Abstract
Tonoplast H+-pyrophosphatases (VPs) mediate vacuolar Na+ sequestration, a process important for salt tolerance of plants. The function of VP in the highly drought- and salt-tolerant perennial Iris lactea under salt stress is unclear. Here, we isolated IlVP from I. lactea and investigated its function in transgenic tobacco. IlVP was found to comprise 771 amino acid residues and showed 88% similarity with Arabidopsis AtVP1. IlVP was mainly expressed in shoots and was up-regulated by salt stress. Overexpression of IlVP enhanced growth of transgenic tobacco plants compared with wild-type (WT) plants exposed to salt stress. Transgenic plants accumulated higher quantities of Na+ and K+ in leaves, stems, and roots under salt stress, which caused higher leaf relative water content and decreased cell membrane damage compared with WT plants. Overall, IlVP encoding a tonoplast H+-pyrophosphatase can reduce Na+ toxicity in plant cells through increased sequestration of ions into vacuoles by enhanced H+-pyrophosphatase activity.
Similar content being viewed by others
Introduction
Plant physiological drought leads to ionic imbalance in cells, depressed functioning of cell membranes and metabolic activity, and even cell death owing to excessive soil Na+ concentrations1. To cope with salinity stress, strategies adopted by plants cells to Na+ compartmentalisation into vacuoles alleviated the cellular Na+ toxicity to maintain osmotic balance using Na+ as a osmoregulation substance, thus to improve salt tolerance of plant2. Previous studies suggested that tonoplast Na+/H+ antiporters (NHXs) could mediate Na+ compartmentation into vacuolar3. The process is driven by electrochemical gradient of protons across tonoplast generated by the H+- ATPase and H+-pyrophosphatase (H+-PPase) in tonoplast4,5. It has been suggested that H+-PPase plays an important role in salt tolerance via the establishment of a transmembrane electrochemical gradient6,7. First cloned from Arabidopsis thaliana, H+-PPase genes have subsequently been cloned from other plants, such as Hordeum vulgare 8, Beta valgaris 9, Pyrus serotina 10, Triticum aestivum 11, Thellungiella halophila 12, and Haloxylon ammodendron 13. H+-PPase activity and transcript levels can vary among different plant species, organ types, growth stages, and Na+ concentrations in nutrient solutions14. In Daucus carota 15, Helianthus annuus 16, Suaeda salsa 17, and Thellungiella halophila 18 subjected to different NaCl concentrations, tonoplast H+-PPase activity was higher in treated plants than in the control, further demonstrating that NaCl may induce an increase in H+-PPase activity. In contrast, however, Matsumoto and Chung19 reported that H+-PPase activity in Hordeum vulgare roots treated with 200 mM NaCl was half that of the control, and similar results were obtained in a study of Mesembryanthemum crystallinum treated with 400 mM NaCl20. These suggest that overexpressing the H+-PPase resulted in enhanced resistance to salt in various transgenic plants linked with the increased Na+ compartmentation into the vacuoles.
Iris lactea Pall. var. chinensis (Fisch.) Koidz., a wild perennial monocotyledonous halophyte, is widely distributed in desert steppe and saline lowland meadows in northern China, Siberian regions, eastern Russia, and Mongolia21. Moreover, this species has attractive leaves and flowers, a wide range abundant seeds, stronger salt and drought tolerance, higher pest and disease resistance, and easy cultivation, which has become a popular groundcover plant for landscape design and park greenspace construction in northern China because of its ornamental foliage and flower22,23. Our previous research showed that the salt sensitive BJCY-ML035 in meadow grassland (37°31′12′′ N, 112°19′00′′ E; altitude 760 m) and the salt tolerant BJCY-ML007 in saline lowland meadow (43°45′15′′ N, 83°10′30′′ E; altitude 1,071 m) were screeed out from the sixteen accessions of I. lactea in northern China by the comprehensive assessment of salinity soils24. Further research suggested that the specific locus ISSR841-220 associated with the VP gene was found in the BJCY-ML007 compared with BJCY-ML03525. However, the role of IlVP in the salt tolerance of I. lactea is still unclear.
