Abstract
Chitin, a polymer of N-acetyl-D-glucosamine (GlcNAc), functions as a major structural component in chitin-containing organism including crustaceans, insects and fungi. Recently, we reported that acidic chitinase (Chia) is highly expressed in mouse, chicken and pig stomach tissues and that it can digest chitin in the respective gastrointestinal tracts (GIT). In this study, we focus on major livestock and domestic animals and show that the levels of Chia mRNA in their stomach tissues are governed by the feeding behavior. Chia mRNA levels were significantly lower in the bovine (herbivores) and dog (carnivores) stomach than those in mouse, pig and chicken (omnivores). Consistent with the mRNA levels, Chia protein was very low in bovine stomach. In addition, the chitinolytic activity of E. coli-expressed bovine and dog Chia enzymes were moderately but significantly lower compared with those of the omnivorous Chia enzymes. Recombinant bovine and dog Chia enzymes can degrade chitin substrates under the artificial GIT conditions. Furthermore, genomes of some herbivorous animals such as rabbit and guinea pig do not contain functional Chia genes. These results indicate that feeding behavior affects Chia expression levels as well as chitinolytic activity of the enzyme, and determines chitin digestibility in the particular animals.
Similar content being viewed by others
Introduction
Chitin, a liner β-1, 4-linked polymer of N-acetyl-D-glucosamine (GlcNAc), is the second most abundant polysaccharide in nature and functions as a major structural polymer in many organisms1. Chitin has been found in organisms living in a wide range of environments ranging from terrestrial to underwater habitats (insects2,3,4, spiders5, fungi6,7,8,9, protists10,11, crabs12,13, lobsters14, shrimps15,16, corals17, mollusk18,19, polychaetes20, diatoms21,22 and freshwater and marine sponges23,24,25). It exists in three polymorphs, α26,27,28, β28, and γ29,30, which differ in the orientation and packing of the chitin molecular chains.
Chitin-containing organisms, in particular insects, have recently become attractive as a potential novel animal feed resource due to their nutritional values, production cost and a low impact on the environment31,32,33.
Chitinases (EC 3.2.1.14; KO 1183) hydrolyze the β-1, 4 glycoside bonds of chitin. Mammals, including mice and humans, do not synthesize chitin but possess two active chitinases, chitotriosidase (Chit1) and acidic chitinase (hereafter referred to as “Chia”; alternative name: acidic mammalian chitinase, AMCase) in their genomes34,35. These mammalian chitinases belong to the family 18 of glycoside hydrolases35,36,37.
The levels of Chit1 are significantly upregulated in Gaucher disease, chronic obstructive pulmonary disease (COPD), Alzheimer’s disease, atherothrombosis, diabetes mellitus, cystic fibrosis as well as in smokers38,39,40,41,42,43,44.
Significant increase in Chia mRNA and protein levels has been detected in an induced asthma mouse model as well as in antigen-induced mouse models of allergic lung inflammation45,46. In addition, it has been shown that there are single nucleotide polymorphisms in human Chia, which are associated with asthma47,48,49.
Recently, it has been shown that Chia can function as a protease-resistant major glycosidase under the gastrointestinal conditions in mouse, chicken and pig50,51,52. However, the gene expression and enzymatic activity level in other mammals are still unknown. Here, we report that high Chia mRNA level in stomach is dependent on the feeding behavior of the animals. Chia mRNA expression levels were much lower in bovine (herbivores) and dog (carnivores) than those in mouse, pig and chicken (omnivores) stomachs. Moreover, the chitinolytic activities of recombinant bovine and dog Chia enzymes were slightly but significantly lower when compared with those of mouse, pig and chicken Chia. Thus, feeding behavior seems to be directly linked to the Chia mRNA expression.
Results
Chia mRNA level is very low in bovine and dog stomach tissues
Previously, Chia mRNA has been reported to be highly expressed in mouse, chicken and pig stomach tissues50,51,52. To compare the chitinase mRNA levels in other livestock and domestic animals, total RNAs from normal bovine and dog tissues were analyzed using a quantitative real-time PCR (qPCR) assay50,51,52,53,54 using a single standard DNA molecule (Supplementary Fig. S1). Pepsinogen55 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH)56 were used as reference genes.
Bovine Chia mRNA levels were highest in lung and liver (Fig. 1a), which are consistent with a previous report57. High levels of Chit1 mRNA were detected in the lung and kidney (Fig. 1a). However, expression of Chia and Chit1 mRNAs were lower than that of GAPDH, a housekeeping gene constitutively expressed in most tissues56,58,59.
Expression of Chia and Chit1 mRNAs in bovine and dog tissues. The evaluation of Chia and Chit1 mRNA expression in bovine (a and b) and dog (c) tissues using a standard DNA containing genes fragments including Chia, GAPDH, pepsinogen (Pep) and Chit1 of bovine and Chia and GAPDH of dog. Both chitinases were quantified by qPCR using the standard DNA. All values are expressed as molecules per 10 ng of total RNA. All mRNA copy numbers were derived based on the same standard dilutions. The upper panel indicates the actual number, whereas the lower panel shows each value on logarithmic scale. *p < 0.05, **p < 0.01. P-values were determined using Student’s t-test.
We also compared the expression levels of the chitinases and reference genes in all four bovine stomachs tissues. The quantitative data are shown in Fig. 1b. Chia mRNA levels were similar in all stomachs, not exceeding those of GAPDH (Fig. 1b). On the other hand, Chit1 mRNA was very low in these tissues (Fig. 1b). Pepsinogen A is an aspartic protease being a major component within the set of gastric enzymes55. This protein is also abundantly present in the mouse, chicken and pig stomachs50,51,52,53. In bovine, pepsinogen A mRNA was predominantly expressed in the fourth stomach (so-called “abomasum”) exceeding GAPDH (Fig. 1b).
