Abstract
Plant specific transcription factors, SQUAMOSA promoter-binding protein-like (SPL), are involved in many biological processes. However, no systematical study has been reported in cotton. In this study, a total of 177 SPL genes were identified, including 29, 30, 59 and 59 SPLs in Gossypium arboreum, G. raimondii, G. barbadense, and G. hirsutum, respectively. These SPL genes were classified into eight phylogenetical groups. The gene structure, conserved motif, and clustering were highly conserved within each orthologs. Two zinc finger-like structures (Cys3His and Cys2HisCys) and NLS segments were existed in all GrSPLs. Segmental duplications play important roles in SPL family expansion, with 20 genes involved in segmental duplications and 2 in tandem duplications, and ten ortholog pairs in syntenic regions between G. raimondii and A. thaliana. Several putative cis-elements, involved in light, stresses and phytohormones response, were found in the promoter regions of GhSPLs, suggesting that plant responses to those environmental changes may be induced through targeting SPL transcription factors. RNA-seq analysis shows that SPL genes were differentially expressed in cotton; some were highly expressed during fiber initiation and early development. Comparing with other plants, SPL genes show subfunctionalization, lost and/or gain functions in cotton during long-term domestication and evolution.
Similar content being viewed by others
Introduction
SQUAMOSA promoter-binding protein-like (SPL), one class of plant-specific transcription factors, have a highly conserved SBP domains (for SQUAMOSA-PROMOTER BINDING PROTEIN) with approximately 78 amino acids in length, and containing an eight Cys or His sequence motif (two Zn-finger like structure) and contained a nuclear localization signal (NLS) motif. The two Zn-finger binding sites consist a Cys3HisCys2HisCys or Cys6HisCys, in which the first zinc finger is Cys3His or Cys4, and second is Cys2HisCys1. SBP1 and SBP2 are the first two members of the SBP/SPL gene family, which were identified in Antirrhinum majus floral meristem involved in the control of early flower development2. Many SPL members were targeted by the miR156/157, and the miR156/SPL module plays important roles in diverse developmental processes in Arabidopsis, including shoot development, the phase change from vegetative growth to reproductive growth, and tolerance to abiotic stresses3,4,5. Various functions of SPL genes were also reported in other plant species, including governing yield-related traits in hexaploid wheat6, redundantly initiating side tillers and affecting biomass yield of energy crop in switchgrass7, regulating floral organ size and ovule production in cotton8, and regulating ovary and fruit development in tomato9.
Genome-wide identification of SPL/SBP-box gene family have been characterized in many plant species, including potato10, soybean11, oilseed rape12, pepper13, peanut14, Chinese cabbage15, citrus16, Prunus mume17, Salvia miltiorrhiza (Danshen)18, castor bean19, Populus trichocarpa20, apple21, grape22, tomato23, Arabidopsis and rice24. Guo et al.25 reported identification and phylogenetic relationship of 120 SPL genes from nine species representing the main green plant lineages: green alga, moss, lycophyte, gymnosperm and angiosperm. However, the identification and functional analysis of SPL gene family is much beyond in cotton than that in other plant species. There is only one report in which Zhang and colleagues26 cloned 24 SPL genes in cotton.
Cotton (Gossypium spp.) is one of the most important economic crops, and provides natural textile fiber and oilseed. G. hirsutum L. (AD1) and G. barbadense L. (AD2) are two tetraploids cultivated species, which were formed about 1–2 million years ago (MYA) through interspecific hybridization and chromosome doubling of A-genome (resembling A2 G. arboreum) and D-genomes (resembling D5 G. raimondii)27,28. Recently, the whole-genome sequences of four cotton species were released, including two allotetraploid species G. hirsutum acc. TM-129 and G. barbadense, cv. Xinhai21 and acc. 3–7930,31 and their two diploid progenitors G. arboreum32 and G. raimondii33,34. Those genome sequences provide a possible to identify SPL genes at a genome-wide level in cotton.
In this study, we identified SPL genes in four cotton species, defined the corresponding relationships and chromosomal locations. We built a phylogenetic tree of the SPL gene family in Gossypium, A. thaliana, O. sativa and P. trichocarpa, and carried out a genome-wide intra- and inter-genomic duplication analysis of G. raimondii and other three plant species. Additionally, we systematically analyzed the gene structure, conserved motif, cis-acting elements and expression pattern of all identified GhSPL genes in G. hirsutum. The results will provide a solid foundation to understand the distribution, structure and evolution of the SPL gene family in cotton, and will contribute to investigate of the detailed functional differentiation and application of these genes in the future.