To test whether the overexpression of IlVP confers improved salt tolerance in plant, we introduced the gene into tobacco to measure and analyse the growth performance and Na+, K+ concentrations in the transgenic tobacco plants and in wild-type (WT) plants subjected to salinity stress. The results indicate that IlVP-mediated compartmentalisation of Na+ into vacuoles may play a key role in salt tolerance of plant. This would provide a potential benefits for generating engineered plants to increased tolerance to salinity conditions.
Results
Isolation and characterisation of IlVP
An 893-bp fragment was first obtained by RT-PCR using degenerate primers P1 and P2 (Supplementary Figure 1). A nucleotide BLAST search revealed that the isolated cDNA fragment shared high sequence homology with many known VPs from other plants (e.g. Oryza sativa), indicating that a partial potential VP had been isolated from I. lactea. Sequences of the 5′ and 3′ ends were obtained by rapid amplification of cDNA ends (RACE), which yielded products of 1,117 bp and 881 bp, respectively. The open reading frame (ORF) of IlVP was 2,316 bp long and encoded a polypeptide protein consisting of 771 amino acid residues (Supplementary Figure 1). The predicted protein had an isoelectric point of 5.16 and a molecular weight of 80.7 kDa. The cDNA sequence of IlVP was submitted to GenBank under accession number KY406740.
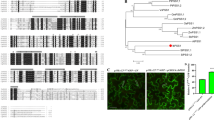
Analysis using the TMpred tool indicated that IlVP contained 14 transmembrane regions (Fig. 1a). Both of N- and C-terminus were located in vacuole. Multiple sequence alignment showed that the PPi binding site sequences were GGG, DVGADLVGK, and DNVGDNVGD, all located in the loop sequence connecting domains TM5 and TM6 in the cytoplasm. The core VP sequence, essential for implementation of proton transfer functions, was highly conserved and corresponded to that of PdVP, OsVP1, and AtVP. Alignment of H+-PPase amino acid sequences of I. lactea and other plant species showed that IlVP was 96%, 93%, and 88% similar to PdVP, OsVP, and AtVP, respectively (Fig. 1b). Phylogenetic analysis indicated that IlVP was most closely related to PdVP and MaVP, and only distantly related to GmVP (Fig. 1b). Consequently, IlVP may have the same function as other plant vacuolar membrane H+-PPases such as AtVP and may play an important role in drought resistance and salt tolerance.
(a) Alignment of amino acid sequences of H+-PPase genes from Iris lactea var. chinensis (IlVP) with those from Phoenix dactylifera (PdVP), Oryza sativa (OsVP), and Arabidopsis thaliana (AtVP). Amino acid sequences enclosed in red frames represent the PPi binding sites and activity domains of H+-PPase. (b) Phylogenetic tree of H+-PPase genes from Iris lactea and other plant species. Genes and GenBank accession numbers are as follows: AtVP (Arabidopsis thaliana, NM_101437), BdVP (Brachypodium distachyon, XM_003564169), CrVP (Chenopodium rubrum, AF533336), CsVP (Citrus sinensis, XM_006474322), EgVP (Eucalyptus grandis, XM_010035677), GmVP (Glycine max, XM_003528254), HbVP (Hevea brasiliensis, AY514019), HvVP (Hordeum vulgare, AK360389), IlVP (Iris lactea, KY406740), MaVP (Musa acuminata, XM_009386846), NtVP (Nicotiana tomentosiformis, XM_009630002), NnVP (Nelumbo nucifera, XM_010246610), OsVP (Oryza sativa, D45383), PdVP (Phoenix dactylifera, XM_008790581), PpVP (Prunus persica, AF367446), PtVP (Populus trichocarpa, XM_006381029), RcVP (Ricinus communis, XM_002530709), SbVP (Sorghum bicolor, HM143921), SiVP (Setaria italic, XM_004964638), SlVP (Solanum lycopersicum, NM_001278976), TcVP (Theobroma cacao, XM_007023235), VvVP (Vitis vinifera, XM_002273171), and ZmVP (Zea mays, BT086232).
