Abstract
Both influenza viral hemagglutinin and neuraminidase can induce protective immune responses in humans. Although the viral hemagglutinin antigens have been quantified in influenza vaccines, the amounts of neuraminidase remain undetermined. Using comprehensive bioinformatics analyses of all neuraminidase sequences, we identified highly conserved and subtype-specific peptide epitopes within each of N1, N2 and type B neuraminidase groups. Mono-specific antibodies generated against these peptides bound to their respective subtype/type only while demonstrating remarkable specificity against the viral neuraminidase sequences without any cross-reactivity with allantoic and cellular proteins. Moreover, the subtype/type-specific antibodies were found not to interfere with one another when a mixture of vaccine samples was analysed. Importantly, immunoassay based on these antibodies can quantitatively determine neuraminidase in commercial trivalent vaccine samples. Analyses of vaccines from eight manufacturers using the same vaccine seeds revealed significant differences in neuraminidase levels. Specifically, while the ratio between neuraminidase and hemagglutinin in some products are found to be close 1/5, other products have a ratio of approximately 1/100, a level which is far below the theoretical ratio between neuraminidase and hemagglutinin in a virus. The antibody-based assays reported here could be of great value for better quality control of both monovalent and trivalent vaccines.
Similar content being viewed by others
Introduction
Influenza remains a serious ongoing threat to public health. The WHO estimates between three to five million cases of severe illness and 250,000 to 500,000 deaths per year are caused by influenza virus. Vaccination is the most effective means to prevent influenza. The surface of influenza virus contains two glycoproteins, hemagglutinin (HA) and neuraminidase (NA), both of which are known to induce protective immune responses in human and animals.
HA is the most abundant glycoprotein, which is essential for receptor-binding and fusion of viral membranes with the host cells. Currently, the trivalent inactivated vaccine (TIV) is the most widely used form of vaccine. It is comprised of inactivated viruses derived from one H1N1, one H3N2 and one type B strain. Each of these HAs is updated annually by the WHO. The potency of TIV is determined by traditional single radial immunodiffusion (SRID) using strain specific antibodies generated every year. In contrast, no standardized assay is available for quantification of NA in the final products of trivalent vaccines.
NA is the second major glycoprotein on the viral surface. In type A virus, NA is classified into 9 subtypes whereas NA in type B virus (NB) has not been subtype-classified. It is well established that NA facilitates the release of newly generated virus from the infected cell membrane by cleaving sialic acid from host cell receptor. It also prevents aggregation of viral particles by cleaving the sialic acid residues present on viral glycoproteins. NA-specific immune responses have been shown to protect the host from influenza1. Specifically, in preclinical studies, animals immunized with NA antigen can be protected from influenza; NA-specific antibodies are by themselves sufficient to afford protection from lethal challenges1,2,3,4,5,6,7,8. Moreover, NA-specific immune responses in humans have also been correlated with protection of subjects from viral infection9,10,11,12,13. It is worth noting that in a recent study with controlled human influenza virus challenge, when compared with HI antibodies, the levels of pre-existing NI antibodies correlated significantly better with respect to reduced disease severity score, decreased symptoms and a shortened duration of virus shedding14.
While the role of NA-induced immune response is unquestionable, the levels of NA-specific antibodies are only modest in people immunized with the standard influenza vaccine containing 15 μg of the hemagglutinin of each strain; yet, an eight-fold increase in the levels of NA-specific antibody was detected when four times concentrated standard vaccines were used, suggesting an increased amount of the NA antigen in vaccine can ensure a better immune response to NA11. These data reinforce the notion that the NA antigens in the influenza vaccines could be increased and better controlled so as to elicit a balanced immune response15,16,17. However, the neuraminidase in the current influenza vaccines is not quantified but only verified for the presence of enzymatic activity. Furthermore, the activity of neuraminidase as a potential potency marker is deemed unreliable since the enzymatic activity levels vary in the vaccine preparations or could be very low or even missing in some of them17,18,19. The variable level of enzymatic activity is likely due to the instability of the protein to the chemical treatments and storage procedures used in vaccine preparations19,20,21, making the determination of enzymatic activity unacceptable as a means for accurate quantification of NA in vaccines22.