Results
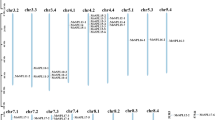
Genome-wide identification of the SPL gene family in Gossypium and their chromosomal distribution
To identify the SQUAMOSA promoter binding protein-like (SPL or SBP) transcription factor genes in cotton, SBP domain (PF03110) was used to search protein database of four cotton species, G. raimondii33, G. arboreum32, G. hirsutum acc. TM-129, and G. barbadense acc. 3–7931 by HMMER35. The candidate SPL genes were verified by the presence of the SBP domain using SMART and CDD36,37. A total of 177 SPL genes were identified in cotton, including 29, 30, 59 and 59 were identified in G. arboretum, G. raimondii, G. hirsutum and G. barbadense, respectively (Table S1). GrSPL genes were named according to the closest orthologs in A. thaliana. Among 17 SPL genes in A. thaliana, AtSPL3, AtSPL4, AtSPL11, AtSPL12, AtSPL15 and AtSPL16 have not orthologs in G. raimondii; and all 30 GrSPLs from the rest of 10 AtSPLs orthologs. AtSPL13 has 5 paralogs in G. raimondii; AtSPL1, AtSPL5 and AtSPL6 have 4 paralogs; AtSPL2, and AtSPL9 have 3 paralogs; AtSPL7, AtSPL8 and AtSPL10 have 2 paralogs; and AtSPL14 only 1 paralogs in G. raimondii. Different paralogs were coded a, b, c, and so on, according to their order of the homologous chromosomes. The corresponding orthologs in G. arboreum, G. hirsutum acc. TM-1 and G. barbadense acc. 3–79 were named as GaSPL, GhSPL, and GbSPL, respectively. The encoded protein lengths of 59 SPL genes varied from 141 to 1083 amino acids, and the predicted molecular weight (Mw) of these SPL proteins ranged from 16.112 to 119.882 kDa in upland cotton G. hirsutum (Table S1).
The 59 GhSPL genes were located on 20 chromosomes, with no SPL gene detected in A5/D5, A6/D6, and A9/D9 homologous chromosomes in G. hirsutum acc. TM-1 (Fig. 1, Table S1). 30 GrSPL genes were located on 11 chromosomes in G. raimondii except on the D5 and D6. GrSPL9b was located on the D9 in G. raimondii, but GhSPL9b_A/GhSPL9b_D were in A4/D4 (scaffold) homologous chromosomes in G. hirsutum. This phenomenon, SPL9b was positioned on the different chromosomes, might be caused by assembly error in the duplicated gene regions, and need to be further confirmed. Furthermore, tandem duplication events were defined as genes separated by five or fewer genes and within 100 Kb region38. Only one gene pairs on D12 (GrSPL10b/GrSPL8b) displayed tandem duplications in G. raimondii (Fig. 1B), and the corresponding tandem duplication (GhSPL10b_D/GhSPL8b_D) was detected on D12 of G. hirsutum (Fig. 1A). On A12, the distance was over 100 Kb (161.95 Kb) between GhSPL10b_A and GhSPL8b_A. Compared with the distribution of SPL genes in G. raimondii and G. hirsutum, SPLs displayed high collinearity in the D-genome of G. raimondii and A-, D-subgenomes of G. hirsutum. Additionally, two pairs of orthologs genes were located in A2/A3 reciprocal translocation section, within GhSPL13b_A and GhSPL2b_A on A3 chromosomes, GhSPL13b_D and GhSPL2b_D on D2.
Phylogenetic analysis and gene duplication observation of SPL gene family
To investigate the phylogenetic relationship of the SPL transcription factor family in cotton, a total of 242 SPLs were used to construct a Neighbour-Joining (N-J) phylogenetic tree by MEGA 7.0 software39. These 242 SPLs included 177 obtained from this research in four cotton species, G. raimondii (30), G. arboretum (29), G. hirsutum (59) and G. barbadense (59); the rest 65 were obtained from three well-studied plant species, including 17 from A. thaliana, 18 from O. sativa and 30 from P. trichocarpa. As shown in Fig. 2, all of the SPL genes were clustered into eight sub-groups (from I to VIII), and each group contained at least one protein from three species (Arabidopsis, rice and poplar) except group VI in which there was no SPL from rice. Additionally, different SPL orthologs were clearly distinguished in cotton. Cotton SPL13, SPL8, SPL9, SPL2, SPL6 and SPL7 were grouped to cluster I–IV, VI and VIII, respectively. Cotton SPL5 and SPL10 were clustered in group V, SPL1 and SPL14 were clustered in group VII.
To reveal SPL duplication events, four whole intra- genomic duplication data files of G. raimondii, A. thaliana, O. sativa and P. trichocarpa, and three inter-genomic duplication data files between G. raimondii and three other species were downloaded from the PGDD database40. All SPL gene duplication events was filtered out (Fig. 3, Table S2). Among 30 SPL genes in G. raimondii, we identified 16 pairs of duplications out of 20 GrSPLs (GrSPL1a/GrSPL1d, GrSPL1c/GrSPL1d, GrSPL2a/GrSPL2b, GrSPL2a/GrSPL2c, GrSPL2b/GrSPL2c, GrSPL5a/GrSPL5c, GrSPL6a/GrSPL6d, GrSPL6b/GrSPL6d, GrSPL7a/GrSPL7b, GrSPL10a/GrSPL10b, GrSPL13a/GrSPL13b, GrSPL13a/GrSPL13e, GrSPL13b/GrSPL13e, GrSPL13c/GrSPL13b, GrSPL13c/GrSPL13d and GrSPL13e/GrSPL2c), involving 7 GrSPL orthologs except GrSPL8, GrSPL9 and GrSPL14. All duplication pairs had Ka/Ks values of less than 1 (ranging 0.16–0.50) (Table S2), suggesting that SNP gene family in G. raimondii had subjected to purifying selection during the long-term evolutionary process. Compared with tandem duplications (only one pair on D12, GrSPL10b/GrSPL8b), segmental duplications played a significant role in expansion of SPL gene family in cotton. As well as, 4, 7 and 25 pairs of duplications were identified in A. thaliana, O. sativa and P. trichocarpa, respectively (Fig. 3, Table S2).