Expression pattern analysis of IlVP
To investigate the tissue-specific expression of IlVP, plants were subjected to 200 mM NaCl for 24 h. In the absence of NaCl, IlVP was constitutively expressed in roots and shoots (Fig. 2a). In the presence of 200 mM NaCl, IlVP transcripts were detected in both organs, with level 7.6 times higher in shoots than in roots (Fig. 2a). Subsequently, I. lactea plants were treated with 0, 25, 50, 100, or 200 mM NaCl for 0, 6, 12, 24, and 48 h. IlVP expression levels in shoots increased significantly as salt concentration and stress duration were increased (Fig. 2b). These findings suggest that IlVP expression is induced by salt. Compartmentalisation of cytoplasmic Na+ into the vacuole would help reduce salt-induced cell damage.
Expression analysis of IlVP in shoots and roots of Iris lactea var. chinensis under different NaCl treatments. The expression levels of IlVP in shoot and root under control (0 mM) and 200 mM were normalized with that in shoot in control. (a) IlVP expression in roots and shoots under control and 200 mM NaCl treatment for 24 h as indicated by quantitative real-time PCR (qRT-PCR); (b) IlVP expression in shoots after treatment with different concentrations of NaCl (0, 25, 50, 100, and 200 mM) for 0, 6, 12, 24, and 48 h as indicated by qRT-PCR. Each bar represents the mean (n = 3), and bars indicate the standard deviation (SD).
Production and molecular characterisation of tobacco plants over-expressing IlVP
To investigate the potential benefit of transferring IlVP into other plant species, we identified eight independent IlVP transgenic tobacco lines by PCR amplification (data not shown). To further examine IlVP transcript levels in transgenic tobacco, we used Northern hybridisation to analyse IlVP expression in young leaves of all PCR-positive lines and WT plants. We observed that the relatively lower expression levels in L4 and the highest expression levels in L18, but had not detected in the WT (Supplementary Figure 2a). We therefore used lines 4 and 18 in the following assay.
Subsequently, we randomly selected four transgenic tobacco lines and determined whether IlVP was introduced into the tobacco genome using Southern hybridisation analysis. As shown in Supplementary Figure 2b, only one band was visible in transgenic tobacco lines 4, 6, 14, and 18 after the hybridisation, whereas line 10 yielded two copies (Supplementary Figure 2b). The Southern blot analysis thus confirmed the integration and expression of IlVP in tobacco.
In addition, membrane proteins of isolation increased markedly the IlVP protein in T4 and T18 by Western blot analysis compared to WT, and the protein level in T18 was higher than in T4 (Supplementary Figure 2c), expecting that heterologous expression of IlVP could enhance salt tolerance in transgenic tobacco.
Effect of NaCl stress on transgenic tobacco growth
The dry weights of both WT and transgenic tobacco plants decreased gradually with increasing NaCl concentration. The weights of the transgenic tobacco plants declined more slowly, and the dry weights were higher in the transgenic tobacco plants than the WT plants (Fig. 3a). In particular, the dry weights of the roots, stems, and leaves were 2.0-, 1.1-, and 2.6-fold higher in T4 and 2.7-, 1.9-, and 3.1-fold higher in T18, respectively, under 200 mM NaCl compared with the WT plants (Fig. 3b,c,d). The growth of the transgenic plants was thus significantly better under salt stress compared with the WT plants, and this growth increased as the relative expression of IlVP increased.
(a) Growth of wild-type and IlVP-transgenic tobacco plants in response to 200 mM NaCl treatment for 7 days. WT: wild type; T4, T18: transgenic tobacco. (b–d) Root, stem and leaf dry weight of wild-type and IlVP-transgenic tobacco plants in response to salt stress, respectively. Each bar represents the mean (n = 7), and error bars indicate the standard deviation (SD). Columns with different letters indicate a significant difference at P < 0.05 (Duncan’s multiple range test).
