Abstract
Polyploidization, a common event during the evolution of different tumours, has been proposed to confer selective advantages to tumour cells by increasing the occurrence of mutations promoting cancer progression and by conferring chemotherapy resistance. While conditions leading to polyploidy in cancer cells have been described, a general mechanism explaining the incidence of this karyotypic change in tumours is still missing. In this study, we tested whether a widespread tumour microenvironmental condition, low pH, could induce polyploidization in mammalian cells. We found that an acidic microenvironment, in the range of what is commonly observed in tumours, together with the addition of lactic acid, induced polyploidization in transformed and non-transformed human cell lines in vitro. In addition, we provide evidence that polyploidization was mainly driven through the process of endoreduplication, i.e. the complete skipping of mitosis in-between two S-phases. These findings suggest that acidic environments, which characterize solid tumours, are a plausible path leading to polyploidization of cancer cells.
Similar content being viewed by others
Introduction
Tetraploidization is a frequent event during cancer evolution. Evidence of widespread presence of polyploid cells comes from the observations that many tumours display a bimodal distribution of chromosome numbers, with the first peak centred on a near-diploid karyotype and the second between that of a triploid and tetraploid genome1,2,3. Even if many cancer cells are chromosomally unstable and tend to gain and lose chromosomes at high frequency4, whole-genome duplication is the most likely explanation for the presence of cancer cells with a ploidy >3 N. Moreover, flow cytometry, cytogenetic and histological analyses from patients or murine models affected by a variety of solid tumours provided experimental evidence that tetraploidization is commonly present in tumour samples even at early stages of its progression5,6,7,8,9,10. Tetraploidization has been positively correlated with cancer evolution and proposed to promote its progression at different stages. Indeed, p53-null tetraploid but not diploid mouse mammary epithelial cells were shown to induce malignant mammary epithelial transformation when transplanted subcutaneously into nude mice11, suggesting that tetraploid cells could help kick-start the transformation process. Moreover, tetraploidization has been shown to confer increased drug resistance to certain classes of cytotoxic drugs or chemotherapy compounds currently used in the clinic12,13 (Tan, Chan and Rancati, unpublished observations), suggesting that they might play an important role also during emergence of drug resistance. Furthermore, it has been shown that genome doubling can lead to the formation of aneuploid cells due to the presence of extra centrosomes and multipolar spindle intermediates14,15. Since aneuploid cells have been associated with tumour progression4, this observation suggests that polyploidization could be an intermediate step in the progression of normal tissues into highly aneuploid cancers displaying chromosomal instability. This hypothesis has been supported by data from colorectal16, breast5, cervical6 and bladder cancers8.
There are currently four known paths leading to tetraploid cells from diploid precursors1: cell-to-cell fusion, cytokinesis failure, mitotic slippage and endoreduplication. While many of these processes occur in normal tissues and are part of the development of specialized cells, some of them have been shown or hypothesized to occur during tumour development. For instance, cell-to-cell fusion, a process in which cell membranes of adjacent cells fuse together, has been reported to increase transformation in vitro17 and occur after viral infection of oncogenic viruses such as the human papilloma virus18. Cytokinesis failure after complete nuclear segregation has been shown to occur in response to common events in cancer progression, such as overexpression of mitotic regulators or presence of lagging chromosomes1. Mitotic slippage was reported in conditions that mimic chemotherapy treatment such as after prolonged incubation with antimitotic drugs19. Lastly, endoreduplication, a process in which cells completely skip mitosis and enter into the subsequent G1 phase, was shown to occur during persistent telomere damage and dysfunction20, which are common steps during tumour evolution.
Despite the above links between polyploidization and transformation, a general mechanism explaining how this karyotypic change occurs during tumour progression is still missing. Since cancer cells display a large intra- and inter-tumour heterogeneity21, we searched for paths leading to tetraploidization in cancer features that are common to different tumours. In single-cell eukaryotes and plants, it has been recently shown that stressful environments could induce karyotypic changes. For example, heat shock in S. cerevisiae and drug stress in C. albicans was shown to induce aneuploidy22,23; insufficient light, cold stress, drought or exposure to pathogens can induce plants to polyploidize various tissues24. A near universal stress found in solid tumours is the presence of an acidic microenvironment25. While non-transformed adult cells have an extracellular pH (pHe) of ~7.4, cancer cells have a lower average pHe of ~6.7–7.125, with pHe as low as 5.8 being reported26. This acidic environment is primarily generated by a combination of two effects. On one hand, cancer cells display an altered metabolism27 and export large amounts of lactate and protons, thereby acidifying the extracellular environment. On the other hand, poor vascularization and blood perfusion of the tumour mass leads to reduced gas exchange and accumulation of H+ ions in the extracellular environment. The combination of these two factors has been hypothesized to be at the basis of the observed reduced pHe in solid tumours27.