Distribution of the intra- and inter-genomic duplications of SPL genes in G. raimondii, A. thaliana, O. sativa and P. trichocarpa. G. raimondii, A. thaliana, O. sativa and P. trichocarpa chromosomes made a circle using CIRCOS. The different species chromosomes and their intra-genomic duplication were depicted with different colors. Intra-genomic duplication between G. raimondii and other three species (A. thaliana, O. sativa and P. trichocarpa) were indicated with red, cyan and forestgreen lines, respectively.
SPL duplication pattern between G. raimondii and three other species, A. thaliana, O. sativa and P. trichocarpa were also analyzed. Among 30 GrSPLs from 10 Arabidopsis SPL orthologs, 19 pairs of duplication events were identified between G. raimondii and A. thaliana, involving 8 GrSPL orthologs, and 14 GrSPL and 12 AtSPL genes, respectively; 22 pairs of duplication events were identified between G. raimondii and P. trichocarpa, involving 5 GrSPL orthologs, and 10 GrSPL and 11 PtSPL genes, respectively; only 5 pairs of duplications were observed in G. raimondii and O. sativa, and this indicated that SPL gene family of Gossypium and O. sativa were less conserved. There were more GrSPL genes or orthologs between G. raimondii and A. thaliana than that between G. raimondii and P. trichocarpa. We compared syntenic map of G. raimondii and A. thaliana (Fig. 3, Table S2), with ten ortholog pairs positions on segmental duplicated blocks including the following: GrSPL1d/AtSPL1; GrSPL2a, 2b/AtSPL11; GrSPL5a/AtSPL4, 5; GrSPL5c/AtSPL3, 5; GrSPL5d/AtSPL5; GrSPL6c/AtSPL6; GrSPL7a, 7b/AtSPL7; GrSPL9a, 9c/AtSPL9, 15; GrSPL13c, 13d/AtSPL13B; and GrSPL14a/AtSPL14, 16.
Gene structure and conserved motif analysis of SPLs in G. hirsutum
With GhSPLs as an example, we analyzed the SPL gene exon/intron structure, conserved motif, and putative cis-acting elements from GhSPLs promoters. An unrooted N-J tree was also constructed only using 59 SPL protein sequences from G. hirsutum (Fig. 4A). The gene structures of 59 GhSPLs were analyzed by GSDS 2.041, and displayed in Fig. 4B. The number of introns of 59 GhSPLs varied from 0 to 9. Nearly half of SPL genes (27 GhSPLs) had two introns (6 GhSPL6, 3 GhSPL8, 6 GhSPL9, 4 GhSPL10 and 8 GhSPL13); 5 GhSPL2 and 8 GhSPL5 had three and one introns, respectively; 14 GhSPLs had nine introns (including 8 GhSPL1, 4 GhSPL7 and 2 GhSPL14); the remaining SPL members GhSPL2a_D, GhSPL8a_A, GhSPL13d_A/D and GhSPL6a_D had 6, 4, 3 and 0 introns, respectively. By comparing the SPL gene structures of G. hirsutum and Arabidopsis, we found that the pattern of exon/intron structures of SPLs in G. hirsutum is quite similar to AtSPLs. In Arabidopsis, AtSPL1, AtSPL7 and AtSPL14 had 9 introns; AtSPL6, AtSPL8 AtSPL9, AtSPL10 and AtSPL13 had 2 introns; AtSPL2 and AtSPL5 had 3 and 1 introns, respectively (supplementary material Fig. S1). This result reveals that different GhSPL orthologs exhibited different exon-intron structures and were similar to Arabidopsis orthologs.
Phylogenetic analysis and gene structure of GhSPLs. (A) The unrooted NJ tree of 59 GhSPL genes was constructed using MEGA 7, and the bootstrap test was performed with 1000 replicates. (B) The exon/intron distribution of GhSPL genes. Exons and introns were represented by black boxes and lines, respectively.
The MEME42 was used to predict motifs of 59 SPL protein sequences in G. hirsutum. 20 motifs, named motifs 1 to 20 were identified (Fig. 5). The length of 20 identified motifs and consensus sequence were listed in supplementary material Table S3, and Logos of 20 conserved motifs are shown in supplementary material Fig. S2. The lengths of those conserved motifs were between 21 (motif 15) and 159 amino acids (motif 4). The number of the conserved motifs in each GhSPL protein varied from 2 to 13. All GhSPL proteins contained motif 1 (two Zn-finger like structure), and 56 GhSPLs contained motif 2 (nuclear localization signal, NLS) except GhSPL5c_D, GhSPL6a_D and GhSPL9b_D. Seven GhSPL5 proteins only had motifs 1 and 2; GhSPL8 had motifs 1, 2 and 14; GhSPL13 had motifs 1, 2 and 15; GhSPL9 and GhSPL10 had motifs 1, 2 14, and 15; other GhSPLs had more motifs, such as GhSPL2 (6 motifs), GhSPL7 (6–7 motifs), GhSPL14 (9–10 motifs) and GhSPL1 (11–13 motifs). Then, multiple alignment of all 59 GhSPL proteins was performed by MAFFT version 7, and presented the SBP domain structures in detail. All GhSPLs exhibit two zinc finger-like structures and NLS segments, with the exception of three SPLs (GhSPL5c_D, GhSPL6a_D and GhSPL9b_D) which lacked NLS. The first Zn-finger like structure (Cys3His), the second Zn-finger like structure (Cys2HisCys) and highly conserved NLS were signed in Fig. 6B. The SBP domain motif logo and protein sequence were showed in Fig. 6A. Therefore, the sequences SBP domain of GhSPLs, two Zn-finger structure and NLS section, were also conserved in cotton, and SPL motif member architecture within each of GhSPL orthologs tend to have a similar number and type of motifs.