Next, gene expression analysis in dog tissues was performed. According to the NCBI genome database, Chit1 gene is not present in the dog genome. Chia mRNA was expressed at relatively high levels in the intestine, kidney, stomach and brain (Fig. 1c). However, it was low in lung and liver (Fig. 1c), where Chia has been reported to be highly expressed in mouse, chicken, pig and human50,51,52,53,54. The GAPDH mRNA levels exceeded those of Chia in all examined dog tissues (Fig. 1c).
Next, the expression levels of the chitinases and the reference genes using the standard DNA (Supplementary Fig. S1) and cDNAs reverse-transcribed from bovine, dog, pig, chicken and mouse stomach total RNAs were compared (Fig. 2). Bovine and dog stomachs expressed Chia mRNA at low levels, 1/60 and 1/6 of GAPDH, respectively. In contrast, Chia mRNA expression was prominent in mouse and chicken stomachs with levels 86 and 156 times higher than GAPDH, respectively. In pig, Chia mRNA was 25 times higher than GAPDH (Fig. 2). These results indicate that bovine (herbivores) and dog (carnivores) express low amounts, while omnivores (mouse, pig and chicken) express excessive amounts of Chia mRNA.
Chia mRNA is highly expressed in omnivores stomach tissue. Expression levels of Chia and gastric genes as well as GAPDH were quantified on the same scale by qPCR using the standard DNA (Supplementary Fig. S1) in bovine, dog, pig, chicken and mouse stomach tissues. Y axis represents molecules per 10 ng of total RNA. Pep, pepsinogen. The upper panel indicates the actual number, whereas the lower panel shows each value on logarithmic scale. Values represent mean ± SD conducted in triplicate. *p < 0.05, **p < 0.01.
Pepsinogen mRNA levels exceeded the GAPDH in tested all stomach tissues (Fig. 2). It was very high in the pig stomach tissue and it was almost 3,000 times higher than that of GAPDH. In the bovine and dog stomach tissues, the pepsinogen mRNA levels were 40 and 120 times higher than that of GAPDH, respectively. These results indicate that, when compared to pig, both Chia and pepsinogen mRNAs are being produced in the bovine and dog stomachs at low levels.
Low protein levels of Chia and pepsinogen in bovine stomach extract
We investigated bovine Chia protein and its chitinolytic activity in artificially created bovine stomach environment at pH 2.0 and 37 °C as described previously50,51,52. Soluble protein fraction was prepared from the fourth bovine stomach (abomasum) in the absence of protease inhibitor and incubated at pH 7.6 or pH 2.0 for up to 60 min. The protein fractions were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), followed by Coomassie Brilliant Blue (CBB) staining (Fig. 3a). At pH 7.6, no changes in the band pattern and intensities were noticed during the 60 min incubation (Fig. 3a,b). In contrast, time-dependent decrease of the soluble proteins with a marked reduction was observed after as early as 5 min of incubation at pH 2.0 (Fig. 3a,b).
Chia and pepsinogen level in the bovine stomach extract. Soluble protein fraction was prepared from bovine abomasum tissue in the absence of protease inhibitor and incubated at pH 7.6 or pH 2.0 for up to 60 min and analyzed by (a) SDS-PAGE, (b) total protein levels quantification, (c) chitinolytic and protease activity measured in bovine and pig stomach extracts at pH 2.0 as described in the Methods and (d) western blot using anti-Chia or anti-pepsin. The images of (a) were cropped from original full-length gel images with same exposure time shown in Supplementary Fig. S2a. The images of (d) were cropped from original full-length gel images shown in Supplementary Fig. S2b and c, respectively. The image of (d) Values in (b) and (c) represent mean ± SD from a single experiment conducted in triplicate. **p < 0.01.
To evaluate the protein levels of Chia and pepsin, soluble protein analysis from bovine and pig stomachs (5.0 μg each) after incubation at pH 2.0 for 10 min was performed. The chitinolytic and pepsin activities in pig were significantly higher than those in the bovine stomach (Fig. 3c). In accordance, the Western blot analysis using anti-Chia and anti-pepsin antibodies showed that these enzymes were undetectable after 10 min of incubation at pH 2.0 in the bovine stomach extract while they were still present in the pig stomach extract (Fig. 3d). These results indicate that, consistently with the mRNA levels, Chia as well as pepsin proteins in bovine stomach are very low (Fig. 1b, 2).
Activity of bovine and dog Chia is lower than that of the omnivorous animals
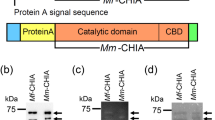
To determine the chitinolytic activity of Chia, bovine, dog, pig, chicken and mouse enzymes were expressed in E. coli as fusion proteins with truncated form of Staphylococcus aureus Protein A and V5-His tag49,60 as described in the Methods (Fig. 4a,b; Supplementary Fig. S3). 4-nitrophenyl N,N′-diacetyl-β-D-chitobioside (4-NP-chitobioside) was used as a substrate.
Omnivores Chia activity is higher than that of carnivores and herbivores. (a) The schematic representations of the E. coli-expressed bovine, dog, pig, chicken and mouse Chia fusion proteins. Chia is a secreted protein with a molecular mass of approximately 50 kDa, which contains an N-terminal catalytic domain (CatD) and a C-terminal chitin-binding domain (CBD). The E. coli-produced proteins contain the affinity tail of Protein A at the N-terminus. E. coli-recombinant proteins contain V5-His at the C-terminus (Supplementary Fig. S3). (b) Western blot analysis of the recombinant proteins using anti-V5 antibody. Arrow highlights the positions of the fusion proteins (Protein A-Chia-V5-His). The image of (d) was cropped from original full-length gel image shown in Supplementary Fig. S4a. (c) Comparison of the chitinolytic activities of Chia proteins using 4-NP-chitobioside. Error bars represent the mean ± SD from a single experiment conducted in triplicate. **p < 0.01.