Effect of NaCl stress on leaf relative water content and plasmalemma permeability
The relative water content is an important physiological indicator of plant growth status. The plant water content can be divided into free and bound water, with the majority present as free water. The free water ratio can reflect plant salt resistance in response to the metabolic situation. With increasing salt concentration, the leaf relative water content distinctly decreased in both the WT and transgenic tobacco plants compared with the control, but that of the transgenic tobacco plants declined more slowly and remained higher than that of the WT plants (Fig. 4). In response to treatment with 200 mM NaCl, the leaf relative water content of the transgenic tobacco plants was 1.05 times higher in T4 and 1.08 times higher in T18 compared with the WT (Fig. 4a). IlVP was therefore found to enhance the salt resistance of the tobacco plants. The water-retention capacity of the transgenic tobacco plants increased significantly under salt stress.
Leaf relative water content (a) and relative membrane permeability (b) of wild-type and IlVP-transgenic tobacco plants in response to salt stress for 7 days. Each bar represents the mean (n = 7), and error bars indicate the standard deviation (SD). Columns with different letters indicate a significant difference at P < 0.05 (Duncan’s multiple range test).
Maintenance of the cell microenvironment and normal metabolism relies on plant cell membranes. The relative plasma membrane permeability of the WT and transgenic tobacco plants increased with NaCl treatment, with a lower increase observed in the transgenic than the WT plants (Fig. 4b). For instance, the relative plasma membrane permeability was 28.3% (T4) and 43.2% (T18) lower than that of the WT plants subjected to 200 mM NaCl treatment for 7 days. Damage to the cell membranes of the transgenic plants under salt stress was thus less severe than that of the WT plants, while the salt resistance of the transgenic plants was higher than that of the WT.
Effect of NaCl stress on Na+ and K+ concentrations
Concentrations of Na+ in the tissues (roots, stems, and leaves) of the WT and transgenic plants (T4 and T18) increased with increasing NaCl concentrations; considerably higher increases were observed in the transgenic compared with the WT plants. Under 200 mM NaCl treatment for 7 days, the Na+ concentrations in the roots, stems, and leaves of T4 were 38.7%, 15.7% and 12.2% higher, and those of T18 were 188.0%, 29.5% and 33.5% higher, respectively, compared with the WT (Fig. 5a,b,c). The K+ concentration in the tissues of transgenic plants was significantly higher than that in the WT plants in the presence of 200 mM NaCl. Although the accumulation of K+ in T4, T18 and WT decreased with external NaCl treatment, the tissues of transgenic plants retained more K+ (Fig. 5d,e,f). Under NaCl treatment, the transgenic lines showed significantly higher concentrations of Na+ and K+ than the WT plants.
Cation concentration in tissues of wild-type and IlVP-transgenic tobacco plants in response to salt stress. Na+ (a–c) and K+ (d–f) concentrations were measured after treatment for 7 days with different NaCl concentrations (0, 50, 100, and 200 mM). Each bar represents the mean (n = 7), and error bars indicate the standard deviation (SD). Columns with different letters indicate a significant difference at P < 0.05 (Duncan’s multiple range test).
Discussion
In a previous study, we determined that I. lactea has strong salt resistance24, and a rapid tissue culture propagation system was subsequently established26. It is well known that tonoplast H+-PPase is involved in the sequestration of Na+ into vacuoles, which contributes to salt tolerance of plants27.
The tonoplast H+-PPase is encoded by a highly hydrophobic, single-subunit protein with a calculated molecular mass of 80 kDa28. In higher plants, H+-PPase cDNA commonly contains a 2,283–2,319 bp ORF encoding 761–773 amino acid residues with a deduced calculated molecular mass of 79–81 kDa29,30,31. Our results showed that IlVP consists of 2,316 bp, encodes a protein of 771 amino acids with a calculated molecular mass of 80.7 kDa, and contains 14 trans-membrane domains. Two contrasting responses in tonoplast H+-PPase activity under salt stress have been reported. Some researchers have reported that H+-PPase activity declines in response to salt treatment19, whereas other studies have shown that NaCl may enhance H+-PPase activity15,16,18. H+-PPase hydrolytic activity in barley roots and leaves has been found to increase under different salt concentrations17. IlVP was mainly expressed in shoots under the NaCl concentrations in our study, and transcript abundance of IlVP in shoots increased gradually with increasing NaCl concentrations (50 and 200 mM). This response is conducive for compartmentalisation of Na+ in the vacuole of the leaf cytoplasm, with a consequent reduction in the salt damage caused to plants32. Overexpression of the H+-PPase gene may enhance trans-membrane electrochemical gradients and improve secondary transport carrier efficiency across the vacuole membrane under salt stress33. A variety of inorganic ions accumulate in the vacuoles to maintain the balance between ionic equilibrium, osmotic equilibrium, and cell turgor-pressure stability; in this way, damage to cells by inorganic ions is reduced, and salt or osmotic stress tolerance of cells is enhanced14. Thus, H+-PPase plays an important role as a proton pump in the process of salt or osmotic stress resistance and adaptation.
