adriaens, T., Branquart, E. & Maes, D. The multicoloured Asian ladybird Harmonia axyridis Pallas (Coleoptera: Coccinellidae), a threat for native aphid predators in Belgium? Belg J Zool 133, 195–196 (2003).
Rice, N. R. et al. Assessment of legume and nonlegume ground covers on Coleoptera: Coccinellidae density for low-input pecan management. American J Alternative Agriculture 13, 111, https://doi.org/10.1017/s0889189300007785 (2009).
Musser, F. R. & Shelton, A. M. Bt sweet corn and selective insecticides: impacts on pests and predators. J Econ Entomol 96, 71–80, https://doi.org/10.1603/0022-0493-96.1.71 (2003).
Wells, M. L., McPherson, R. M., Ruberson, J. R. & Herzog, G. A. Coccinellids in cotton: Population response to pesticide application and feeding response to cotton aphids (Homoptera: Aphididae). Environ Entomol 30, 785–793, https://doi.org/10.1603/0046-225x-30.4.785 (2001).
Michaud, J. P. Biological control of Asian citrus psyllid, Diaphorina citri (Hemiptera: Psyllidae) in Florida: A preliminary report. Entomol News 113, 216–222 (2002).
Mcclure, M. S. Potential of the Asian Predator, Harmonia-Axyridis Pallas (Coleoptera, Coccinellidae), to Control Matsucoccus-Resinosae Bean and Godwin (Homoptera, Margarodidae) in the United-States. Environ Entomol 16, 224–230, https://doi.org/10.1093/ee/16.1.224 (1987).
Koch, R. L. The multicolored Asian lady beetle, Harmonia axyridis: a review of its biology, uses in biological control, and non-target impacts. J Insect sci 3, 32, https://doi.org/10.1093/jis/3.1.32 (2003).
Brown, P. M. J. et al. The global spread of Harmonia axyridis (Coleoptera: Coccinellidae): distribution, dispersal and routes of invasion. BioControl 56, 623–641, https://doi.org/10.1007/s10526-011-9379-1 (2011).
Koch, R. L., Hutchison, W. D., Venette, R. C. & Heimpel, G. E. Susceptibility of immature monarch butterfly, Danaus plexippus (Lepidoptera: Nymphalidae: Danainae), to predation by Harmonia axyridis (Coleoptera: Coccinellidae). BioControl 28, 265–270, https://doi.org/10.1016/S1049-9644(03)00102-6 (2003).
Roy, H., Brown, P. & Majerus, M. In An ecological and societal approach to biological control 295–309 (Springer, 2006).
Cottrell, T. E. Suitability of exotic and native lady beetle eggs (Coleoptera: Coccinellidae) for development of lady beetle larvae. BioControl 31, 362–371, https://doi.org/10.1016/j.biocontrol.2004.06.004 (2004).
Sato, S., Yasuda, H. & Evans, E. W. Dropping behaviour of larvae of aphidophagous ladybirds and its effects on incidence of intraguild predation: interactions between the intraguild prey, Adalia bipunctata (L.) and Coccinella septempunctata (L.), and the intraguild predator, Harmonia axyridis Pallas. Ecol Entomol 30, 220–224, https://doi.org/10.1111/j.0307-6946.2005.00688.x (2005).
Cottrell, T. E. & Shapiro-Ilan, D. I. Susceptibility of a native and an exotic lady beetle (Coleoptera: Coccinellidae) to Beauveria bassiana. J Invertebr Pathol 84, 137–144, https://doi.org/10.1016/j.jip.2003.09.003 (2003).
Shapiro-Ilan, D. I. & Cottrell, T. E. Susceptibility of lady beetles (Coleoptera: Coccinellidae) to entomopathogenic nematodes. J Invertebr Pathol 89, 150–156, https://doi.org/10.1016/j.jip.2005.04.002 (2005).
Vilcinskas, A., Stoecker, K., Schmidtberg, H., Rohrich, C. R. & Vogel, H. Invasive harlequin ladybird carries biological weapons against native competitors. Science 340, 862–863, https://doi.org/10.1126/science.1234032 (2013).