A few alternative approaches have been explored to quantify NA in vaccines. Specifically, the use of strain specific antibodies in ELISA and immunochromatography has been reported20,21. However, these antibodies are strain specific and need to be made for the NA of each viral strain, which in practice would incur significant challenges and resources due to the rapid mutation rate of NA. Although physicochemical methods can quantify the HA proteins without the use of antibody-based reagents23,24,25,26,27,28,29,30,31,32 there has been no report on the quantitative detection of the NA proteins in trivalent vaccines with the exception of isotope dilution mass spectrometry (IDMS), which was used to analyse the NA activity in an H1N1 vaccine33. Clearly, lack of appropriate reagents or methods could have significantly impeded NA quantification22.
In a previous study, we developed a universal antibody against the most conserved epitope (222–230) of all influenza NA34. As the epitope targeted by the universal antibody is nearly 100% conserved in all NAs of both type A and type B virus, immunoassays based on this universal antibody can be used to quantify monovalent vaccine. Understandably, this NA universal antibody is not suitable for trivalent vaccines because the antibody would bind to all NA proteins in the vaccine preparations. To address the challenge in quantifying NA in trivalent vaccines, we set out to develop type/subtype-specific universal antibodies targeting the most conserved sequences in N1, N2 and NB, respectively. We report here that quantitative immunoassays based on these antibodies could be successfully used to analyze NA antigen contents in both monovalent and trivalent vaccines.
Results
Selection of peptides and generation of antibodies against N1, N2 and NB
Following our analysis of the public database, a set of 15 amino acid consensus sequences were identified for each subtype in a procedure used to maximize conservation and surface accessibility as described previously35,36. Sequence QHPELTGLDCIRPCF (408–422), designated as CNA1, was selected for N1, RTLLMNELGVPFHLG (159–173), designated as CNA2, for N2, and STFQKALLISPHRFG (90–104), designated as CNAB, for NB (N2 numbering). These identified peptides for N1, N2 or NB are remarkably conserved within their respective groups, i.e., CNA1 was found to be 94% conserved within N1 subtype, CNA2 97% for N2 subtype, and CNAB 99.79% for all type B neuraminidase. Importantly, these peptides share no similarity to other groups except their own.
The selected peptides were synthesized, modified with a 6-aminocaproic-cysteine linker and conjugated to a KLH carrier protein before used to immunize rabbits to produce mono-specific antibodies35.
Preparation and Characterization of Antibodies
The mono-specific antibodies against CAN1, CAN2 and CANB were obtained after 4 rounds of immunization of rabbits. We first attempted to use an indirect ELISA by coating the plates with vaccine samples to determine Ab titers, yet no NA antibody was detected, which is likely due to that the amount of NA antigens in the vaccines being too low to be detected. To overcome this problem, GST fusion protein expressing the epitopes were constructed and used as the coating antigen in ELISA. The titer of all three antibodies against fusion proteins were above 1: 100,000 (Supplementary Fig. S1).
We next determined the specificity of the monospecific antibodies. GST-fusion proteins or peptides were used as the coating antigen in an indirect ELISA. As shown in Fig. 1, CAN1, CAN2 and CAN/B monospecific antibodies bound strongly to their respective epitopes in the GST fusion proteins; the binding between the antibodies and peptide epitopes in GST-fusion protein is generally better than that between the antibodies and the unconjugated free peptides (Fig. 1). This observation is likely due to the unconjugated peptides having their epitopes masked upon coating to the plates.
Binding of the antibody to the diverse strains/type of influenza viruses
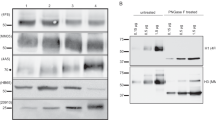
In order to determine whether the mono-specific antibodies exclusively bind to corresponding type/subtype of NA, WB assays were performed using all nine subtype of type NA and one type B NA. Allantoic fluids containing the viruses were used directly to determine the specificity of antibodies against the viral sequences. As shown in Fig. 2, all three antibodies can bind to their corresponding type/subtype of NA, with no significant cross-reactivity to any other type of NA or the allantoic fluid proteins.
Binding of CAN1/2/B Antibodies to various type/subtypes NA proteins. Allantoic fluids containing viruses were fractionated in SDS-PAGE. Four gels were run and transferred to 4 membranes in parallel, followed by detection of the NA proteins using the CAN1 antibodies (panel A), CAN2 antibodies (panel B), CANB antibodies (panel C). Rabbit polyclonal anti-NP (nucleoprotein) proteins of influenza A viruses were used as control (panel D). Note: Anti-NP antibody against type A cannot bind to the NP of type B viruses. Original pictures see Supplementary Figure S3.