Recombinant mouse and chicken Chia had the highest activity at pH 2.0 and the pig enzyme at pH 2.0–4.0 (Fig. 4c), in accordance with our previous reports49,50,51,52,60,61. Dog Chia had optimal activity at pH 2.0 while the bovine enzyme is most active at around pH 4.0 (Fig. 4c). However, both showed lower chitinolytic activity as compared to the mouse, chicken and pig Chia with activity levels at 1/6 (bovine) and 1/4 (dog) of that of the pig Chia at pH 4.0 and at pH 2.0, respectively (Fig. 4c).
Recombinant bovine and dog Chia proteins can degrade chitin substrates under the artificial GIT conditions
Even though the recombinant bovine and dog Chia proteins have low chitinolytic activity, we investigated, whether they do possess the ability to degrade polymeric chitin under gastrointestinal tract (GIT) conditions. Colloidal and crystalline chitin were incubated with bovine (Fig. 5a,b) and dog (Fig. 5c,d) fusion proteins under conditions mimicking GIT environment. The enzymes released mainly (GlcNAc)2 from the colloidal (Fig. 5a,c) as well as the crystalline (Fig. 5b,d) substrate. These results imply that both bovine and dog Chia are able to degrade chitin in the GIT.
Chitin substrates and chitin-containing organisms are degraded by recombinant Chia enzymes. Recombinant bovine and dog Chia proteins were incubated at 37 °C for 16 hours under GIT-like environment in the presence of pepsin or trypsin/chymotrypsin. Degradation products generated by incubation of (a and c) colloidal or (b and d) crystalline chitin in the GIT-mimicking conditions were analyzed by FACE. Arrow indicates the positions of (GlcNAc)2. Recombinant bovine and dog Chia proteins can degrade chitin substrates under the artificial GIT conditions. The images of (a–d) were cropped from original full-length gel images with same exposure time shown in Supplementary Fig. S5.
Functional and pseudo Chia genes adapted by the feeding behavior
As shown above, Chia mRNA level was significantly lower in the bovine (herbivore) in comparison to omnivores (Fig. 2). The NCBI Gene search showed that genomes of some herbivorous animals, such as rabbit and guinea pig, do not contain Chia genes (Fig. 6a). To further examine whether rabbit and guinea pig possess sequences similar to Chia genes in their genomes, we searched the regions between DENN/MADD Domain Containing 2D (DENND2D) and Pitchfork (PIFO) genes by NCBI blast search. These genes are conserved in mouse, rabbit, guinea pig, pig and human and are used as landmark genes in the vicinity of the Chia gene. DENND2D is a candidate tumor suppressor62,63, while PIFO regulates cilia disassembly64. Although we found vestigial Chia genes in both rabbit and guinea pig, they lack protein-coding abilities resulting from change to inactive pseudogenes (Fig. 6a).
The Chia gene evolution affected by the feeding behavior, which may determine chitin digestibility. (a) Schematic representation of the Chia genes as well as neighboring marker genes. Rabbit and guinea pig (herbivorous animals), do not have Chia gene in their genomes. Chia-like regions can be found between Dennd2d and PIFO. The vestigial genes lacking protein-coding abilities were found in the regions. (b) The relationship between feeding behavior and chitin digestibility in major livestock as well as laboratory and domestic animals based on Chia transcriptional levels in the stomach, chitinolytic activity and pseudogenization.
Based on the Chia transcription level and chitinolytic activity in stomach as well as pseudogenization, we summarized the relationship between the feeding behavior and chitin digestibility in major livestock as well as laboratory and domestic animals as shown in Fig. 6b. The chitin adaptability seems to be higher in omnivores with sufficient levels of Chia mRNAs and proteins in their stomach tissues as compared to carnivores and herbivores with low levels and activity of Chia.
Discussion
In previous reports, we have shown that mouse, chicken and pig stomach tissues express high levels of Chia mRNA and their translation products can degrade chitin substrates into (GlcNAc)2 under GIT conditions50,51,52. These results led to a speculation that all animals express large amounts of Chia enzyme in their stomach tissues. Here we reported that this observation is not reflected in all animals and is restricted to the omnivores. The Chia mRNA level in the stomach is strongly related to the feeding behaviors. Also, its chitinolytic activity is correlated to Chia transcription levels. Furthermore, some herbivores such as rabbit and guinea pig do not have functional Chia genes in their genomes. These results imply that feeding behaviors determine the Chia mRNA levels and its specific activity as well as Chia gene conservation during the evolution.
Bovine is a ruminant with four stomachs. Chia mRNA was expressed at low levels in all stomachs when compared with other animals. Although dog is a monogastric animal, Chia mRNA level was also very low in the stomach as well as in other tested tissues. Since dogs do not possess Chit1 gene in the genome, chitinases might to play different roles than in other mammals.
Pepsin has been detected in variety of animal stomach tissues and studied extensively55,65. As far as we know, comparison of their levels among multiple mammalian species has never been reported. As shown in Fig. 2, pepsinogen is a major transcript exceeding GAPDH in pig, chicken and mouse stomach tissues. However, the expression of pepsinogen in bovine and dog stomachs was much lower than in other animals (Fig. 2). Based on these results, we suggest that expression of pepsinogen as well as Chia may be controlled by their feeding behavior in the stomach.
Chitin digestibility can be estimated to some extent by Chia gene expression level in stomach as well as its chitinolytic activity. Our data showed that specific activity of Chia is as follows: mouse: chicken: pig: bovine: dog = 1.0: 0.7: 0.9: 0.25: 0.1. When those mRNA levels (number of molecules) are multiplied by its specific activities, the ratio is roughly: 140,000: 113,000: 6,000: 10: 1. These results clearly indicate that there are substantial differences in Chia mRNA levels and chitinase activities between omnivores, herbivores and carnivores stomach tissues and that chitin-digestibility is primarily determined by Chia mRNA levels.