We therefore tested whether acidic microenvironments could trigger polyploidization as a stress response in mammalian cells. In this paper, we report that lactic acidosis alone induced tetraploidization in transformed and non-transformed human cell lines in vitro. Using the Fluorescent Ubiquitination-based Cell Cycle Indicator (FUCCI) system, we provide evidence that tetraploidization mostly occurs through the process of endoreduplication. Since acidosis is a hallmark of solid tumours, these observations suggest that acidic microenvironments are a potential common route to polyploidization of cancer cells in different solid tumours.
Results
Optimised conditions for acidic tissue culture media
Current pre-made liquid cell-culture media contain a buffer system that maintains a stable pH between ~7.0 and ~7.4 in a 5% CO2 incubator. The buffer system is based on the equilibrium between an optimised amount of sodium bicarbonate and atmospheric CO2. In presence of increased H+ ions and acidic conditions, the pH is readily stabilised back at ~7.0–7.4 (Fig. 1a) by increasing concentration of HCO3− ions in the media. Therefore media using this buffer system are not suitable for growth of mammalian cells at pH ranges different from ~7.0–7.4. We thus had to replace sodium bicarbonate with a buffer system that can stably maintain acidic pH levels.
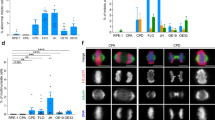
A combination of 10 mM HEPES and 10 mM PIPES is an optimised buffer system for acidic growth conditions. (a) pH levels of McCoy’s 5A media containing sodium bicarbonate before (right after pH titration, left column) and at the end (right column) of a 8-hour incubation in a 5% CO2 incubator. N = 3 independent replicates, unpaired t-test, p-values: *** < 0.001. (b) pH levels of 10 mM HEPES, 10 mM PIPES buffered media. 3 different starting pH levels were tested, and media was incubated in a 5% CO2 incubator for the indicated time period. The slight pH decrease of the media upon 5% CO2 incubation could be ascribed to CO2 solubilization and consequent carbonic acid formation (c) Relative frequency of diploid/polyploid (top panel) and euploid/aneuploid (bottom panel) cells after 1 day of incubation in standard or HEPES/PIPES buffer media in the indicated cell lines. N = 3 biological replicates, >100 cells per replicate were analysed for the top panel; 50 cells per replicate were analysed for the bottom panel; statistical analysis: unpaired t-test. (d) Fold increase in number of DLD-1 cells incubated with either standard or HEPES/PIPES buffer media over 24 hours. 5 × 105 cells were seeded and grown for one day before exposure with the indicated media. Standard media refers to media with sodium bicarbonate buffer system. N = 3 biological replicates, SEM are depicted as error bars. Unpaired t-test, n.s.: not significant.
HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) and PIPES (1,4- piperazinediethanesulfonic acid) are commonly-used, well-tolerated buffering agents with a buffering capacity in the range of pH ~6.8–8.2 and pH ~6.1–7.5, respectively. We thereby selected a buffer system that relies on a combination of these two chemicals at a concentration not known to be toxic for mammalian cells (see Material and Methods). This buffer system stably maintained the pH of the media (Fig. 1b, especially at lower pH) and had no noticeable effects on cell morphology (Supplementary Fig. S1), on karyotypic stability (Fig. 1c) or on cellular growth over a 24-hour incubation (Fig. 1d) when tested at pH 7.4. We therefore concluded that this buffer system was non-toxic, maintained a fairly stable pH and was suitable for use throughout this study. Moreover, to confirm the generality of our findings, all experiments presented in Figs 2 and 3 have also been repeated with media that utilised a 15 mM Bis-Tris (2,2-Bis(hydroxymethyl)-2,2′,2″-nitrilotriethanol) buffer system with qualitatively similar results (Supplementary Fig. S2, Supplementary Fig. S3).
Acidic environments increase the proportion of polyploid cells. (a) Diagram illustrating the different treatment regimens. 106 cells were seeded and grown for one day before a 24-hour exposure with the indicated media. 24-hour after the release from stress, cells were harvested for metaphase spread preparation. (b) Percentage of polyploid cells in the tested cell lines after exposure to indicated regimens. N ≥ 3 biological replicates, > 100 cells per replicate were analysed; SEM are depicted as error bars. Unpaired t-test, p-values: * < 0.05, ** < 0.01, *** < 0.001, n.s.: not significant.