We next analyzed binding of the antibodies to influenza reference antigens used for the potency determination of commercial influenza vaccines. In agreement with results of antibody binding to the viruses presented in Fig. 2, all three antibodies demonstrated type/subtype-specificity when four reference antigens were tested (Fig. 3). Taken together, these results indicate that these antibodies are highly specific for the viral NA sequences within their respective type/subtype.
Binding of subtype-specific antibodies to their respective reference antigens derived from multiple viral strains. The reference antigens used are described in Table S1. Lane 1–4: 4 strains of H1N1; Lane 5–8: 4 strains of H3N2; Lane 9–12: 4 strains of type B vaccines. The samples were fractionated on SDS-PAGE (1 μg HA/lane). Four gels were run and transferred to 4 membranes in parallel, followed by detection of the NA proteins in W.B. using the CAN1 (panel A) and CAN2 antibodies (panel B) and CANB antibodies (panel C). Rabbit polyclonal anti-NP (nucleoprotein) proteins of influenza A viruses were used as control (panel D). Note: Anti-NP antibody against type A cannot bind to the NP of type B viruses.
Antibody-based assay for NA quantification of products from eight manufacturers
Having succeeded in determining the type/subtype specificity of the mono-specific antibodies, we next investigated whether these antibodies could be used to quantitatively analyze the contents of NA in vaccine preparations. We employed a slot blot for NA quantification as this method has been shown to be simple assay capable of quantifying HA in a high throughput manner37.
We first analyzed the specificity of the NA subtype-specific antibody-based slot blot. To this end, vaccine reference antigens of H1N1 (A/California/7/2009), H3N2 (A/Hong Kong/4801/2014) and B (B/Brisbane/60/2008) were loaded on the membrane based on the HA concentration of 2 μg/ml. CAN1, CAN2 and CANB antibodies were added to react with the blotted proteins; we found that CAN1 could only bind to H1N1, CAN2 to H3N2 and CANB to type B vaccine, a result in agreement with that obtained by WB as described in Figs 2 and 3 (supplement information Fig. S2).
Because current vaccines are mainly trivalent formulations, we next determined potential interference in these final products. Monovalent and trivalent preparations (2 μg/ml HA from each strain) were compared in the slot blot. As shown in Fig. 4, the NA antigens in monovalent and trivalent detected with their respective NA antibodies were comparable, indicating that presence of three different NA antigens did not interfere with the ability of antibodies in binding to their respective NA.
Specificity and interference of NA slot-blot. Samples were diluted to final concentration of HA 2 μg/ml (trivalent samples contain 2 μg/ml HA from each strain). Each sample was tested in six replicates. The NA contents of trivalent samples were normalized and reported as percentage of monovalent samples.
We next developed the standard curve using the reference antigens for quantitative analyses of vaccine samples. As presented in Fig. 5A, the concentration of the NA antigen references from H1N1 was proportional to the signal intensity developed in slot blot, with curve fitting very well with the 4-PL model (R2 = 0.99) as shown in Fig. 5B. Therefore, the standard curve was used for subsequent analyses of vaccine samples.
Development of standard curve for NA quantification using slot blot. Panel A: quantitative detection of NA using CAN1 antibodies against N1: the N1 antigens at various concentrations were loaded onto NC membrane. The membrane was then incubated with the CAN1 antibodies, followed by detection with anti-rabbit IgG peroxidase conjugate. Similar results were obtained with CAN2 and CANB antibody binding to their corresponding NA antigens (not shown in the figure). Panel B: standard curve for the quantification of N1: the currently accepted 4-parameter logistic (4-PL) model was employed for the calibration curve fitting in the immunoassays. Original pictures see Supplementary Figure S4.
To assess the reproducibility, three lots vaccine preparations were analyzed in three separate experiments and results for the measured potencies were compared. As presented in Table 1, little variation was observed between runs, as demonstrated by the low standard deviation values and the coefficient of variance values between 2.4% and 14.3%.