In this report, we also compared specific activities of recombinant mouse, chicken, pig, bovine and dog Chia proteins. Although the primary sequences share about 70–79% identity, the chitinolytic activities of mouse, chicken and pig Chia were 4-to-10 times higher than those of bovine and dog enzymes. These results suggest that several non-conserved amino acid residues may influence the Chia activity and pH dependency. Furthermore, only the bovine Chia’s chitinolytic activity was highest at pH 4.0 (other species at pH 2.0). These results indicate that herbivorous stomach may have been adapted to express Chia with optimal activity in the less acidic condition.
Sequence changes and expression pattern of gene often determine the response of the organism to environmental stimuli66. For example, ruminant-like species have remarkably high concentrations of lysozyme c (EC 3.2.1.17) in the mucosa of the true stomach with differences in the time-, pH- and ionic strength-dependence of the rate of bacterial lysis when compared to conventional mammalian lysozymes c67. Similarly, the umami taste receptor gene, Tas1r1, has been shown to become a pseudogene in giant panda during a dietary switch to bamboo due to the relaxation of the functional constraint and ORF-disrupting substitutions68. The gene expression and it enzymatic activity of human Chia is affected by an insertion in the 5′ UTR69, promoter polymorphisms47 and nonsynonymous substitutions48,49. This knowledge allowed us to speculate that changing feeding behavior in some herbivores and carnivores had a major effect on the conservation of coding or non-coding region of the Chia genes.
Organisms containing chitin have been investigated due to their potential as alternative animal diets32,70,71,72,73. In pilot trials, shrimp shell chitin and dried mealworm (Tenebrio molitor) larvae were added to corn-based feeds for chicken and porcine diets, respectively74,75. Both studies showed that such supplementation is safe with effect on survival rate, growth performance, nutrient digestibility and blood profiles. A recent study reported that certain insect-eating primates underwent similar physiological adaptation as observed in mouse, chicken and pig as well as insectivorous bats76 as for the Chia enzymes utilization for chitin digestion in the insect exoskeletons77.
In this report, we show that, comparing to carnivores and herbivores, omnivores possess high ability of chitin digestion in their GIT. Before introduction of chitin-containing organisms to animal feeds, further research is needed on nutritive quality and safety evaluation of the potential by-products from such materials72,78,79.
Bovine stomachs contain high levels of cellulases (major digestive enzyme in herbivores), mainly produced by microorganisms80,81. Bovines often ingest insects abundantly present in their diet, e.g. with grass. Since the levels of Chia mRNA in the bovine stomach tissues are very low, it is possible that GIT bacteria supplement the chitinases, similarly to cellulase. Bovine GIT contains billions of bacteria in symbiosis82. Thus, bacterial chitinases may indeed play an important role in chitin digestion in bovines and possibly dogs. This hypothesis warrants further scrutiny.
Ingestion of insects by dogs is also not an uncommon event and chitin is usually well-tolerated. However, health issues can be caused by overfeeding. In “chitin overdose” cases in dogs or bovines, a mixture of lyophilized (recombinant or natural Chia from pig or chicken) chitinases or a probiotic treatment83,84 by chitinase-producing microbes could represent a potential treatment. Using such agents, chitin-containing organisms could potentially be utilized for non-omnivorous animals’ diets.
Methods
Bovine, pig and chicken stomach tissues
Frozen bovine, pig and chicken stomach tissues were purchased from Funakoshi Co., Ltd (Tokyo, Japan). The tissues were dissected and quickly frozen on dry ice and kept at −80 °C.
Total RNA and cDNA preparation
The Bovine Total RNA Panel and the Dog Total RNA Panel were purchased from Zyagen (San Diego, CA, USA) to examine the distribution of transcripts in various tissues. Mouse Total RNA Master Panel and Pig Total RNA panel were also purchased from Takara Bio USA, Inc. (Mountain View, CA, USA) and Zyagen, respectively. Mouse and pig stomach total RNAs from the panels were used to synthesize cDNAs for analyzing the level of Chia mRNA. In addition, total RNA was isolated from the frozen bovine or chicken stomach tissue using TRIzol Reagent (Thermo Fisher Scientific, Waltham, MA, USA) per manufacturer’s instructions. Total RNAs were reverse-transcribed into cDNA essentially as described previously50,51,52.
Selection of primer pairs for qPCR
Primers for qPCR were designed using PrimerQuest Input (Integrated DNA Technologies, Coralville, IA, USA) and evaluated their suitability based on whether they gave single products, as reflected by a single melting temperature (Tm). PCR reactions (final volume 13 µL) contained 2× SYBR Green Master Mix (Brilliant II SYBR Green QPCR Master Mix, Agilent, Santa Clara, CA, USA), 2.7 ng of bovine or dog cDNA or appropriate amount of the external standards (see below) and 2.5 pmol of primers for Chia, Chit1, pepsinogen and GAPDH. The PCR reactions were performed using Mx3005P QPCR System (Agilent) as follows: an initial denaturation and polymerase activation step for 10 min at 95 °C, followed by 40 cycles of denaturation at 95 °C for 30 sec, annealing at 55 °C for 30 sec and polymerization at 72 °C for 10 sec. Melting curves were generated after amplification. The primers’ sequences are listed in Supplementary Table S1.
Construction of the standard DNA and qPCR
Construction of the 18 genes standard DNA containing the genes of major proteins in stomach was described previously50,51,52. The standard DNA (Supplementary Fig. S1) was synthesized and inserted into pTAKN-2 vector by Eurofins Genomics (Tokyo, Japan). The standard DNA was prepared by PCR from the plasmid DNA using the forward primer 5′- GCTGCTGGTATCTCCAACAT -3′ and the revers primer 5′- TGGGCGTGGCTCAGGTAT -3′ and used for qPCR. qPCR was performed using standard DNA and cDNAs reverse-transcribed from total RNAs from animals essentially as described previously50,51,52.