Conserved motifs of GhSPL proteins. Based on the GhSPL protein sequences, the conserved motifs were identified using MEME (suite 4.11.4), and each motif is indicated with a colored box numbered (1 to 20) at the bottom. Motif 1 and motif 2 were two Zn-finger like structure and nuclear localization signal (NLS). Motif 7 contained the miR156/miR157 recognition element as a target site for the miR156/miR157 in 3′ UTR.
In conclusion, members belonging to the same GhSPL orthologs had a similar gene structure, motif architecture, tended to cluster together in phylogenetic tree.
The cis-acting elements analysis of GhSPL gene promoter regions
The upstream sequences of 59 GhSPL genes (2500 bp upstream of the initiation codon) were used for cis-acting element prediction by PlantCARE43. A total of 42 types of putative cis-elements involved in light were present in the promoters of GhSPLs, including 23 light partial responsive elements (I-box, GAG-motif, GATA-motif, TCT-motif, GA-motif, CATT-motif, TCCC-motif, chs-CMA1a, chs-CMA2a, Gap-box, Box II, LAMP-element, L-box, etc), 6 light responsive elements (Box I, GT1-motif, Sp1, 3-AF1 binding site, MNF1 and AAAC-motif), and other light responsive elements, such as Box 4, G-box, ACE, ATCT-motif, MRE, AE-box, as-2-box, AT1-motif, and ATC-motif (Table S4). Other major cis-elements also include elements responsible to stress response [such as defense and stress (TC-rich repeats), WRKY binding site (W box), heat (HSE), drought (MBS), low-temperature (LTR) and wound (WUN-motif)], and phytohormone response [such as auxin (TGA-element and AuxRR-core), abscisic acid (ABRE), ethylene (ERE), gibberellin (P-box, TATC-box and GARE-motif), salicylic acid (TCA-element) and MeJA (CGTCA-motif and TGACG-motif)] (Fig. 7, Table S4). This suggests the important roles of GhSPL genes in biological processes as well as response to abiotic stresses and phytohormones in cotton.
Expression profiles of SPL genes in G. hirsutum
In order to understand the putative functions of GhSPL genes, we analyzed the expression profiles of all the identified 59 SPLs by using the currently available RNA-seq data of G. hirsutum acc. TM-129, including 12 different tissues and organs: root, stem, leaf, petal, stamen, −3, 0 or 3 DPA ovules, and 5, 10, 20 or 25 DPA fibers. A heat map expression of GhSPLs was showed by Mev4.9.0 in Fig. 8. Nine SPL genes GhSPL1b_A/D, GhSPL1c_A/D, GhSPL1d_A/D, GhSPL14a_A/D and GhSPL7b_D were highly expressed in all tissues. Six GhSPL2 were highly expressed in stem, leaf, petal, −3, 0 and 3 DPA ovules. GhSPL7a_A/D and GhSPL7b_A were highly expressed in stem and leaf. In the GhSPL5 orthologs, GhSPL5b_A were highly expressed in stem and −3 DPA ovules; GhSPL5c_A were highly expressed in −3, 0 and 3 DPA ovules; GhSPL5c_D were highly expressed in 3 DPA ovules and 5 DPA fibers; GhSPL5d_D were highly expressed in petal; GhSPL5a_A/D, GhSPL5b_D and GhSPL5d_A were low expressed in all tissues. In the GhSPL6 orthologs, GhSPL6d_A/D were highly expressed in 3, 5 and 10 DPA fibers, GhSPL6a_A/D, GhSPL6b_A/D and GhSPL6c_A/D were low expressed in all tissues. In the GhSPL8 and GhSPL9 orthologs, GhSPL8a_A/D and GhSPL9a_A/D were highly expressed from −3 to 3 DPA ovules; GhSPL9b_A/D were highly expressed in petal; GhSPL8b_A/D and GhSPL9c_A/D were low expressed in all tissues. In the GhSPL10 orthologs, GhSPL10b_A/D were highly expressed in 5 DPA fibers; and GhSPL10a_A/D were low expressed in all tissues. In the GhSPL13 orthologs, GhSPL13a_A/D were highly expressed in leaf, −3, 0 and 3 DPA ovules; GhSPL13b_A/D and GhSPL13e_A/D were highly expressed in leaf; GhSPL13d_A/D were highly expressed in 0 and 3 DPA ovules. However, GhSPL13c_A/D were no detected in all tested tissues.
Expression patterns of GhSPL genes in different tissues and organs in G. hirsutum acc. TM-1. Twelve tissues and organs were root, stem, leaf, petal, stamen, −3, 0 or 3 DPA ovules, 5, 10, 20 and 25 DPA fibers. The color represents GhSPLs expression levels: Log2 (FPKM). The phylogenetic relationship was showed on the left.