Hammond, S. M., Caudy, A. A. & Hannon, G. J. Post-transcriptional gene silencing by double-stranded RNA. Nat Rev Genet 2, 110–119, https://doi.org/10.1038/35052556 (2001).
Hannon, G. J. RNA interference. Nature 418, 244–251, https://doi.org/10.1038/418244a (2002).
Niimi, T., Kuwayama, H. & Yaginuma, T. Larval RNAi applied to the analysis of postembryonic development in the ladybird beetle, Harmonia axyridis. J Insect Biotechnol Sericology 74, 95–102, https://doi.org/10.11416/jibs.74.95 (2005).
Roberts, A. F., Devos, Y., Lemgo, G. N. & Zhou, X. Biosafety research for non-target organism risk assessment of RNAi-based GE plants. Front Plant Sci 6, 958, https://doi.org/10.3389/fpls.2015.00958 (2015).
Linghua, X. et al. The coming of RNA-based pest controls. J Plant Prot 42, 673–690, https://doi.org/10.13140/RG.2.1.1878.4081 (2015).
Baum, J. A. et al. Control of coleopteran insect pests through RNA interference. Nat Biotechnol 25, 1322–1326, https://doi.org/10.1038/nbt1359 (2007).
Du, W. & Zeng, F. Identification of Development-Related Genes in the Ovaries of Adult Harmonia axyridis (Pallas) Lady Beetles Using a Time- Series Analysis by RNA-seq. Sci Rep 6, 39109, https://doi.org/10.1038/srep39109 (2016).
Shi, Z. et al. Two novel soluble trehalase genes cloned from Harmonia axyridis and regulation of the enzyme in a rapid changing temperature. Comp Biochem Physiol B Biochem Mol Biol 198, 10–18, https://doi.org/10.1016/j.cbpb.2016.03.002 (2016).
Tang, B. et al. Transcriptome analysis and identification of induced genes in the response of Harmonia axyridis to cold hardiness. Comp Biochem Physiol D Genomics Proteomics 22, 78–89, https://doi.org/10.1016/j.cbd.2017.01.004 (2017).
VanGuilder, H. D., Vrana, K. E. & Freeman, W. M. Twenty-five years of quantitative PCR for gene expression analysis. BioTechniques 44, 619–626, https://doi.org/10.2144/000112776 (2008).
Ginzinger, D. G. Gene quantification using real-time quantitative PCR: An emerging technology hits the mainstream. Exp Hematol 30, 503–512, https://doi.org/10.1016/S0301-472x(02)00806-8 (2002).
Heid, C. A., Stevens, J., Livak, K. J. & Williams, P. M. Real time quantitative PCR. Genome Res 6, 986–994, https://doi.org/10.1101/gr.6.10.986 (1996).
Bustin, S. A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 29, 23–39, https://doi.org/10.1677/jme.0.0290023 (2002).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3, RESEARCH0034, https://doi.org/10.1186/gb-2002-3-7-research0034 (2002).
Lourenco, A. P., Mackert, A., Cristino, A. D. & Simoes, Z. L. P. Validation of reference genes for gene expression studies in the honey bee, Apis mellifera, by quantitative real-time RT-PCR. Apidologie 39, 372–U333, https://doi.org/10.1051/apido:2008015 (2008).
Macabelli, C. H. et al. Reference gene selection for gene expression analysis of oocytes collected from dairy cattle and buffaloes during winter and summer. PloS one 9, e93287, https://doi.org/10.1371/journal.pone.0093287 (2014).
Li, R. et al. Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PloS one 8, e53006, https://doi.org/10.1371/journal.pone.0053006 (2013).
Thellin, O. et al. Housekeeping genes as internal standards: use and limits. J Biotechnol 75, 291–295, https://doi.org/10.1016/S0168-1656(99)00163-7 (1999).
Pan, H. et al. Selection of Reference Genes for RT-qPCR Analysis in the Monarch Butterfly, Danaus plexippus (L.), a Migrating Bio-Indicator. PloS one 10, e0129482, https://doi.org/10.1371/journal.pone.0129482 (2015).