Acidic-triggered polyploid cells carried diplochromosomes. (a) Representative metaphase spreads showing a diploid cell (left) and a polyploid cell without (middle) or with (right) diplochromosomes. The inset in the bottom of the right panel shows magnification of two couples of diplochromosomes. All images were acquired using the same magnification; scale bar: 10 µm. (b) Scatter plots displaying the percentage of cells with diplochromosomes among all polyploid cells versus the percentage of polyploid cells among all analysed cells, following exposure to the indicated regimens. The large error bars in some data points are due to the low total number of polyploid cells in that sample. R2 is reported in each panel. N ≥ 3 biological replicates, >100 cells per replicate were analysed; error bars represent SEM.
Lactic acidosis induced polyploidization in transformed and non-transformed cell lines
To test if karyotypic changes could be triggered by acidic environments, cells were exposed to acidic (pH 6.4 or 6.7) or control (pH 7.4) media for 24 hours. Subsequently, cells were harvested for chromosome counting 24 hours after they were returned to standard culturing conditions. Since acidic tumour environments can contain up to 40 mM of lactic acid28, the same regimen was applied in presence or absence of 25 mM of lactic acid (Fig. 2a). It should be noted that the lactic acid was added before titration of the media, thus the pH of the experimental media is as stated. This stress regimen was tested on multiple chromosomally stable human cell lines (such as DLD-1, HCT-15 or HCT-116 cancer cells, or the non-transformed retinal epithelial cells RPE-1), all of which were either diploid or pseudodiploid.
Chromosome counting on metaphase spreads showed that exposure to different acidic environments in presence or absence of lactic acid increased the proportion of polyploid cells (defined as cells with ≥66 chromosomes, see Materials and Methods) in the majority of tested cell lines (Fig. 2b). Though this observation is in contrast with a previous report showing that acidosis per se does not trigger polyploidization29, we note that the cell culturing conditions used in our study are different and have been optimised for pH stabilization of the media. While addition of lactic acid by itself did not change the cellular karyotype (Fig. 2b, compare pH 7.4 lane vs. pH 7.4 + 25 mM lactic acid lane), it often led to an increased amount of polyploid cells when combined with lower pH levels (Fig. 2b, see DLD-1, HCT-15 and RPE-1). This observation suggests that lactate molecules in the tumour microenvironment might work as an active signal to trigger polyploidization more than just contributing to this karyotypic change by lowering the pH. In contrast, the application of this stress regimen in presence or absence of lactic acid did not alter the proportion of aneuploid cells (defined as cells with a non-modal chromosome count of <66 chromosomes, Supplementary Fig. S4), suggesting that polyploidization is not the result of an increased chromosome instability.
Polyploidization arose from endoreduplication events
Endoreduplication is a process by which cells undergo two rounds of DNA replication without entering mitosis and dissolving centromeric cohesion30,31. Following endoreduplication, metaphase spreads contain diplochromosomes, which are chromosomal structures characterised by four sister chromatids held together (Fig. 3a). Metaphase spread analysis after acid treatment showed that increasing percentages of polyploidization were accompanied by an increase of polyploid cells carrying diplochromosomes (Fig. 3b), suggesting that polyploidization was mostly occurring through endoreduplication. To confirm this, we performed live-cell imaging on cell cycle progression of cells exposed to lactic acidosis using FUCCI. The FUCCI system relies on fragments of specific cell cycle proteins tagged with different fluorophores and therefore cells expressing this construct show different fluorescence colours at different stages of cell cycle progression32,33. Specifically for the implemented system that we utilised in this study, G1 cells appear red as they express mCherry-hCdt1 (hCdt1 amino acid residues 30/120), G2/M cells appeared green as they express mAG-hGeminin (hGeminin amino acid residues 1/110), while S phase cells are yellow as they express a combination of the two proteins. Upon endoreduplication, cells will cycle from G2 to G1 (from green to red fluorescence) without physically rounding up or separating (indicating that no mitosis occurred).