Discussion
In the past couple of years, SPL transcription factors have been attracting attention from the scientific community. Genome-wide identification of SPL gene family has been reported in several plant species. The number of SPLs varies from species to species. For instance, there are 15 SPLs in potato, pepper, peanut, citrus, Prunus mume, danshen, castor bean and tomato10,13,14,16,17,18,19,23, 17–20 in grape, rice and Arabidopsis22,24, 27–30 in Chinese cabbage, Populus and apple15,20,21, 41 in soybean11, and 58 in oilseed rape12. However, no genome-wide identification of SPL gene family has been reported in cotton although there are four cotton species sequenced. In this study, we reported for the first time the genome-wide identification of SPL genes and systematically investigated the functional structure of SPL transcription factor family. Based on our results, we identified 29, 30 59 and 59 SPL genes in G. arboretum, G. raimondii, G. hirsutum and G. barbadense, respectively (Table S1). The number of SPLs in A or D genome diploid cotton were similar to Populus, and an amount of SPLs of allotetraploid cotton species were very close to oilseed rape B. napus (AACC, 2n = 38). To compare the number of SPL genes and corresponding relationships in four cotton species, we found that there were a typical polyploidization phenomenon. All 29 SPLs in diploid A-genome (G. arboretum) and 30 SPLs in D-genome (G. raimondii) could be found their homologous genes in allotetraploid genomes (G. hirsutum AADD, 20 = 52). Only SPL6a was found two homologous genes in four cotton species, GrSPL6a and GhSPL6a_D in the D-genome (G. raimondii) and D-subgenome of G. hirsutum, respectively, and no unique genes were found in the A-genome G. arboreum, A-subgenome of G. hirsutum, and A- and D- subgenome of G. barbadense. Additionally, phylogenetic analysis of SPL proteins in various species showed that green alga were grouped together, and other SPLs were classified into 6–7 groups21,22. In this study, phylogenetic tree of 242 SPL proteins from four cotton species (G. raimondii, G. arboretum, G. hirsutum and G. barbadense), A. thaliana, O. sativa and P. trichocarpa, showed that all SPL genes were clustered into eight groups (I–VIII) (Fig. 2). Among 177 cotton SPLs from 10 Arabidopsis SPLs orthologs, each kinds of orthologs were clustered together.
Segmental duplications play an important role in the gene expansion of SPL transcription factor gene family. Many segmental duplication gene pairs were found in SPL gene family in plants20,21,22. In this study, we identified one pair of tandem duplication (GrSPL10b/GrSPL8b) and 16 pairs of segmental duplications involved 20 SPL genes in G. raimondii. There were 4, 7 and 25 pairs of duplications in A. thaliana, O. sativa and P. trichocarpa, respectively. Additionally, the duplication pattern of SPLs between G. raimondii and other three species were analysis; 5, 19 and 22 pairs of duplication events were identified between G. raimondii and O. sativa, G. raimondii and A. thaliana, G. raimondii and P. trichocarpa, respectively. There were 10 ortholog pairs in syntenic regions between G. raimondii and A. thaliana (Fig. 3, Table S2). Inter-genomic duplication events between Arabidopsis and other two species were identified in previous study, including eleven SPLs ortholog pairs between apple and Arabidopsis21, and nine SPLs ortholog pairs between grape and Arabidopsis22.
By comparing the number of introns of SPLs gene in cotton and Arabidopsis, we found that different SPL orthologs contained different gene structures, including 9 introns in SPL1, SPL7 and SPL14; 2 introns in SPL6, SPL8, SPL9, SPL10 and SPL13; 3 introns in SPL2 had; 1 introns in SPL5. As well as different cotton SPL orthologs shared similar motifs. GhSPL5 had motifs 1 and 2; GhSPL8 had motifs 1, 2 and 14; GhSPL13 had motifs 1, 2 and 15; GhSPL9 and GhSPL10 had motifs 1, 2 14, and 15; GhSPL1, GhSPL2, GhSPL7 and GhSPL14 had more motifs. Among 20 motifs, motif 1 was two Zn-finger like structure, existed in all 59 GhSPL proteins. Motif 2 was nuclear localization signal (NLS), and 56 GhSPLs contained this motif except GhSPL5c_D, GhSPL6a_D and GhSPL9b_D. As showed in Fig. 6B, SBP conserved domain, two zinc finger-like structures and NLS segments, were shared by all GrSPLs. In addition, we speculate that different SPL orthologs probably play similar function between cotton and Arabidopsis, due to the presence of similar exon/intron structure and conserved motifs.