Having developed the standard curve for N1, N2 and NB quantification, we next analyzed NA antigens in eight commercial influenza vaccines for the 2016–2017 flu season. We found that the NA levels vary dramatically among N1, N2 and NB, with N1 being the highest and NB the lowest no matter which manufacturers’ products were analyzed (Table 2). Although the NA levels could be relatively close in several lots of products from a given manufacturer, the amount of NA varies drastically between the eight different manufacturers. For example, NA amounts of H1N1 in manufacturer A were consistently higher than other seven producers’ products whereas manufactures E, F and H have lower NA amounts, with F and H generally being the lowest. This is true regardless N1, N2 or NB was compared. Interestingly, we were able to obtain six vaccine lots from manufacturer A for NA quantification; we found NA contents, irrespective of the vaccine strains (H1N1, H3N2 and Type B), were fairly consistent and generally higher than the vaccines from other manufacturers, particularly for N1 of H1N1 which is more than 5 μg/ml or roughly one-fifth of the HA amounts. This ratio of 1/5 between NA and HA approaches the expected ratio of 1:4 in some native viral strains38. As only two or three lots were analyzed from other producers, it is hard for us to draw any conclusion even though it appears that the variations in NA levels were less pronounced than that between the manufacturers. However, compared to producers A or D, F and H have nearly 10–20 times less NA in their products or NA/HA ratio was only 1/100–2/100, far below the theoretical ratio of NA/HA (25/100) in an influenza virus38. Taken together, these data indicate that manufacturing process could dramatically affect the levels of NA in the current influenza vaccines.
Discussion
HA and NA are two major surface proteins on influenza virus particles, both of which are well known to elicit a protective immune response. However, only HA is quantitatively determined and serve as the potency marker in the current inactivated influenza vaccines, whereas NA is not quantified but only verified for the presence of neuraminidase activity in vaccine bulks. The measurement NA activity is unreliable for quantitative determination of NA as its activity could be dramatically affected in the manufacturing process such as chemical treatment, e.g. formalin inactivation and ether treatment for virus splitting. Clearly, a simple method capable of quantifying the NA proteins would be of great use for vaccine quality control.
In our previous report, two highly conserved regions in virtually all influenza viruses NA were identified through extensive bioinformatics analyses, one of which was found to be a nearly 100% conserved among all type A and B influenza strains. Universal antibodies against this epitope could bind all nine subtypes of NA in type A as well as all NA in type B34. Although the universal anti-NA antibodies could be used to quantify monovalent vaccines or used for in-process control, they cannot be used to determine NA antigens in the current trivalent or quadrivalent vaccines. Thereby, it would be essential to develop an immunoassay capable of quantitatively differentiating the NA type and subtypes in the current seasonal vaccines.
Several lines of evidences prompted us to develop this NA quantification assay. First, like HA, NA is also known to be evolving in an unpredictable fashion, making it necessary to develop antibodies which require no need for annual updating. Second, the antibodies must be able to distinguish NAs in the trivalent vaccines. Third, the NA should be determined by a simple assay in a high throughput manner. Fourth, there has been no report on the analyses of NA variations in trivalent vaccines from various manufacturers.
To achieve these goals, we first conducted comprehensive bioinformatics analyses of all NA strains in the public database. Interestingly, all consensus sequences within NA subtypes found by Shannon entropy analysis are located in the head region of NA, which is formed by six identical antiparallel β-sheets (motifs) organized in the form of a propeller-like structure. Although the conserved sequences of both N1 and N2 subtype were successfully identified on motif 1, 2 and 6, respectively, only those sequences from motif 6 of N1 and sequences from motif 1 of N2 were found to be dissimilar with other subtypes, an observation further verified by multiple alignment and Shannon entropy analysis. Thus, the epitope with the greatest differential abundance (i.e. conserved in each of N1, N2 and NB, but absent in other NA subtypes) was finally selected. Specifically, amino acid 408–422 was chosen for N1, amino acid 159–173 for N2 and amino acid 90–104 for NB. Indeed, mono-specific antibodies targeting these peptides were confirmed in subsequent experiments to bind to their corresponding NA type/subtype only and have no cross reactivity to other subtype NA or allantoic proteins.
The potential of the NA antibody-based assay for NA quantification was explored using eight manufacturers’ products. While the variation of NA in different virus strains is not surprising, drastic difference of NA in products from eight manufacturers is noteworthy as they use the same seed strains provided by the WHO for vaccine production. Some products consistently have NA/HA ratio close to that in a native virus (1:4)38, whereas others contain 10–20 fold less of NA in the final products. It remains unclear as to how the NA proteins were lost during the manufacturing process, given each producer may have different manufacturing process. Hopefully, with simple detection method like this, it could be easier to conduct in-process control. Nevertheless, these findings reveal that manufacturing process can significantly influence NA amounts, and it might be worth considering incorporation of this type of assay for better quality control. Unlike the HA antibodies which need to be annually updated, the NA antibodies reported here may be of practical value for vaccine developers, industry and regulatory agencies.