In control media, FUCCI-tagged DLD-1 cells displayed a typical cell cycle progression. Initially, red G1-phase cells progressed to yellow S-phase and then to green G2-phase cells before undergoing mitotic rounding up and cell division (Fig. 4a and Supplementary Video S1). The duration of the cell cycle was qualitatively comparable with untagged DLD-1 cells (data not shown). When FUCCI-tagged DLD-1 cells were imaged during continuous exposure to lactic acidosis stress, we noticed several changes. Firstly, there was a delay in the cell cycle progression; for example the cell marked with a yellow arrowhead in Fig. 4b divided at 41:00 despite being in G2/M for at least 30 hours. Secondly, upon cell division, a large proportion of cells either arrested in G1 or underwent cell death; for example the cells indicated with yellow arrowheads undergo cell death at 56:00 (Fig. 4b–d and Supplementary Video S2). Interestingly, among the cells that re-entered another cell cycle in presence of lactic acidosis, the majority of them underwent endoreduplication and progressed from G2 to G1 without undergoing mitosis (Fig. 4e,f and Supplementary Video S3). We then asked whether cells that have undergone endoreduplication re-enter a new cell cycle and divide when the conditions are shifted back to standard (control) media. To this end, cells that had been exposed to lactic acid for 23 hours and 50 minutes were shifted to standard conditions until the end of the imaging experiment. Upon release from the acidic stress, the majority of cells that underwent endoreduplication restarted their cell cycle (Fig. 4g and Supplementary Video S4). For example, the indicated cell in Fig. 4g entered G2/M at 46:25 roughly 20 hours after the shift back to standard media and underwent tripolar mitosis at 83:00. These observations suggested that lactic acidosis induces polyploidization mostly though endoreduplication. As a result, polyploid cells arising from endoreduplication could progress through mitosis upon stress release and could become chromosomally unstable by undergoing aberrant mitosis. We also noted that, upon release from stress, ~25% of S- or G2-phase cells that were previously exposed to acidic environments underwent endoreduplication (Fig. 4h-i and Supplementary Video S5), suggesting that exposure to acidic environments could have long-lasting effects on the genome instability of exposed cells.
Acidic-driven polyploidization occurred through endoreduplication. (a) Highlighted cell completes cell division in control media. (b) Cell cycle progression in lactic acidosis media. White arrowheads: cell with a lengthened cell cycle followed by G1 arrest of at least ~32 hours (from 24:35 to 56:00). Yellow arrowheads: daughter cells at 41:00 with a lengthened cell cycle followed by cell death ~15 hours after cell division (56:00). (c) Cell cycle length quantification (from G1 to cytokinesis) in control or lactic acidosis media. Control N = 86, stress N = 7. (d) Quantification of cell fate following cell division in control or lactic acidosis media. Cells were classified as not entering the cell cycle if they remained in G1 phase for at least 24 hours after cell division. N = 2 biological replicates, ≥45 cells were analysed per condition per replicate. (e) The indicated cell undergo endoreduplication in presence of lactic acidosis stress. (f) Frequency of cells undergoing cell division or endoreduplication in control or lactic acidosis media. N = 2 biological replicates, ≥50 cells were analysed per condition per replicate. (g) Following release into standard media (23:50), the indicated endoreduplicated cell re-entered the cell cycle (46:25) and underwent multipolar mitosis (83:00). (h) Arrowheads: S/G2-phase cell at the time of media replacement from lactic acidosis to control (23:45) that underwent endoreduplication resetting to G1 (31:45) and progressed through a novel cell cycle (G2-phase at 62:45). (i) Quantification of S- and G2-phase cells undergoing cell division or endoreduplication in their first cell cycle upon release from lactic acidosis. For lactic acidosis, cells were scored after the release from lactic acidosis (24:00). For control, cells unexposed to lactic acidosis media were scored from 24:00 after the beginning of the imaging experiment. N = 2 biological replicates, ≥25 cells were analysed per condition per replicate. Unpaired t-test, p-values: * < 0.05, ** < 0.01, *** < 0.001; error bars represent SEM. Time displayed in hh:mm, with 00:00 corresponding to beginning of video. Scale bar: 50 µm. Full videos are provided as Supplementary Videos S1–5.