Several cis-acting elements were found in the promoter regions of GhSPLs (Fig. 7, Table S4), which suggests that SPL transcription factors may be regulated by light, stresses and/or phytohormones. All the identified SPL gene show a development- and tissue-dependent expression patterns (Fig. 8). GhSPL1 and GhSPL14 orthologs were highly expressed in all tested developmental stage and tissues. Some SPL genes were highly expressed in certain tissues, and others were low expressed in all tested tissues in the same orthologs. GhSPL2, GhSPL5b_A/D, GhSPL5c_A/D, GhSPL6d_A/D, GhSPL8a_A/D, GhSPL9a_A/D, GhSPL10b_A/D, GhSPL13a_A/D, GhSPL13d_A/D were highly expressed in −3, 0 and 3 DPA ovules or 5, 10 DPA fibers. This result suggests that cotton SPL gene family may play an important role during fiber initiation, and cotton paralog genes possibly existed subfunctionalization, lost functions, even gained new functions. To date, there are only two expression and function study of SPL genes in cotton8,26. Liu et al.8 reported the expression level of GhSPLs and two MADS-box genes (orthologs of AtAGL6 and SITDR8) were repressed in the miR157 over-expression cotton lines. Hypothesized that the miR157/SPL may regulate floral organ size and ovule production in cotton. Zhang et al.26 reported that GhSPL3 and GhSPL18 might be involved in the development of leaves and second shoots, and promoting flowering by overexpression target genes in Arabidopsis plants.
MicroRNAs (miRNAs) may also involve in SPL-regulated gene networks. Among 17 SPLs in Arabidopsis, 10 were putative targets of miR156/1574,44. 11 of 19 SPLs in rice45, 18 of 28 SPLs in Populus20, 17 of 41 SPLs in soybean11 were reported to be potential targeted by certain miR156. In this study, we also found that 31 of 59 identified SPLs were potentially targeted by miR156 in upland cotton, which are from 6 different orthologs (GhSPL2, GhSPL6, GhSPL9, GhSPL10 and GhSPL13). Interestingly, motif 7 was existed in those SPLs, and it is a potential target site for the miR156/miR157. In Arabidopsis and rice, motif contains miR156 recognition element was also reported in all miR156-targeted SPLs4,25,45. Thus, SPL gene function analysis mainly through significantly represses the SPL transcriptions by over-expression of miR156/miR157. Arabidopsis as an important model plant species, the majority of AtSPL genes have been well functionally characterized. SPL2, SPL9, SPL10, SPL11, SPL13 and SPL15 contribute shoot development and the phase transition from vegetative growth to reproductive growth5. SPL3, SPL4 and SPL5 primarily promote floral induction and/or floral meristem identity, by SOC1-SPL module control flowering time46, and act synergistically with FT-FD module to induce flowering under LDs47. SPL3, SPL9 and SPL10 are involved in lateral root growth48. SPL1 and SPL12 confer plant thermotolerance at the reproductive stage49. SPL7 regulates the Cu deficiency response50,51. SPL8 acts in concert to secure male fertility and regulates gynoecium differential patterning52,53,54. Based on the high conservation of SPL gene family between cotton and Arabidopsis, we speculate that SPL gene family in cotton may involve in the timing of vegetative and reproductive phase change, root growth, leaf development, fertility, fiber initiation development, response to stresses and yield (Fig. 9). However, the detailed function of each SPL transcription factor in cotton remains to be investigated. This is a genome-wide analysis of SPL gene family in cotton, which will provide the overall and useful information for well functional analysis in the future.
Materials and Methods
Sequence sources
The sequences of four sequenced cotton species, G. raimondii, G. arboreum, G. hirsutum acc. TM-1, and G. barbadense acc. 3–79, were downloaded from http://www.phytozome.net/, http://cgp.genomics.org.cn, http://mascotton.njau.edu.cn/, and http://cotton.cropdb.org/cotton/download/data.php, respectively. The SPL protein sequence data were obtained for A. thaliana, O. sativa and P. trichocarpa from the Plant Transcription Factor Databases55 (Plant TFDB v4.0, planttfdb.cbi.pku.edu.cn/), the General Feature Format (GFF) file Arabidopsis Information Resource (TAIR release 10, http://www.arabidopsis.org), the Rice Genome Annotation Project Database (RGAP release 7, http://rice.plantbiology.msu.edu/index.shtml), and (ftp://plantgenie.org/Data/PopGenIE/Populus_trichocarpa/v2.2/), respectively. The gene name and ID were listed in the supplementary materials (Table S5), including 17, 18, and 30 known SPL genes in A. thaliana, O. sativa and P. trichocarpa, respectively.
Identification of SPL transcription factor family in cotton and their chromosomal mapping
SBP domain (PF03110) for SQUAMOSA-PROMOTER BINDING PROTEIN was downloaded from Pfam56 (http://pfam.xfam.org/), and was employed to identify all possible SPL genes in four cotton species using HMMER (v3.1b2)35 (http://hmmer.org) with the e-value < 1e-10. Each candidate SPL gene was further confirmed using SMART36 (http://smart.embl-heidelberg.de/) and CDD37 (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The theoretical pI (isoelectric point) and molecular weight of the GhSPLs were investigated within Expasy57 (http://web.expasy.org/protparam/).
The physical location of the SPLs in G. hirsutum and G. raimondii were fetched from the corresponding GFF files. MapInspect (http://mapinspect.software.informer.com/) was used to visualize the distribution of the SPL genes in Gossypium genome.
Phylogenetic and gene duplication
A phylogenetic tree was constructed using ClustalW alignment and the Neighbor-Joining (NJ) method in MEGA 7.0 software39 (https://mega.nz/), with 1000 replicates boot- strap test. The genome-wide intra- and inter-genomic duplication files of G. raimondii, A. thaliana, O. sativa and P. trichocarpa were downloaded from the PGDD40 (http://chibba.agtec.uga.edu/duplication), and the visualization was carried out with the CIRCOS tool58 (http://circos.ca/). The ratios of Ka/Ks were used to assess the selection pressure for duplication genes.