Zhang, S. et al. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 555, 393–402, https://doi.org/10.1016/j.gene.2014.11.038 (2015).
Hruz, T. et al. RefGenes: identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genomics 12, 156, https://doi.org/10.1186/1471-2164-12-156 (2011).
Gutierrez, L. et al. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6, 609–618, https://doi.org/10.1111/j.1467-7652.2008.00346.x (2008).
Taylor, S., Wakem, M., Dijkman, G., Alsarraj, M. & Nguyen, M. A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods 50, S1–5, https://doi.org/10.1016/j.ymeth.2010.01.005 (2010).
Bustin, S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55, 611–622, https://doi.org/10.1373/clinchem.2008.112797 (2009).
Maroniche, G. A., Sagadin, M., Mongelli, V. C., Truol, G. A. & del Vas, M. Reference gene selection for gene expression studies using RT-qPCR in virus-infected planthoppers. Virol J 8, 308, https://doi.org/10.1186/1743-422x-8-308 (2011).
Scharlaken, B. et al. Reference gene selection for insect expression studies using quantitative real-time PCR: The head of the honeybee, Apis mellifera, after a bacterial challenge. J Insect Sci 8, https://doi.org/10.1673/031.008.3301 (2008).
Veazey, K. J. & Golding, M. C. Selection of stable reference genes for quantitative rt-PCR comparisons of mouse embryonic and extra-embryonic stem cells. PloS one 6, e27592, https://doi.org/10.1371/journal.pone.0027592 (2011).
Pan, H., Yang, X., Siegfried, B. D. & Zhou, X. A Comprehensive Selection of Reference Genes for RT-qPCR Analysis in a Predatory Lady Beetle, Hippodamia convergens (Coleoptera: Coccinellidae). PloS one 10, e0125868, https://doi.org/10.1371/journal.pone.0125868 (2015).
Yang, C. et al. Selection of reference genes for RT-qPCR analysis in a predatory biological control agent, Coleomegilla maculata (Coleoptera: Coccinellidae). Sci Rep 5, 18201, https://doi.org/10.1038/srep18201 (2015).
Yang, C. et al. Selection of Reference Genes for RT-qPCR Analysis in Coccinella septempunctata to Assess Un-intended Effects of RNAi Transgenic Plants. Front Plant Sci 7 (2016).
Raaijmakers, M. H. G. P., van Emst, L., de Witte, T., Mensink, E. & Raymakers, R. A. P. Quantitative assessment of gene expression in highly purified hematopoietic cells using real-time reverse transcriptase polymerase chain reaction. Exp Hematol 30, 481–487, https://doi.org/10.1016/S0301-472x(02)00787-7 (2002).
Radonic, A. et al. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun 313, 856–862, https://doi.org/10.1016/j.bbrc.2003.11.177 (2004).
Vilcinskas, A., Mukherjee, K. & Vogel, H. Expansion of the antimicrobial peptide repertoire in the invasive ladybird Harmonia axyridis. Proc Biol Sci 280, 20122113, https://doi.org/10.1098/rspb.2012.2113 (2013).
Zi, Q. et al. Molecular cloning and cold-induced expression of trehalose-6-phosphate synthase gene in Harmonia axyridis (Coleoptera: Coccinellidae). Acta Entomol Sinica 55, 651–658 (2012).
Andersen, C. L., Jensen, J. L. & Orntoft, T. F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64, 5245–5250, https://doi.org/10.1158/0008-5472.CAN-04-0496 (2004).
Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol Lett 26, 509–515, https://doi.org/10.1023/B:Bile.0000019559.84305.47 (2004).
Silver, N., Best, S., Jiang, J. & Thein, S. L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol 7, 33, https://doi.org/10.1186/1471-2199-7-33 (2006).
Tang, B. et al. Trehalase in Harmonia axyridis (Coleoptera: Coccinellidae): effects on beetle locomotory activity and the correlation with trehalose metabolism under starvation conditions. Appl Entomol Zool 49, 255–264, https://doi.org/10.1007/s13355-014-0244-4 (2014).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408, https://doi.org/10.1006/meth.2001.1262 (2001).