Discussion
Though polyploidy has been positively correlated with early as well as late stages of tumour evolution11,12,13 and contributes to the rampant aneuploid population in cancers14,15, we still miss a general mechanism describing how cancer cells increase their ploidy. Here, we showed that in vitro exposure of transformed or non-transformed human cells to fluctuating acidic environments was sufficient to trigger genome doubling through the process of endoreduplication. We also demonstrated that some of the acidic-triggered polyploid cells underwent multipolar mitosis, likely causing chromosome instability. Given the fact that acidosis is a general feature of solid tumours, these observations suggest that in vivo acidic tumour microenvironments are a potential common trigger towards polyploidization and aneuploidization. Moreover, tetraploidization related to cancer cells has been shown to result from either viral-mediated infection17, presence of lagging chromosome at the metaphase plate34, persistent DNA damage due to telomere attrition20 and dysregulation of mitotic proteins (i.e.: Mad2, Aurora A) or tumour suppressors (i.e.: Adenomatous Polyposis Coli)1. The acidic-driven tetraploidization described in this manuscript adds to these observations and is conceptually different from what previously reported. Indeed, the acidic-driven tetraploidization does not rely on any intrinsic cellular factors, such as mutations or chromosome alignment issues. As such, exposure to acidic environments is sufficient to drive tetraploidization regardless of the genetic background or chromosome instability status of the exposed cells. As tumours form, acidic microenvironments are generated as a result of an increased lactate secretion from anaerobic glycolysis and decreased blood perfusion. It is known that acidification of the microenvironment contributes to tumour evolution by promoting angiogenesis and degradation of the extracellular matrix27. The findings in this study provide an additional connection between acidosis and tumour evolution. Acidic environments could trigger polyploidization and subsequent generation of aneuploid cells, which in turn have both been shown to promote tumour evolution and drug resistance4,13. On possible implications for chemotherapy, a few recent reports indicated that an increased pH in tumour masses inhibit or delay the formation of metastasis35,36,37. Since centrosome amplification, a hallmark of polyploid cells, has been shown to induce cellular invasion38, we speculate that treatments aiming at reducing the acidification of the tumour microenvironment might curb metastasis by decreasing the incidence of polyploid cells in the tumour.
While p53 was shown to restrain the growth of polyploid cells39, we were able to detect endoreduplication-induced tetraploid cells using metaphase spreads in a non-transformed RPE-1 cell line with intact p53 signalling cascade. This observation suggests that at least some polyploid cells formed through the process of endoreduplication might proliferate despite a wild-type p53 signalling cascade. Endoreduplication occurs during the development of specialized cells such as trophoblast giant cells in mammals40. Therefore we speculate that endoreduplication-derived polyploid cells might be insensitive to the p53-mediated G1 arrest as endoreduplication occurs normally during development. Future studies will be required to follow up the long-term fate of polyploid cells derived from endoreduplication events in presence or absence of an active p53 signalling cascade. Confirmation of the ability of endoreduplication-derived polyploid cells to proliferate despite an intact p53 signalling cascade could have important implications in our understanding of what triggers p53 activation in polyploid cells formed by other mechanisms. Moreover, if polyploidization in cancer cells is mostly driven by endoreduplication, our observations would imply that the p53 status of tumour cells does not affect the proliferation capacity of tetraploid cells. This would extend the generality of our finding and make them applicable to virtually all solid tumours.
Lastly, the finding that cells can endoreduplicate even after the acidic stress is removed suggests that endoreduplication could be boosted by fluctuating acidic environments, which are typical of tumours. Moreover, factors contributing to the observed tetraploidization might have long-lasting effects on cell cycle progression. The time needed for the transcriptional activation of effector genes or the accumulation or degradation of a molecular signal to a certain threshold might be possible explanations for the temporal lag seen between the acidic stimulus and tetraploidization. The process of endoreduplication has been well studied in normal development, e.g. the cyclin E-Cdk2 cycles of trophoblast giant cells41. It would be interesting to investigate the possibility that the same pathways are hijacked by an acidic microenvironment. Alternatively, it has also been seen that endoreduplication can be induced through drugs (e.g. DNA hypomethylating agent 5-azaC42, phosphatase 2A inhibitor okadaic acid43, arsenite44) and disruption of the cell cycle45,46,47. Further investigation in these areas could provide hints to the molecular mechanism that underlies this phenomenon of acidosis-induced endoreduplication.
Materials and Methods
Cell lines and cell culture conditions
Colorectal carcinoma cell lines HCT-116 (CCL-247), HCT-15 (CCL-225), DLD-1 (CCL-221) and hTERT RPE-1 (CRL-4000) were obtained from American Type Culture Collection and were passaged in standard conditions and recommended standard media (DLD-1 and HCT-15: RPMI1640; HCT-116: McCoy’s 5A; RPE-1: DMEM/F-12 mix) unless otherwise specified. Experimental media was prepared from powdered media (RPMI1640, Gibco 31800; McCoy’s 5A, Sigma M4892; DMEM/F-12, Thermo 12500), with the addition of 10 mM HEPES (Sigma) and 10 mM PIPES (Sigma) or 15 mM Bis-Tris (Sigma), 10% FBS (Gibco), and additional supplements to match the formula of the pre-made liquid media. 25 mM L-(+)-lactic acid (Sigma) was optionally added.