Plant salt resistance is enhanced by excessive expression of tonoplast H+-PPase. Overexpression of SaVP1 in Arabidopsis enhances tolerance to drought and salt stresses; this overexpression also results in the up-regulation of several K+ and Ca2+ channel/transporter genes that show a function similar to that of vacuolar H+-PPase from other plants33. Overexpression of KfVP1 increases salt and drought tolerance of Arabidopsis 31. The AVP1 protein content in AVP1-transgenic Arabidopsis seedlings was significantly higher than that of WT plants34. Recovery of salt resistance can be achieved by overexpression of Arabidopsis AVP1 in yeast salt-sensitive enal mutants35. In one study, inorganic ion accumulation was higher in the roots and leaves of AVP1 genetically modified alfalfa (Medicago sativa) than in WT plants, and the leaf osmotic potential of transgenic plants was reduced. In addition, the salt and drought resistance of the transgenic plants was significantly enhanced36. PvVP1 has been transferred to non-halophytic grass, thus providing a feasible basis to improve the salt resistance of Paspalum vaginatum 37. Overexpression of the H+-PPase gene provides a stimulus for ion compartmentalisation, thereby maintaining the balance between ionic balance and osmotic equilibrium within the cell and enhancing plant salt resistance10. In the present study, using the anti-AtVP found that exogenous IlVP protein was largely expressed in transgenic plants by Western blots analysis compared to WT. This suggested the salt tolerance of the IlVP transgenic tobacco plants was higher than that of the WT plants. Compared with the WT plants, the physiological indices of the transgenic tobacco plants showed greater stability and slower changes, which indicated that the physiological system of the transgenic tobacco plants had not been severely damaged under NaCl stress. In addition, overexpression of IlVP enhanced the accumulation of Na+ in tissues. On the one hand, these differences might be ascribed to enhanced sequestration of Na+ into the vacuole and maintained the balance between K+ and Na+, and increased the osmotic regulation ability, because of overexpressing of H+-PPase38,39,40. On the other hand, potassium is required for plant growth, tropisms, cell expansion, enzyme activity, ion homeostasis and stomatal movements41. Our study showed that the K+ concentrations in transgenic tobacco were higher than in the WT plants under salt stress. The increased accumulation of potassium is likely to be overexpression of the H+-PPase resulted in the enhanced K+ uptake and the release of organic acids, which contribute to an increased rhizosphere acidification and to enhanced phosphorus uptake, and thus improved salt tolerance in transgenic plants42.
Conclusions
IlVP was cloned from I. lactea. IlVP expression was observed mainly in shoots, and its transcript abundance changed with increasing salt concentration and duration of exposure.
Phenotypes, the leaf relative water content, relative plasma membrane permeability, and concentrations of Na+ and K+ in roots, stems, and leaves were measured in response to treatment with different NaCl concentrations for 7 days. The transgenic tobacco plants displayed enhanced tolerance to NaCl stress compared with the WT plants. These results suggest that overexpression of IlVP in tobacco plants enhances sequestration of Na+ into vacuoles to alleviate Na+ toxicity in the cytoplasm, further maintaining cellular K+ and Na+ homeostasis and cell membrane stability, thereby enhancing tobacco salt tolerance.