Finally, it is worthy to mention that although there are mounting evidences supporting the role of NA in inducing protective immune responses, HA remains to be the critical antigens. Indeed, recombinant HA vaccine products have been licensed for human vaccination39. While the amount of HA per dose in the recombinant vaccine (45 µg) is three time more than that in the egg-based inactivated vaccine (15 µg), the difference in the HA amounts between rHA and current inactivated vaccine remains to be fully understood in terms of induction of immune responses. Moreover, other forms of vaccines are also being explored including virus-like particle-based vaccines; the VLPs under development are reported to vary in the compositions of the viral proteins40,41. Future work should be conducted to determine whether incorporation of NA in these new generations of products may improve the immunogenicity of these products. The availability of tools for NA quantification could facilitate the development of newer generations of vaccines. Interested investigators may contact the authors to request the antibody reagents for their research work.
Materials and Methods
Viruses and vaccines
Table S1 lists the viruses, human vaccines and vaccine reference antigens used in the characterization of the antibodies. All the virus strains used in this study were the same as reported previously34. The viruses were propagated in embryonated chicken eggs. The flu vaccine reference antigens were kindly provided by the National Institute for Biological Standards and Control (NIBSC), Potters Barr, UK and Therapeutic Goods Administration (TGA), Symonston ACT 2609, Australia.
The seasonal influenza vaccines used in this study are derived from 3 virus strains recommended by WHO for the 2016–2017 Northern Hemisphere, i.e., A/California/7/2009(H1N1), A/Hong Kong/4801/2014(H3N2) and B/Brisbane/60/2008 (B).
Identify the conserved but unique epitopes in NA type/subtype
To identify universally conserved regions specific to the N1, N2 or NB, a bioinformatics approach was employed as previously described35. In brief, all NA sequences from NCBI influenza virus resource (http://www.ncbi.nih.gov/ genomes/FLU/FLU.htm) were downloaded separately for each NA subtype and NB, with redundant or identical sequences deleted. A multiple alignment was then performed for each subtype followed by extraction of the consensus sequences from full-gene genome, in which the conservation index were sorted by the degree of variation and determined by calculating the Shannon entropy for each position of amino acid35. Geneious 7.0.6 software was then used to confirm the bioinformatics findings and ensure the uniqueness in one subtype and universality of the selected peptide within its own subtype36.
Preparation of peptides and their conjugates for immunization
The current trivalent inactivated influenza vaccines are derived from one strain of A/H1N1, A/H3N2 and B type of influenza virus, respectively. Based on bioinformatics analyses, one peptide was chosen for each of the three groups N1, N2 and NB (more details in Results). These peptides were synthesized, modified with a 6-aminocaproic-cysteine linker and conjugated to a KLH carrier protein; the peptide-conjugates were then used to immunize rabbits for the preparation of type/subtype-specific universal antibodies as previously described35.
Expression of fusion proteins containing epitopes
The GST fusion proteins containing each of CAN1, CAN2 and CANB epitopes were prepared for characterization of the antibodies. In short, forward and reverse oligonucleotides (Table S2) were synthesized according to the sequence of the peptide. After annealing, the double stranded polynucleotides were inserted into the pET19b-GST vector using the BamH1/Xho1 restriction sites. The pET19b-GST-CAN1/2/B recombinant plasmids were transformed into competent Escherichia coli BL21 (DE3) to express proteins. The bacterially expressed proteins were purified by His Gel (GE Healthcare, Uppsala, Sweden) according to the manufacturer’s instructions.
ELISA
96-well plates (Costar, Kennebunk, ME) were coated at 4 °C overnight with 100 μl/well of CAN1, CAN2 or CANB peptides or their respective fusion proteins at 1 μg/ml in 0.01 M bicarbonate buffer (pH 9.6). The plates were washed 5 times by PBS-T, followed by blocking with PBS containing 5% skim milk for 1 h at room temperature. The type/subtype specific universal antibodies was then added, followed by incubation for 1 h at room temperature before HRP conjugated goat anti rabbit IgG antibody was added. After incubation for 1 h at room temperature, the plates were washed 5 times before Tetramethylbenzidine (TMB) substrate was added for colorimetric development. Optical density at 450 nm was measured by a Multiskan Ascent microplate reader (Thermo Scientific, Shanghai, China). Unless specified, pre-immunization serum was used as negative control.