Gene structure and conserved motif
The exon/intron structures of GhSPLs were drawn using GSDS 2.041 (http://gsds.cbi.pku.edu.cn/), through inputting genes GFF files. MEME (suite 4.11.4)42 (http://meme-suite.org/) was employed to identify conserved motifs of GhSPLs with the following parameters: the maximum number of motifs 20, and optimum width from 6 to 250. In addition, SBP domain was presented alone using Multiple alignment program MAFFT version 7 (http://mafft.cbrc.jp/alignment/server/).
Promoter regions cis-acting elements analysis
The promoter sequences (2500 bp upstream of the initiation codon “ATG”) of 59 GhSPL genes were extracted from genome sequences of G. hirsutum. The PlantCARE43 (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/search_CARE.html) were used to find putative cis-acting elements.
Expression pattern analysis
To analyze the expression patterns of GhSPL genes, we used RNA-seq data of G. hirsutum acc. TM-129, including root, stem, leaf, petal, stamen, −3, 0 or 3 DPA ovules, 5, 10, 20 and 25 DPA fibers. The expression levels of GhSPL genes were calculated using Log2 (FPKM), fragments per kilobase of exon per million fragments mapped. Expression patterns were display in Mev4.9.0 (https://sourceforge.net/projects/mev-tm4/), and clustered by hierarchical clustering model.
References
Yamasaki, K. et al. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J Mol Biol. 337, 49–63 (2004).
Klein, J., Saedler, H. & Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol Gen Genet. 250, 7–16 (1996).
Cui, L. G., Shan, J. X., Shi, M., Gao, J. P. & Lin, H. X. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 80, 1108–1117 (2014).
Wang, H. & Wang, H. The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Mol Plant. 8, 677–688 (2015).
Xu, M. et al. Developmental functions of miR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet. 12, e1006263 (2016).
Zhang, B. et al. Functional conservation and divergence among homoeologs of TaSPL20 and TaSPL21, two SBP-box genes governing yield-related traits in hexaploid wheat. Plant Physiol. pii: pp.00113 (2017).
Wu, Z. et al. Switchgrass SBP-box transcription factors PvSPL1 and 2 function redundantly to initiate side tillers and affect biomass yield of energy crop. Biotechnol Biofuels. 9, 101 (2016).
Liu, N. et al. MicroRNA 157-targeted SPL genes regulate floral organ size and ovule production in cotton. BMC Plant Biol. 17, 7 (2017).
Ferreira e Silva, G. F. et al. microRNA156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. Plant J. 78, 604–618 (2014).
Kavas, M., Kızıldoğan, A. K. & Abanoz, B. Comparative genome-wide phylogenetic and expression analysis of SBP genes from potato (Solanum tuberosum). Comput Biol Chem. 67, 131–140 (2017).
Tripathi, R. K., Goel, R., Kumari, S. & Dahuja, A. Genomic organization, phylogenetic comparison, and expression profiles of the SPL family genes and their regulation in soybean. Dev Genes Evol. 227, 101–119 (2017).
Cheng, H. et al. Genomic identification, characterization and differential expression analysis of SBP-box gene family in Brassica napus. BMC Plant Biol. 16, 196 (2016).
Zhang, H. X. et al. Genome-Wide Identification and Analysis of the SBP-Box Family Genes under Phytophthora capsici Stress in Pepper (Capsicum annuum L.). Front Plant Sci. 7, 504 (2016).
Li, M. et al. Cloning and characterization of SPL-family genes in the peanut (Arachis hypogaea L.). Genet Mol Res. 15, https://doi.org/10.4238/gmr.15017344 (2016).
Tan, H. W., Song, X. M., Duan, W. K., Wang, Y. & Hou, X. L. Genome-wide analysis of the SBP-box gene family in Chinese cabbage (Brassica rapa subsp. pekinensis). Genome. 58, 463–477 (2015).
Shalom, L. et al. Molecular characterization of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) gene family from Citrus and the effect of fruit load on their expression. Front Plant Sci. 6, 389 (2015).
Xu, Z. et al. Identification and expression analysis of the SQUAMOSA promoter-binding protein (SBP)-box gene family in Prunus mume. Mol Genet Genomics. 290, 1701–1715 (2015).
Zhang, L. et al. Genome-wide analysis and molecular dissection of the SPL gene family in Salvia miltiorrhiza. J Integr Plant Biol. 56, 38–50 (2014).
Zhang, S. D. & Ling, L. Z. Genome-wide identification and evolutionary analysis of the SBP-box gene family in castor bean. PLoS One. 9, e86688 (2014).
Li, C. & Lu, S. Molecular characterization of the SPL gene family in Populus trichocarpa. BMC Plant Biol. 14, 131 (2014).
Li, J. et al. Genome-wide identification and analysis of the SBP-box family genes in apple (Malus × domestica Borkh.). Plant Physiol Biochem. 70, 100–114 (2013).
Hou, H. et al. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family genes in grape. PLoS One. 8, e59358 (2013).
Salinas, M., Xing, S., Höhmann, S., Berndtgen, R. & Huijser, P. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta. 235, 1171–1184 (2012).