Metaphase spreads
KaryoMAX Colcemid (Gibco) was used for metaphase spread preparation. Following incubation with Colcemid for 2 to 4 hours, cells were harvested by trypsinization and washed once in PBS. Cells were gently resuspended in ~100 μl of PBS, and 75 mM potassium chloride (Kanto) was slowly added up to a volume of 10 ml. The samples were then incubated for 10 min in a 37 °C water bath. Following that, 1 ml of Carnoy solution (3:1 Methanol:Acetic acid) was added to each sample, and samples were centrifuged for 10 min at 1,000 rpm. Supernatant was discarded and cells were gently resuspended in a small amount of remaining supernatant. 10 ml of Carnoy solution was slowly added to the samples and cells were fixed for at least 1 hour at room temperature (RT). Samples were centrifuged for 10 min at 1,000 rpm, supernatant was discarded, and cells were fixed again with 10 ml of fresh Carnoy for at least 1 hour at RT. Samples were then centrifuged and pellets were resuspended in a suitable volume (~0.5–1.5 ml) of fixative. 20 μl of this suspension was drawn up and dropped from a height (10–30 cm) onto glass slides and allowed to air-dry. DNA was then stained with Giemsa (Gibco) and imaged. To obtain polyploid cell proportions, chromosome spreads were quantified as polyploid if the chromosome count was ≥66 or as diploid if the chromosome count was <66. To obtain aneuploid cell proportions, only chromosome spreads of diploid cells were quantified and cells were classified as aneuploid if the chromosome count was not the modal number of the specific cell line or as euploid if the chromosome count matched the modal number. DLD-1, HCT-15, RPE-1 display a modal chromosome count of 46, whereas HCT-116 of 45.
FUCCI
FUCCI plasmids were provided by the Miyawaki lab and characterised previously48. Each FUCCI plasmid (mCherry-hCdt(30/120)/pCSII-EF-MCS, mAG-hGeminin(1/110)/pCSII-EF-MCS) was co-transfected into 293T cells with suitable packaging vectors for viral expression. Harvested viruses were sequentially transduced into DLD-1 cells. FACS (Fluorescence-activated cell sorting) was used to select cells expressing fluorophore of interest at each stage.
Imaging
Metaphase spreads were visualised with an Imager.Z2 (Zeiss) and imaged with a CoolCube 1 (Metasystems) and analysed using ImageJ. Live-cell imaging was performed on an IX81 (Olympus) fitted with a CO2 Module S (Pecon) and a JCS controller (Shinko) for CO2 and temperature control respectively, and imaged with a CoolSnap HQ (Photometrics). Live-cell imaging videos were recorded up to 120 hours. Images were acquired every 5 min. Exposures were as follow: Phase contrast 30 ms, GFP 100 ms and RFP 100 ms. 20× magnification lenses were used for the acquisition of the live-imaging videos.
References
Davoli, T. & de Lange, T. The causes and consequences of polyploidy in normal development and cancer. Annual review of cell and developmental biology 27, 585–610, https://doi.org/10.1146/annurev-cellbio-092910-154234 (2011).
Storchova, Z. & Kuffer, C. The consequences of tetraploidy and aneuploidy. Journal of cell science 121, 3859–3866, https://doi.org/10.1242/jcs.039537 (2008).
Mitelman, F., Johansson, B. & Mertens, F. (Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer (2017). http://cgap.nci.nih.gov/Chromosomes/Mitelman).
Giam, M. & Rancati, G. Aneuploidy and chromosomal instability in cancer: a jackpot to chaos. Cell Division 10, 3, https://doi.org/10.1186/s13008-015-0009-7 (2015).
Dutrillaux, B., Gerbault-Seureau, M., Remvikos, Y., Zafrani, B. & Prieur, M. Breast cancer genetic evolution: I. Data from cytogenetics and DNA content. Breast Cancer Res Treat 19, 245–255 (1991).
Olaharski, A. J. et al. Tetraploidy and chromosomal instability are early events during cervical carcinogenesis. Carcinogenesis 27, 337–343, https://doi.org/10.1093/carcin/bgi218 (2006).
Ottesen, G. L. Carcinoma in situ of the female breast. A clinico-pathological, immunohistological, and DNA ploidy study. APMIS. Supplementum, 1–67 (2003).
Shackney, S. E. et al. Origins and clinical implications of aneuploidy in early bladder cancer. Cytometry 22, 307–316, https://doi.org/10.1002/cyto.990220407 (1995).
Tanaka, T., Mori, H., Takahashi, M. & Williams, G. M. DNA content of hyperplastic and neoplastic acinar cell lesions in rat and human pancreas. Journal of experimental pathology 1, 315–326 (1984).