Western blot
NA antigens from all nine subtypes of type A influenza virus and Type B influenza virus were used to determine the type/subtype specificity of the NA antibodies by Western blot (WB). In brief, all allantoic fluids directly from eggs inoculated with influenza viruses were fractionated on sodium dodecyl sulfate (SDS)-12% polyacrylamide gels (PAGE), followed by transferring the samples to PVDF membrane (Millipore, Beijing, China). The membrane was then blocked with 5% skim milk in PBS at 37 °C for 1 h, followed by incubation for 1 h at 37 °C with NA-antisera as described above. Peroxidase-conjugated goat anti-rabbit immunoglobulin(IgG) (Sigma, St. Louis, MO) was added for an additional incubation of 1 h at room temperature, followed by chemiluminescent detection using ECL prime (GE health, Buckinghamshire, UK).
NA antigen reference preparation
The NA reference materials were prepared by SDS-PAGE as reported previously42. In brief, a monovalent bulk material was treated with glycosidase PNGase F (New England Biolabs, Ipswich, MA) at 37 °C for 16 h in the buffer provided by the manufacturer in the presence of 1% NP 40; the samples were then subjected to SDS-PAGE separation. The position of NA band in SDS-PAGE was confirmed by mass spectrometry. Protein quantification were conducted by densitometric analyses of the protein bands using a high resolution scanner and Launch Doc-ITLS software (for UVP Bio Doc-It Imaging System, Upland. CA, USA). The amounts of NA were determined based on the ratio of NA protein to the total proteins42.
Slot blot
Slot blot was conducted as described previously36. Briefly, the vaccines and the NA references were treated with 4 M urea (final concentration) to dissolve the aggregates which could form during the storage. The NA reference antigens in serial dilutions ranging from 15 to 2000 ng/ml were used to make a standard curve. Vaccine samples were tested in duplicate. After the reference materials and samples were loaded on membrane, type/subtype-specific antibodies were used as primary Ab incubation, followed by a 45 min incubation at room temperature with a 1/15,000 dilution of the ECL HRP-conjugated sheep anti-rabbit IgG in5% milk/TBS-Tween 0.1% buffer. After the addition of the SuperSignal West Dura Extended Duration substrate for 5 min, the blots were exposed for 10 to 60 s in a Bio-Rad ChemiDocTMMPImaging System. The ImageLab 4.0 software was used to measure the density of the bands.
Data analysis
Unless specified, results are presented as mean ± SD with n = 3 and the currently accepted 4-parameter logistic (4-PL) model was employed for the calibration curve fitting in the immunoassays by SoftMax Pro v5 (Molecular Device, Sunnyvale, CA).
Data availability statement
Any materials, data and associated protocols in this publication would be promptly available to readers without undue qualifications in material transfer agreements. The antibody reagents reported in this paper will be available to scientific community for research use only.
References
Sylte, M. J. & Suarez, D. L. Influenza neuraminidase as a vaccine antigen. Curr Top Microbiol Immunol 333, 227–241 (2009).
DiNapoli, J. M. et al. Newcastle disease virus-vectored vaccines expressing the hemagglutinin or neuraminidase protein of H5N1 highly pathogenic avian influenza virus protect against virus challenge in monkeys. J Virol 84, 1489–1503 (2010).
Johansson, B. E., Grajower, B. & Kilbourne, E. D. Infection-permissive immunization with influenza virus neuraminidase prevents weight loss in infected mice. Vaccine 11, 1037–1039 (1993).
Rott, R., Becht, H. & Orlich, M. The significance of influenza virus neuraminidase in immunity. J Gen Virol 22, 35–41 (1974).
Sandbulte, M. R. et al. Cross-reactive neuraminidase antibodies afford partial protection against H5N1 in mice and are present in unexposed humans. PLoS Med 4, e59 (2007).
Schulman, J. L., Khakpour, M. & Kilbourne, E. D. Protective effects of specific immunity to viral neuraminidase on influenza virus infection of mice. J Virol 2, 778–786 (1968).