Yang, Z. et al. Comparative study of SBP-box gene family in Arabidopsis and rice. Gene. 407, 1–11 (2008).
Guo, A. Y. et al. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene. 418, 1–8 (2008).
Zhang, X. et al. Genomic organization, differential expression, and functional analysis of the SPL gene family in Gossypium hirsutum. Mol Genet Genomics. 290, 115–26 (2015).
Wendel, J. F. New World tetraploid cottons contain Old World cytoplasm. Proc Natl Acad Sci USA 86, 4132–4136 (1989).
Chen, Z. et al. Entire nucleotide sequences of Gossypium raimondii and G. arboreum mitochondrial genomes revealed A-genome species as cytoplasmic donor of the allotetraploid species. Plant Biol (Stuttg). 19, 484–493 (2017).
Zhang, T. et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 33, 531–537 (2015).
Liu, X. et al. Gossypium barbadense genome sequence provides insight into the evolution of extra-long staple fiber and specialized metabolites. Sci Rep. 5, 14139 (2015).
Yuan, D. et al. The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci Rep. 5, 17662 (2015).
Li, F. et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet. 46, 567–572 (2014).
Paterson, A. H. et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 492, 423–427 (2012).
Wang, K. et al. The draft genome of a diploid cotton Gossypium raimondii. Nat Genet. 44, 1098–1103 (2012).
Finn, R. D., Clements, J. & Eddy, S. R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–37 (2011).
Letunic, I., Doerks, T. & Bork, P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–260 (2015).
Marchler-Bauer, A. et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, D222–226 (2015).
Wang, L. et al. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 10, 282 (2010).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 33, 1870–1874 (2016).
Lee, T. H., Tang, H., Wang, X. & Paterson, A. H. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 41, D1152–1158 (2013).
Hu, B. et al. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 31, 1296–1297 (2015).
Bailey, T. L., Williams, N., Misleh, C. & Li, W. W. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–373 (2006).
Lescot, M. et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–527 (2002).
Wu, G. & Poethig, R. S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target. SPL3. Development. 133, 3539–3547 (2006).
Xie, K., Wu, C. & Xiong, L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 142, 280–293 (2006).
Jung, J. H., Ju, Y., Seo, P. J., Lee, J. H. & Park, C. M. The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 69, 577–588 (2012).
Jung, J. H., Lee, H. J., Ryu, J. Y. & Park, C. M. SPL3/4/5 Integrate developmental aging and photoperiodic signals into the FT-FD Module in Arabidopsis flowering. Mol Plant. 9, 1647–1659 (2016).
Yu, N., Niu, Q. W., Ng, K. H. & Chua, N. H. The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 83, 673–685 (2015).
Chao, L.M. et al. Arabidopsis transcription factors SPL1 and SPL12 confer plant thermotolerance at reproductive stage. Mol Plant. pii: S1674-2052(17)30099-0 (2017).
Garcia-Molina, A., Xing, S. & Huijser, P. A conserved KIN17 curved DNA-binding domain protein assembles with SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE7 to adapt Arabidopsis growth and development to limiting copper availability. Plant Physiol. 164, 828–840 (2014).
Garcia-Molina, A., Xing, S. & Huijser, P. Functional characterisation of Arabidopsis SPL7 conserved protein domains suggests novel regulatory mechanisms in the Cu deficiency response. BMC Plant Biol. 14, 231 (2014).
Unte, U. S. et al. SPL8, an SBP-box gene that affects pollen sac development in. Arabidopsis. Plant Cell. 15, 1009–1019 (2003).
Xing, S., Salinas, M., Höhmann, S., Berndtgen, R. & Huijser, P. miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell. 22, 3935–3950 (2010).
Xing, S. et al. SPL8 and miR156-targeted SPL genes redundantly regulate Arabidopsis gynoecium differential patterning. Plant J. 75, 566–577 (2013).
Jin, J. et al. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–1045 (2017).
Finn, R. D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–230 (2014).
Wilkins, M. R. et al. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 112, 531–552 (1999).
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009).
Author information
Authors and Affiliations
Contributions
C.C. and B.Z. designed the experiments. C.C. performed the study, analyzed the data and drafted the manuscript. C.C., W.G. and B.Z. reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cai, C., Guo, W. & Zhang, B. Genome-wide identification and characterization of SPL transcription factor family and their evolution and expression profiling analysis in cotton. Sci Rep 8, 762 (2018). https://doi.org/10.1038/s41598-017-18673-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-18673-4
This article is cited by
-
Identification of Alfalfa SPL gene family and expression analysis under biotic and abiotic stresses
Scientific Reports (2023)
-
Genome-wide identification and characterization of the SPL gene family and its expression in the various developmental stages and stress conditions in foxtail millet (Setaria italica)
BMC Genomics (2022)
-
Genome-wide identification and expression analysis of the SPL transcription factor family and its response to abiotic stress in Quinoa (Chenopodium quinoa)
BMC Genomics (2022)
-
The PyPIF5-PymiR156a-PySPL9-PyMYB114/MYB10 module regulates light-induced anthocyanin biosynthesis in red pear
Molecular Horticulture (2021)
-
Systematic Identification, Evolution and Expression Analysis of the SPL Gene Family in Sugarcane (Saccharum spontaneum)
Tropical Plant Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.