Deitch, A. D., Miller, G. J. & deVere White, R. W. Significance of abnormal diploid DNA histograms in localized prostate cancer and adjacent benign prostatic tissue. Cancer 72, 1692–1700 (1993).
Fujiwara, T. et al. Cytokinesis failure generating tetraploids promotes tumourigenesis in p53-null cells. Nature 437, 1043–1047, https://doi.org/10.1038/nature04217 (2005).
Castedo, M. et al. Selective resistance of tetraploid cancer cells against DNA damage-induced apoptosis. Ann N Y Acad Sci 1090, 35–49, https://doi.org/10.1196/annals.1378.004 (2006).
Kuznetsova, A. Y. et al. Chromosomal instability, tolerance of mitotic errors and multidrug resistance are promoted by tetraploidization in human cells. Cell cycle (Georgetown, Tex.) 14, 2810–2820, https://doi.org/10.1080/15384101.2015.1068482 (2015).
Ganem, N. J., Godinho, S. A. & Pellman, D. A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278–282, https://doi.org/10.1038/nature08136 (2009).
Silkworth, W. T., Nardi, I. K., Scholl, L. M. & Cimini, D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One 4, e6564, https://doi.org/10.1371/journal.pone.0006564 (2009).
Dewhurst, S. M. et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov 4, 175–185, https://doi.org/10.1158/2159-8290.CD-13-0285 (2014).
Duelli, D. M. et al. A virus causes cancer by inducing massive chromosomal instability through cell fusion. Curr Biol 17, 431–437, https://doi.org/10.1016/j.cub.2007.01.049 (2007).
Hu, L. et al. Human papillomavirus 16 E5 induces bi-nucleated cell formation by cell-cell fusion. Virology 384, 125–134, https://doi.org/10.1016/j.virol.2008.10.011 (2009).
Gascoigne, K. E. & Taylor, S. S. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell 14, 111–122, https://doi.org/10.1016/j.ccr.2008.07.002 (2008).
Davoli, T., Denchi, E. L. & de Lange, T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell 141, 81–93, https://doi.org/10.1016/j.cell.2010.01.031 (2010).
Burrell, R. A. et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 494, https://doi.org/10.1038/nature11935 (2013).
Chen, G., Bradford, W. D., Seidel, C. W. & Li, R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature 482, 246–250, https://doi.org/10.1038/nature10795 (2012).
Selmecki, A. M., Dulmage, K., Cowen, L. E., Anderson, J. B. & Berman, J. Acquisition of Aneuploidy Provides Increased Fitness during the Evolution of Antifungal Drug Resistance. PLOS Genetics 5, e1000705, https://doi.org/10.1371/journal.pgen.1000705 (2009).
Scholes, D. R. & Paige, K. N. Plasticity in ploidy underlies plant fitness compensation to herbivore damage. Mol Ecol 23, 4862–4870, https://doi.org/10.1111/mec.12894 (2014).
Webb, B. A., Chimenti, M., Jacobson, M. P. & Barber, D. L. Dysregulated pH: a perfect storm for cancer progression. Nat Rev Cancer 11, 671–677, https://doi.org/10.1038/nrc3110 (2011).
Tannock, I. F. & Rotin, D. Acid pH in tumours and its potential for therapeutic exploitation. Cancer Res 49, 4373–4384 (1989).
Gatenby, R. A. & Gillies, R. J. Why do cancers have high aerobic glycolysis? Nat Rev Cancer 4, 891–899, https://doi.org/10.1038/nrc1478 (2004).
Walenta, S. et al. High lactate levels predict likelihood of metastases, tumour recurrence, and restricted patient survival in human cervical cancers. Cancer Res 60, 916–921 (2000).
Dai, C., Sun, F., Zhu, C. & Hu, X. Tumour environmental factors glucose deprivation and lactic acidosis induce mitotic chromosomal instability–an implication in aneuploid human tumours. PLoS One 8, e63054, https://doi.org/10.1371/journal.pone.0063054 (2013).
Levan, A. & Hauschka, T. S. Endomitotic reduplication mechanisms in ascites tumours of the mouse. J Natl Cancer Inst 14, 1–43 (1953).
Gatti, M., Rizzoni, M., Palitti, F. & Olivieri, G. Studies on induced aberrations in diplochromosomes of Chinese hamster cells. Mutation research 20, 87–99 (1973).
Newman, R. H. & Zhang, J. Fucci: street lights on the road to mitosis. Chemistry & biology 15, 97–98, https://doi.org/10.1016/j.chembiol.2008.02.003 (2008).