Bovine stomach extract preparation
Soluble fraction was prepared from bovine fourth stomach tissues (0.2 g) by homogenization followed by centrifugation at 15,000 g for 10 min at 4 °C50,51,52. The supernatants were used as the stomach extract and pre-incubated at 37 °C for 0, 10, 20, 40 or 60 min at pH 7.6 or pH 2.0. After incubation at pH 2.0 and 37 °C, the solutions were neutralized. Total protein levels quantification was carried as described previously50,51.
Pig stomach extract preparation
Soluble fraction was prepared from pig stomach tissues as described previously52. After incubation at pH 2.0 and 37 °C for 10 min, the solutions were neutralized. Protein concentration was determined as described above.
SDS-polyacrylamide gel electrophoresis and western blot
The obtained protein fractions were analyzed using standard SDS-polyacrylamide gel electrophoresis (PAGE), followed by CBB staining. Western blot analysis was performed using polyclonal anti-mouse C terminal Chia50,54 or polyclonal goat anti-pepsin52, as described previously50,52.
Chitinase enzymatic assays
Chitinase enzyme activity was determined with 4-nitrophenyl-N, N′-diacetylchitobioside (Sigma-Aldrich, St. Louis, MO, USA) as a substrate in McIlvaine’s buffer (0.1 M citric acid and 0.2 M Na2HPO4; pH 2.0 to pH 8.0) or 0.1 M Gly-HCl buffer (pH 1.0 to pH 3.0) at 37 °C for 1 hour as described previously49,54. Chia unit definition was also reported previously60.
Pepsin enzymatic assays
Pepsin activity was measured using hemoglobin from bovine blood (Sigma-Aldrich) as the substrate as described previously50.
E. coli expression vectors
Coding regions of mature form of bovine, dog, pig and chicken Chia cDNAs were amplified from the corresponding animal’s cDNAs by PCR using KOD Plus DNA polymerase (Toyobo Co., Ltd, Osaka, Japan) and oligonucleotide primers (Eurofins Genomics) anchored with the restriction sites for EcoRI and XhoI (Supplementary Table S2) as described previously60. Amplified cDNA was digested with EcoRI and XhoI and cloned into the same sites of the pEZZ18/pre-Protein A-Chia-V5-His60. The entire nucleotide sequence of the resulting plasmid DNA (pEZZ18/Chia/V5-His) was confirmed by sequencing (Eurofins Genomics). The pEZZ18/pre-Protein A-mouse Chia-V5-His was prepared as described previously60. Expression of these plasmid DNA in E. coli cells led to the production of the mature Protein A-Chia-V5-His (Fig. 4a; Supplementary Fig. S3).
Preparation of the recombinant Chia proteins expressed in E. coli
Using the plasmid DNAs (the pEZZ18/pre-Protein A-Chia-V5-His), E. coli BL21 (DE3) (Merck Millipore, Tokyo, Japan) was transformed to express pre-Protein A-Chia-V5-His proteins. Transformed E. coli BL21 (DE3) strains were grown in 1.5 L LB medium containing 100 µg/mL ampicillin at 37 °C for 18 h. Cells were harvested by centrifugation at 7,000 g for 20 min at 4 °C. The recombinant protein was prepared from E. coli and purified by IgG Sepharose (GE Healthcare, Piscataway, NJ, USA) chromatography as described previously49,60. The protein-containing fractions were desalted using PD MidiTrap G-25 (GE Healthcare) equilibrated with the TS buffer [20 mM Tris-HCl (pH 7.6), 150 mM NaCl and a protease inhibitor (Complete, Roche, Basel, Switzerland)]. Protein A-mouse Chia-V5-His was also prepared as described previously60. The recombinant products were detected by Western blot using anti-V5-HRP monoclonal antibody (Thermo Fisher Scientific).
Degradation of colloidal and crystalline chitin substrates
Colloidal chitin and crystalline chitin were prepared from shrimp shell α-chitin (Sigma-Aldrich) and used as substrates to determine the chitinase activity. The degree of deacetylation (DD) of chitin was determined by elemental analysis. The elemental analysis of chitin was performed at the Analytical Center of the Tokyo University of Pharmacy and Life Sciences. Shrimp shell chitin (2.1% of DD) was powdered in a Wiley mill (Thomas Scientific, Swedesboro, NJ, USA) to 250 μm particle size and used as crystalline chitin. Also, it was incubated in concentrated HCl at 40 °C for 30 min and following filtration using a fused-in fritted glass disc (SIBATA SCIENTIFIC TECHNOLOGY LTD, Saitama, Japan), washed extensively with water to attain neutral pH and used as colloidal chitin. All enzymatic reactions using colloidal chitin (at a final concentration of 1 mg/mL) or crystalline chitin (1 mg/reaction) as substrates were incubated in a volume of 50 µL containing recombinant bovine (0.01 mU) or dog Chia (0.03 mU) in the presence of pepsin (0.6 µg) or trypsin/chymotrypsin (0.6 µg) as described previously52. N-acetyl chitooligoaccharides (Seikagaku Corporation, Tokyo, Japan) were used as a standard.
Statistical analysis
Biochemical data were compared by Student’s t-test.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Wysokowski, M. et al. Poriferan chitin as a versatile template for extreme biomimetics. Polymers 7, 235–265 (2015).
Neville, A. C., Parry, D. A. & Woodhead-Galloway, J. The chitin crystallite in arthropod cuticle. J. Cell Sci. 21, 73–82 (1976).