Takahashi, Y. et al. Protective immunity afforded by inactivated H5N1 (NIBRG-14) vaccine requires antibodies against both hemagglutinin and neuraminidase in mice. J Infect Dis 199, 1629–1637 (2009).
Webster, R. G., Laver, W. G. & Kilbourne, E. D. Reactions of antibodies with surface antigens of influenza virus. J Gen Virol 3, 315–326 (1968).
Murphy, B. R., Kasel, J. A. & Chanock, R. M. Association of serum anti-neuraminidase antibody with resistance to influenza in man. N Engl J Med 286, 1329–1332 (1972).
Beutner, K. R. et al. Evaluation of a neuraminidase-specific influenza A virus vaccine in children: antibody responses and effects on two successive outbreaks of natural infection. J Infect Dis 140, 844–850 (1979).
Cate, T. R. et al. A high dosage influenza vaccine induced significantly more neuraminidase antibody than standard vaccine among elderly subjects. Vaccine 28, 2076–2079 (2010).
Couch, R. B., Kasel, J. A., Gerin, J. L., Schulman, J. L. & Kilbourne, E. D. Induction of partial immunity to influenza by a neuraminidase-specific vaccine. J Infect Dis 129, 411–420 (1974).
Kilbourne, E. D. et al. Purified influenza A virus N2 neuraminidase vaccine is immunogenic and non-toxic in humans. Vaccine 13, 1799–1803 (1995).
Memoli, M. J. et al. Evaluation of Antihemagglutinin and Antineuraminidase Antibodies as Correlates of Protection in an Influenza A/H1N1 Virus Healthy Human Challenge Model. Mbio 7, e00417–00416 (2016).
Brett, I. C. & Johansson, B. E. Immunization against influenza Avirus: comparison of conventional inactivated, live-attenuated and recombinant baculovirus produced purified hemagglutinin and neuraminidase vaccines in a murine model system. Virology 339, 273–280 (2005).
Johansson, B. E., Pokorny, B. A. & Tiso, V. A. Supplementation of conventional trivalent influenza vaccine with purified viral N1 and N2 neuraminidases induces a balanced immune response without antigenic competition. Vaccine 20, 1670–1674 (2002).
Gillim-Ross, L. & Subbarao, K. Can immunity induced by the human influenza virus N1 neuraminidase provide some protection from avian influenza H5N1 viruses? PLoS Med 4, e91 (2007).
Wood, J. M., Schild, G. C. & Newman, R. W. A sensitive, single-radial diffusion autoradiographic zone size enhancement technique for the assay of influenza haemagglutinin. J Gen Virol 47, 355–363 (1980).
Gulati, U. et al. Mismatched hemagglutinin and neuraminidase specificities in recent human H3N2 influenza viruses. Virology 339, 12–20 (2005).
Gerentes, L., Kessler, N. & Aymard, M. A sensitive and specific ELISA immunocapture assay for rapid quantitation of influenza A/H3N2 neuraminidase protein. J Virol Methods 73, 185–195 (1998).
Tanimoto, T. et al. Estimation of the neuraminidase content of influenza viruses and split-product vaccines by immunochromatography. Vaccine 23, 4598–4609 (2005).
Bright, R. A., Neuzil, K. M., Pervikov, Y. & Palkonyay, L. WHO meeting on the role of neuraminidase in inducing protective immunity against influenza infection, Vilamoura, Portugal, September 14, 2008. Vaccine 27, 6366–6369 (2009).
Bucher, D. J., Li, S. S., Kehoe, J. M. & Kilbourne, E. D. Chromatographic isolation of the hemagglutinin polypeptides from influenza virus vaccine and determination of their amino-terminal sequences. Proc Natl Acad Sci USA 73, 238–242 (1976).
Calam, D. H. & Davidson, J. Isolation of influenza viral proteins by size-exclusion and ion-exchange high-performance liquid chromatography: the influence of conditions on separation. J Chromatogr 296, 285–292 (1984).
Phelan, M. A. & Cohen, K. A. Gradient optimization principles in reversed-phase high-performance liquid chromatography and the separation of influenza virus components. J Chromatogr 266, 55–66 (1983).
Kapteyn, J. C. et al. Haemagglutinin quantification and identification of influenza A&B strains propagated in PER.C6 cells: a novel RP-HPLC method. Vaccine 24, 3137–3144 (2006).