Sakaue-Sawano, A., Kobayashi, T., Ohtawa, K. & Miyawaki, A. Drug-induced cell cycle modulation leading to cell-cycle arrest, nuclear mis-segregation, or endoreplication. BMC cell biology 12, 2, https://doi.org/10.1186/1471-2121-12-2 (2011).
Shi, Q. & King, R. W. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature 437, 1038–1042, https://doi.org/10.1038/nature03958 (2005).
Ibrahim-Hashim, A. et al. Systemic buffers inhibit carcinogenesis in TRAMP mice. The Journal of urology 188, 624–631, https://doi.org/10.1016/j.juro.2012.03.113 (2012).
Ibrahim Hashim, A. et al. Reduction of metastasis using a non-volatile buffer. Clinical & experimental metastasis 28, 841–849, https://doi.org/10.1007/s10585-011-9415-7 (2011).
Robey, I. F. et al. Bicarbonate increases tumour pH and inhibits spontaneous metastases. Cancer Res 69, 2260–2268, https://doi.org/10.1158/0008-5472.can-07-5575 (2009).
Godinho, S. A. et al. Oncogene-like induction of cellular invasion from centrosome amplification. Nature 510, 167–171, https://doi.org/10.1038/nature13277 (2014).
Aylon, Y. & Oren, M. p53: guardian of ploidy. Molecular oncology 5, 315–323, https://doi.org/10.1016/j.molonc.2011.07.007 (2011).
Ullah, Z., Lee, C. Y., Lilly, M. A. & DePamphilis, M. L. Developmentally programmed endoreduplication in animals. Cell cycle (Georgetown, Tex.) 8, 1501–1509, https://doi.org/10.4161/cc.8.10.8325 (2009).
Zielke, N., Edgar, B. A. & DePamphilis, M. L. Endoreplication. Cold Spring Harbor perspectives in biology 5, a012948, https://doi.org/10.1101/cshperspect.a012948 (2013).
Mateos, S., Dominguez, I., Pastor, N., Cantero, G. & Cortes, F. The DNA demethylating 5-azaC induces endoreduplication in cultured Chinese hamster cells. Mutation research 578, 33–42, https://doi.org/10.1016/j.mrfmmm.2005.02.001 (2005).
Ghosh, S., Paweletz, N. & Schroeter, D. Effects of okadaic acid on mitotic HeLa cells. Journal of cell science 103(Pt 1), 117–124 (1992).
Yih, L. H. & Lee, T. C. Induction of C-anaphase and diplochromosome through dysregulation of spindle assembly checkpoint by sodium arsenite in human fibroblasts. Cancer Res 63, 6680–6688 (2003).
Sorensen, C. S. et al. Nonperiodic activity of the human anaphase-promoting complex-Cdh1 ubiquitin ligase results in continuous DNA synthesis uncoupled from mitosis. Molecular and cellular biology 20, 7613–7623 (2000).
Vidwans, S. J. et al. Sister chromatids fail to separate during an induced endoreplication cycle in Drosophila embryos. Curr Biol 12, 829–833 (2002).
Chang, B. D. et al. p21Waf1/Cip1/Sdi1-induced growth arrest is associated with depletion of mitosis-control proteins and leads to abnormal mitosis and endoreduplication in recovering cells. Oncogene 19, 2165–2170, https://doi.org/10.1038/sj.onc.1203573 (2000).
Sakaue-Sawano, A. et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498, https://doi.org/10.1016/j.cell.2007.12.033 (2008).
Acknowledgements
We thank Dr. Hiroyuki Miyoshi and Dr. Atsushi Miyawaki at RIKEN for kindly providing the FUCCI plasmids. We also thank the Rancati and Pavelka labs for helpful discussions, Graham Wright and the IMB Microscopy Unit for assistance. This work was supported by an A*STAR Investigatorship awarded to G.R. (1437a00119).
Author information
Authors and Affiliations
Contributions
Z.T. and G.R. conceptualised the project, planned experiments, and wrote the manuscript; Z.T., V.C., A.C. and E.L. performed and analysed experiments; Z.T., A.C. and G.R. revised the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tan, Z., Chu, D., Chan, Y. et al. Mammalian Cells Undergo Endoreduplication in Response to Lactic Acidosis. Sci Rep 8, 2890 (2018). https://doi.org/10.1038/s41598-018-20186-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-20186-7
This article is cited by
-
Tumor acidosis-induced DNA damage response and tetraploidy enhance sensitivity to ATM and ATR inhibitors
EMBO Reports (2024)
-
Cytogenomic characteristics of murine breast cancer cell line JC
Molecular Cytogenetics (2021)
-
A translational program that suppresses metabolism to shield the genome
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.