Liu, S. et al. Extraction and characterization of chitin from the beetle Holotrichia parallela Motschulsky. Molecules 17, 4604–4611 (2012).
Merzendorfer, H. Insect chitin synthases: a review. J. Comp. Physiol. B 176, 1–15 (2006).
Kaya, M., Seyyar, O., Baran, T., Erdogan, S. & Kar, M. A physicochemical characterization of fully acetylated chitin structure isolated from two spider species: with new surface morphology. Int. J. Biol. Macromol. 65, 553–558 (2014).
Garcia Mendoza, C. & Novaes Ledieu, M. Chitin in the new wall of regenerating protoplasts of Candida utilis. Nature 220, 1035 (1968).
Blumenthal, H. J. & Roseman, S. Quantitative estimation of chitin in fungi. J. Bacteriol. 74, 222–224 (1957).
Elorza, M. V., Rico, H. & Sentandreu, R. Calcofluor white alters the assembly of chitin fibrils in Saccharomyces cerevisiae and Candida albicans cells. J. Gen. Microbiol. 129, 1577–1582 (1983).
Muzzarelli, R. A. A. et al. Current views on fungal chitin/chitosan, human chitinases, food preservation, glucans, pectins and inulin: A tribute to Henri Braconnot, precursor of the carbohydrate polymers science, on the chitin bicentennial. Carbohydr. Polym. 87, 995–1012 (2012).
Das, S. & Gillin, F. D. Chitin synthase in encysting Entamoeba invadens. Biochem. J. 280(Pt 3), 641–647 (1991).
Campos-Gongora, E., Ebert, F., Willhoeft, U., Said-Fernandez, S. & Tannich, E. Characterization of chitin synthases from Entamoeba. Protist 155, 323–330 (2004).
Richards, A. G. Studies on Arthropod Cuticle. III. The Chitin of Limulus. Science 109, 591–592 (1949).
Giraud-Guille, M. M. Fine structure of the chitin-protein system in the crab cuticle. Tissue Cell 16, 75–92 (1984).
Ando, Y., Fukada, E. & Glimicher, M. J. Piezoelectricity of chitin in lobster shell and apodeme. Biorheology 14, 175–179 (1977).
Horst, M. N. The biosynthesis of crustacean chitin by a microsomal enzyme from larval brine shrimp. J. Biol. Chem. 256, 1412–1419 (1981).
Rodde, R., Einbu, A. & Vårum, K. M. A seasonal study of the chemical composition and chitin quality of shrimp shells obtained from northern shrimp (Pandalus borealis). Carbohydr. Polym. 71, 388–393 (2008).
Bo, M. et al. Isolation and identification of chitin in the black coral Parantipathes larix (Anthozoa: Cnidaria). Int. J. Biol. Macromol. 51, 129–137 (2012).
Weiss, I. M. & Schonitzer, V. The distribution of chitin in larval shells of the bivalve mollusk Mytilus galloprovincialis. J. Struct. Biol. 153, 264–277 (2006).
Weiss, I. M., Schonitzer, V., Eichner, N. & Sumper, M. The chitin synthase involved in marine bivalve mollusk shell formation contains a myosin domain. FEBS Lett. 580, 1846–1852 (2006).
Gaill, F., Shillito, B., Lechaire, J. P., Chanzy, H. & Goffinet, G. The chitin secreting system from deep sea hydrothermal vent worms. Biol. Cell 76, 201–204 (1992).
Durkin, C. A., Mock, T. & Armbrust, E. V. Chitin in diatoms and its association with the cell wall. Eukaryot Cell 8, 1038–1050 (2009).
Brunner, E. et al. Chitin-based organic networks: An integral part of cell wall biosilica in the diatom Thalassiosira Pseudonana. Angew. Chem. Int. Ed. 48, 9724–9727 (2009).
Ehrlich, H. et al. First evidence of the presence of chitin in skeletons of marine sponges. Part II. Glass sponges (Hexactinellida: Porifera). J. Exp. Zool. B Mol. Dev. Evol. 308, 473–483 (2007).
Ehrlich, H. et al. Identification and first insights into the structure and biosynthesis of chitin from the freshwater sponge Spongilla lacustris. J. Struct. Biol. 183, 474–483 (2013).
Wysokowski, M. et al. Isolation and identification of chitin in three-dimensional skeleton of Aplysina fistularis marine sponge. Int. J. Biol. Macromol. 62, 94–100 (2013).
Calström, D. The crystal structure of α-chitin (poly-N-acetyl-D-glucosamine). J. Biophys. Biochem. Cytol. 3, 669–683 (1957).
Sikorski, P., Hori, R. & Wada, M. Revisit of alpha-chitin crystal structure using high resolution X-ray diffraction data. Biomacromolecules 10, 1100–1105 (2009).
Minke, R. & Blackwell, J. The structure of alpha-chitin. J. Mol. Biol. 120, 167–181 (1978).
Rudall, K. & Kenchington, W. The chitin system. Biol. Rev. 49, 597–636 (1973).
Kaya, M. et al. On chemistry of gamma-chitin. Carbohydr Polym 176, 177–186 (2017).
Herrero, M. & Thornton, P. K. Livestock and global change: emerging issues for sustainable food systems. Proc. Natl. Acad. Sci. USA 110, 20878–20881 (2013).
Kupferschmidt, K. Buzz food. Science 350, 267–269 (2015).
van Huis, A. Edible insects are the future? Proc. Nutr. Soc. 75, 294–305 (2016).
Lee, C. G. et al. Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu. Rev. Physiol. 73, 479–501 (2011).
Koch, B. E., Stougaard, J. & Spaink, H. P. Keeping track of the growing number of biological functions of chitin and its interaction partners in biomedical research. Glycobiology 25, 469–482 (2015).
Bueter, C. L., Specht, C. A. & Levitz, S. M. Innate sensing of chitin and chitosan. PLoS Pathog. 9, e1003080 (2013).