Garcia-Canas, V., Lorbetskie, B. & Girard, M. Rapid and selective characterization of influenza virus constituents in monovalent and multivalent preparations using non-porous reversed-phase high performance liquid chromatography columns. J Chromatogr A 1123, 225–232 (2006).
Kapteyn, J. C. et al. HPLC-based quantification of haemagglutinin in the production of egg- and MDCK cell-derived influenza virus seasonal and pandemic vaccines. Vaccine 27, 1468–1477 (2009).
Garcia-Canas, V., Lorbetskie, B., Bertrand, D., Cyr, T. D. & Girard, M. Selective and quantitative detection of influenza virus proteins in commercial vaccines using two-dimensional high-performance liquid chromatography and fluorescence detection. Anal Chem 79, 3164–3172 (2007).
Harvey, R., Wheeler, J. X., Wallis, C. L., Robertson, J. S. & Engelhardt, O. G. Quantitation of haemagglutinin in H5N1 influenza viruses reveals low haemagglutinin content of vaccine virus NIBRG-14 (H5N1). Vaccine 26, 6550–6554 (2008).
Luna, L. G., Williams, T. L., Pirkle, J. L. & Barr, J. R. Ultra performance liquid chromatography isotope dilution tandem mass spectrometry for the absolute quantification of proteins and peptides. Anal Chem 80, 2688–2693 (2008).
Garcia-Canas, V. et al. Approach to the profiling and characterization of influenza vaccine constituents by the combined use of size-exclusion chromatography, gel electrophoresis and mass spectrometry. Biologicals 38, 294–302 (2010).
Solano, M. I. et al. Quantification of Influenza Neuraminidase Activity by Ultra-High Performance Liquid Chromatography and Isotope Dilution Mass Spectrometry. Anal Chem 89, 3130–3137 (2017).
Gravel, C. et al. Qualitative and quantitative analyses of virtually all subtypes of influenza A and B viral neuraminidases using antibodies targeting the universally conserved sequences. Vaccine 28, 5774–5784 (2010).
Chun, S. et al. Universal antibodies and their applications to the quantitative determination of virtually all subtypes of the influenza A viral hemagglutinins. Vaccine 26, 6068–6076 (2008).
Gravel, C. et al. Development and applications of universal H7 subtype-specific antibodies for the analysis of influenza H7N9 vaccines. Vaccine 33, 1129–1134 (2015).
Li, C. et al. A simple slot blot for the detection of virtually all subtypes of the influenza A viral hemagglutinins using universal antibodies targeting the fusion peptide. Nat Protoc 5, 14–19 (2010).
Air, G. M. & Laver, W. G. The neuraminidase of influenza virus. Proteins 6, 341–356 (1989).
Quan, F. S., Lee, Y. T., Kim, K. H., Kim, M. C. & Kang, S. M. Progress in Developing Virus-like Particle Influenza Vaccines. Expert Review of Vaccines 15, 1–13 (2016).
Pushko, P. et al. Influenza virus-like particles comprised of the HA, NA, and M1 proteins of H9N2 influenza virus induce protective immune responses in BALB/c mice. Vaccine 23, 5751–5759 (2005).
Thompson, C. M., Petiot, E., Lennaertz, A., Henry, O. & Kamen, A. A. Analytical technologies for influenza virus-like particle candidate vaccines: challenges and emerging approaches. Virology Journal 10, 141–141 (2013).
Li, C. et al. Application of deglycosylation and electrophoresis to the quantification of influenza viral hemagglutinins facilitating the production of 2009 pandemic influenza (H1N1) vaccines at multiple manufacturing sites in China. Biologicals 38, 284–289 (2010).
Acknowledgements
This work was supported Ministry of Science and Technology, PR China (grant nos 2015ZX09101044 to J.W.; 2013DFA31680 to C.L.) and by the Canadian Regulatory Strategy for Biotechnology:(GVD, X.L.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
K.X., Z.J., C.G., and B.J. performed the experiments. K.X., C.L. and G.V.D. performed data analysis. C.L., G.V.D., X.L. and J.W. coordinated the study. K.X., C.L., G.V.D., X.L. and J.W. designed the project. C.L., G.V.D., X.L. and J.W. prepared the paper. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, K., Li, C., Gravel, C. et al. Universal type/subtype-specific antibodies for quantitative analyses of neuraminidase in trivalent influenza vaccines. Sci Rep 8, 1067 (2018). https://doi.org/10.1038/s41598-017-18663-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-18663-6
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.