Rodrigues, T. B. et al. Validation of reference housekeeping genes for gene expression studies in western corn rootworm (Diabrotica virgifera virgifera). PloS one 9, e109825, https://doi.org/10.1371/journal.pone.0109825 (2014).
Shi, X. Q. et al. Validation of reference genes for expression analysis by quantitative real-time PCR in Leptinotarsa decemlineata (Say). BMC Res Notes 6, 93, https://doi.org/10.1186/1756-0500-6-93 (2013).
Tan, Y., Zhou, X. R. & Pang, B. P. Reference gene selection and evaluation for expression analysis using qRT-PCR in Galeruca daurica (Joannis). Bull Entomol Res 107, 359–368, https://doi.org/10.1017/S0007485316000948 (2017).
Rodrigues, T. B., Dhandapani, R. K., Duan, J. J. & Palli, S. R. RNA interference in the Asian Longhorned Beetle:Identification of Key RNAi Genes and Reference Genes for RT-qPCR. Sci Rep 7, 8913, https://doi.org/10.1038/s41598-017-08813-1 (2017).
Lord, J. C., Hartzer, K., Toutges, M. & Oppert, B. Evaluation of quantitative PCR reference genes for gene expression studies in Tribolium castaneum after fungal challenge. J Microbiol Methods 80, 219–221, https://doi.org/10.1016/j.mimet.2009.12.007 (2010).
Sang, W., He, L., Wang, X. P., Zhu-Salzman, K. & Lei, C. L. Evaluation of Reference Genes for RT-qPCR in Tribolium castaneum (Coleoptera: Tenebrionidae) Under UVB Stress. Environ Entomol 44, 418–425, https://doi.org/10.1093/ee/nvv010 (2015).
Wang, Y., Wang, Z. K., Huang, Y., Liao, Y. F. & Yin, Y. P. Identification of suitable reference genes for gene expression studies by qRT-PCR in the blister beetle Mylabris cichorii. J Insect Sci 14, 94, https://doi.org/10.1093/jis/14.1.94 (2014).
Acknowledgements
This work was supported by Biotechnology Risk Assessment Grant Program Competitive Grant No. 2011-33522-30749 from the USDA National Institute of Food and Agriculture. The information reported in this paper (No. 18-08-005) is part of a project of the Kentucky Agricultural Experiment Station and is published with the approval of the Director. These agencies had no role in study design, data collection/analysis, manuscript preparation, or the decision to publish.
Author information
Authors and Affiliations
Contributions
X.G.Z., X.W.Y., and H.P.P. conceived and designed research. X.W.Y. and H.P.P. conducted experiments. X.G.Z. and L.Y. contributed reagents and analytical tools. X.W.Y. and H.P.P. analyzed data. X.W.Y. and X.G.Z. wrote the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, X., Pan, H., Yuan, L. et al. Reference gene selection for RT-qPCR analysis in Harmonia axyridis, a global invasive lady beetle. Sci Rep 8, 2689 (2018). https://doi.org/10.1038/s41598-018-20612-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-20612-w
This article is cited by
-
Identifying optimal reference genes for gene expression studies in Eurasian spruce bark beetle, Ips typographus (Coleoptera: Curculionidae: Scolytinae)
Scientific Reports (2022)
-
Oral RNAi of diap1 results in rapid reduction of damage to potatoes in Henosepilachna vigintioctopunctata
Journal of Pest Science (2021)
-
Development of a universal endogenous qPCR control for eukaryotic DNA samples
Plant Methods (2020)
-
Evaluation of suitable reference genes for normalization of quantitative real-time PCR analysis in rice plants under Xanthomonas oryzae pv. oryzae--infection and melatonin supplementation
Food Production, Processing and Nutrition (2020)
-
Selection of reference genes for normalization of RT-qPCR data in gene expression studies in Anthonomus eugenii Cano (Coleoptera: Curculionidae)
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.