Bussink, A. P., Speijer, D., Aerts, J. M. & Boot, R. G. Evolution of mammalian chitinase(-like) members of family 18 glycosyl hydrolases. Genetics 177, 959–970 (2007).
Hollak, C. E., van Weely, S., van Oers, M. H. & Aerts, J. M. Marked elevation of plasma chitotriosidase activity. A novel hallmark of Gaucher disease. J. Clin. Invest. 93, 1288–1292 (1994).
Letuve, S. et al. Lung chitinolytic activity and chitotriosidase are elevated in chronic obstructive pulmonary disease and contribute to lung inflammation. Am. J. Pathol. 176, 638–649 (2010).
Watabe-Rudolph, M. et al. Chitinase enzyme activity in CSF is a powerful biomarker of Alzheimer disease. Neurology 78, 569–577 (2012).
Artieda, M. et al. Serum chitotriosidase activity is increased in subjects with atherosclerosis disease. Arterioscler. Thromb. Vasc. Biol. 23, 1645–1652 (2003).
Sonmez, A. et al. Chitotriosidase activity predicts endothelial dysfunction in type-2 diabetes mellitus. Endocrine 37, 455–459 (2010).
Livnat, G. et al. Duplication in CHIT1 gene and the risk for Aspergillus lung disease in CF patients. Pediatr. Pulmonol. 49, 21–27 (2014).
Seibold, M. A. et al. Chitotriosidase is the primary active chitinase in the human lung and is modulated by genotype and smoking habit. J. Allergy Clin. Immunol. 122, 944–950 e943 (2008).
Zhu, Z. et al. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science 304, 1678–1682 (2004).
Reese, T. A. et al. Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature 447, 92–96 (2007).
Bierbaum, S. et al. Polymorphisms and haplotypes of acid mammalian chitinase are associated with bronchial asthma. Am. J. Respir. Crit. Care Med. 172, 1505–1509 (2005).
Seibold, M. A. et al. Differential enzymatic activity of common haplotypic versions of the human acidic mammalian chitinase protein. J. Biol. Chem. 284, 19650–19658 (2009).
Okawa, K. et al. Loss and gain of human acidic mammalian chitinase activity by nonsynonymous SNPs. Mol. Biol. Evol. 33, 3183–3193 (2016).
Ohno, M. et al. Acidic mammalian chitinase is a proteases-resistant glycosidase in mouse digestive system. Sci. Rep. 6, 37756 (2016).
Tabata, E. et al. Gastric and intestinal proteases resistance of chicken acidic chitinase nominates chitin-containing organisms for alternative whole edible diets for poultry. Sci. Rep. 7, 6662 (2017).
Tabata, E. et al. Protease resistance of porcine acidic mammalian chitinase under gastrointestinal conditions implies that chitin-containing organisms can be sustainable dietary resources. Sci. Rep. 7, 12963 (2017).
Ohno, M., Tsuda, K., Sakaguchi, M., Sugahara, Y. & Oyama, F. Chitinase mRNA levels by quantitative PCR using the single standard DNA: acidic mammalian chitinase is a major transcript in the mouse stomach. PLoS One 7, e50381 (2012).
Ohno, M. et al. Quantification of chitinase mRNA levels in human and mouse tissues by real-time PCR: species-specific expression of acidic mammalian chitinase in stomach tissues. PLoS One 8, e67399 (2013).
Kageyama, T. Pepsinogens, progastricsins, and prochymosins: structure, function, evolution, and development. Cell. Mol. Life Sci. 59, 288–306 (2002).
Zainuddin, A., Chua, K. H., Abdul Rahim, N. & Makpol, S. Effect of experimental treatment on GAPDH mRNA expression as a housekeeping gene in human diploid fibroblasts. BMC Mol. Biol. 11, 59 (2010).
Suzuki, M., Morimatsu, M., Yamashita, T., Iwanaga, T. & Syuto, B. A novel serum chitinase that is expressed in bovine liver. FEBS Lett. 506, 127–130 (2001).
Kouadjo, K. E., Nishida, Y., Cadrin-Girard, J. F., Yoshioka, M. & St-Amand, J. Housekeeping and tissue-specific genes in mouse tissues. BMC Genomics 8, 127 (2007).
Dabek, J., Wilczok, J., Kulach, A. & Gasior, Z. Altered transcriptional activity of gene encoding GAPDH in peripheral blood mononuclear cells from patients with cardiac syndrome X - an important part in pathology of microvascular angina? Arch. Med. Sci. 6, 709–712 (2010).
Kashimura, A. et al. Protein A-mouse acidic mammalian chitinase-V5-His expressed in periplasmic space of Escherichia coli possesses chitinase functions comparable to CHO-expressed protein. PLoS One 8, e78669 (2013).
Boot, R. G. et al. Identification of a novel acidic mammalian chitinase distinct from chitotriosidase. J. Biol. Chem. 276, 6770–6778 (2001).
Hibino, S. et al. Reduced expression of DENND2D through promoter hypermethylation is an adverse prognostic factor in squamous cell carcinoma of the esophagus. Oncol. Rep. 31, 693–700 (2014).
Sakha, S., Muramatsu, T., Ueda, K. & Inazawa, J. Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci. Rep. 6, 38750 (2016).
Kinzel, D. et al. Pitchfork regulates primary cilia disassembly and left-right asymmetry. Dev. Cell 19, 66–77 (2010).
Richter, C., Tanaka, T. & Yada, R. Y. Mechanism of activation of the gastric aspartic proteinases: pepsinogen, progastricsin and prochymosin. Biochem. J. 335(Pt 3), 481–490 (1998).
Gerstein, M. & Zheng, D. The real life of pseudogenes. Sci. Am. 295, 48–55 (2006).
Dobson, D. E., Prager, E. M. & Wilson, A. C. Stomach lysozymes of ruminants. I. Distribution and catalytic properties. J. Biol. Chem. 259, 11607–11616 (1984).
Zhao, H., Yang, J. R., Xu, H. & Zhang, J. Pseudogenization of the umami taste receptor gene Tas1r1 in the giant panda coincided with its dietary switch to bamboo. Mol. Biol. Evol. 27, 2669–2673 (2010).
Birben, E. et al. The effects of an insertion in the 5′UTR of the AMCase on gene expression and pulmonary functions. Respir. Med. 105, 1160–1169 (2011).
Latunde-Dada, G. O., Yang, W. & Vera Aviles, M. In vitro iron availability from insects and sirloin beef. J. Agric. Food Chem. 64, 8420–8424 (2016).
Oonincx, D. G. & de Boer, I. J. Environmental impact of the production of mealworms as a protein source for humans - a life cycle assessment. PLoS One 7, e51145 (2012).
Van Huis, A. et al. Edible insects: future prospects for food and feed security. FAO Forestry Paper 171, 1–201 (2013).
Bays, H. E. et al. Chitin-glucan fiber effects on oxidized low-density lipoprotein: a randomized controlled trial. Eur. J. Clin. Nutr. 67, 2–7 (2013).
Gernat, A. G. The effect of using different levels of shrimp meal in laying hen diets. Poult. Sci. 80, 633–636 (2001).
Jin, X. H., Heo, P. S., Hong, J. S., Kim, N. J. & Kim, Y. Y. Supplementation of dried mealworm (Tenebrio molitor larva) on growth performance, nutrient digestibility and blood profiles in weaning pigs. Asian-Australas J Anim Sci 29, 979–986 (2016).
Strobel, S., Roswag, A., Becker, N. I., Trenczek, T. E. & Encarnacao, J. A. Insectivorous bats digest chitin in the stomach using acidic mammalian chitinase. PLoS One 8, e72770 (2013).
Janiak, M. C., Chaney, M. E. & Tosi, A. J. Evolution of acidic mammalian chitinase genes (CHIA) is related to body mass and insectivory in primates. Mol. Biol. Evol., in press (2017).
Van Huis, A. Potential of insects as food and feed in assuring food security. Annu. Rev. Entomol. 58, 563–583 (2013).
Veldkamp, T. & Bosch, G. Insects: a protein-rich feed ingredient in pig and poultry diets. Animal Frontiers 5, 45–50 (2015).
Brulc, J. M. et al. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc. Natl. Acad. Sci. USA 106, 1948–1953 (2009).
Hess, M. et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331, 463–467 (2011).
Deusch, S. et al. A structural and functional elucidation of the rumen microbiome influenced by various diets and microenvironments. Front. Microbiol. 8, 1605 (2017).
Manninen, T. J., Rinkinen, M. L., Beasley, S. S. & Saris, P. E. Alteration of the canine small-intestinal lactic acid bacterium microbiota by feeding of potential probiotics. Appl. Environ. Microbiol. 72, 6539–6543 (2006).
Uyeno, Y., Shigemori, S. & Shimosato, T. Effect of probiotics/prebiotics on cattle health and productivity. Microbes Environ. 30, 126–132 (2015).
Acknowledgements
We are grateful to Haruko Miyazaki and Nobuyuki Nukina for their encouragement, to Kazuaki Okawa, Shotaro Honda, Masahiro Kimura, Natsumi Yamashita, Yasutada Imamura and Rieko Oyama for valuable suggestions. This work was supported by a Grant from the Science Research Promotion Fund of the Promotion and Mutual Aid Corporation for Private Schools of Japan (to M.S. and F.O.); by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) (grant numbers 15J10960, 16K07699 to M.O. and F.O., respectively); by the Project Research Grant from the Research Institute of Science and Technology, Kogakuin University (to F.O.); and in part by a grant of the Strategic Research Foundation Grant-aided Project for Private Universities (S1411005 to M.S. and F.O.) from the Ministry of Education, Culture, Sport, Science and Technology, Japan.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: E.T., A.K., A.K., H.M., R.M., Y.H., S.W., M.O., M.S., Y.S., V.M., P.O.B., F.O. Performed research: E.T., A.K., AK., H.M., R.M., Y.H., S.W., M.O., V.M., P.O.B., F.O. Analyzed data: E.T., A.K., A.K., H.M., R.M., Y.H., S.W., M.O., M.S., Y.S., V.M., F.O. Wrote the paper: E.T., V.M., P.O.B., F.O. Contributed to the critical appraisal of the paper and approved the final version: E.T., A.K., A.K., H.M., R.M., Y.H., S.W., M.O., M.S., Y.S., V.M., P.O.B., F.O.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tabata, E., Kashimura, A., Kikuchi, A. et al. Chitin digestibility is dependent on feeding behaviors, which determine acidic chitinase mRNA levels in mammalian and poultry stomachs. Sci Rep 8, 1461 (2018). https://doi.org/10.1038/s41598-018-19940-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-19940-8
This article is cited by
-
Chitin and omega-3 fatty acids in edible insects have underexplored benefits for the gut microbiome and human health
Nature Food (2023)
-
Seascape genomics of common dolphins (Delphinus delphis) reveals adaptive diversity linked to regional and local oceanography
BMC Ecology and Evolution (2022)
-
Full-fat insect meals in ruminant nutrition: in vitro rumen fermentation characteristics and lipid biohydrogenation
Journal of Animal Science and Biotechnology (2022)
-
Occupational exposure to gasoline in gasoline station male attendants promotes M1 polarization in macrophages
Environmental Science and Pollution Research (2022)
-
The effect of dietary supplementation with silkworm pupae meal on gastrointestinal function, nitrogen retention and blood biochemical parameters in rabbits
BMC Veterinary Research (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.