Materials and Methods
Material culture and main experimental reagents
Seeds of I. lactea Pall. var. chinensis (Fisch.) Koidz. were collected from the National Experiment Station of Precision Agriculture, Xiao Tang Shan, China, located approximately 55 km from Beijing (39°34′ N, 116°28′ E). Plump seeds were sterilised with sodium hypochlorite solution (5%) for 5 min, rinsed thoroughly with distilled water, incubated in 40 °C water for 56 h, and then germinated on moistened filter paper for 10 days at 25 °C in the dark. After plumule emergence, uniform seedlings were transferred to plastic containers (19 cm long, 13.5 cm wide and 7.5 cm high) containing modified Hoagland’s solution (2 mM KNO3, 1 mM NH4H2PO4, 0.5 mM Ca(NO3)2·4H2O, 0.5 mM MgSO4·7H2O, 60 µM Fe-citrate, 92 µM H3BO3, 18 µM MnCl2·4H2O, 1.6 µM ZnSO4·7H2O, 0.6 µM CuSO4·5H2O, and 0.7 µM (NH4)6-Mo7O24·4H2O) for 5 weeks. The nutrient solution was renewed every 3 days. All seedlings were grown in the same chamber under a day/night cycle of 16 h/8 h at 25 °C/18 °C, a relative humidity of 50%–60%, and 600 µmol m−2 s−1 photosynthetically active radiation.
Cloning of IlVP
A pair of degenerate primers, P1 and P2, were designed based on VP gene sequences in GenBank. Six-week-old seedlings of I. lactea were treated with 100 mM NaCl for 24 h. After treatment, fresh roots (100 mg) were washed in sterile water and then ground in liquid nitrogen. Total RNA was extracted using a Takara RNA extraction kit. After synthesis of cDNA using a First-Strand PrimeScript RTase cDNA synthesis kit, reverse transcription and PCR amplification were conducted. The PCR protocol was as follows: 94 °C for 2 min, followed by 30 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 1 min, and a final step of 72 °C for 10 min. The amplified fragment was sequenced and analysed using the BLAST tool (http://www.ncbi.nlm.nih.gov/BLAST). IlVP 5′- and 3′-ends were obtained using Clontech SMARTer RACE and Takara 3′-Full RACE kits in accordance with the manufacturers’ instructions and the gene-specific primers P3 and P4, respectively (Supplementary Table 1). The ORF of IlVP was amplified using a Tks Gflex DNA Polymerase PCR kit with primers P5 and P6 (Supplementary Table 1).
DNA sequence and phylogenetic analyses
The IlVP sequence was analysed, and the coding regions were predicted using DNAMAN 6.0 software. IlVP sequence homology analysis and phylogenetic tree construction was carried out using MEGA 6.0 software. The isoelectric point and molecular mass were predicted using the online Compute pI/Mw tool (http://web.expasy.org/compute_pi/).
Expression analysis of IlVP
Six-week-old seedlings of I. lactea var. chinensis were treated with 200 mM NaCl for 24 h. After washing the roots in sterile water, 200 mg of fresh roots and shoots were ground separately in liquid nitrogen. Total RNA was extracted using a Takara RNA extraction kit, after which cDNA was synthesised using a PrimeScript RT Reagent Kit with gDNA Eraser. In addition, 6-week-old seedlings of I. lactea were treated with 0, 25, 50, 100, and 200 mM NaCl for 0, 6, 12, 24, and 48 h; total RNA was extracted, cDNA was synthesised as described above, and the expression of IlVP in shoots was analysed. Quantitative real-time RT-PCR (qRT-PCR) was performed using SYBR Premix Ex Taq II (Perfect Real Time) (Takara) on a StepOnePlus Real-Time PCR system (ABI) to monitor the amplification of each cDNA fragment. The qRT-PCR amplification was performed using the IlVP-specific primer pair P7 and P8. Actin amplified with the primer pair A1 and A2 was used as an internal reference in the qRT-PCR analysis. All experiments were carried out with three biological replicates.
The qRT-PCR protocol consisted of three steps: predenaturation at 95 °C for 30 s; PCR amplification for 40 cycles of 95 °C for 5 s and 60 °C for 1 min; and finally, dissociation, consisting of 95 °C for 10 s, 65 °C for 5 s, and 95 °C for 5 s.
Construction of plant expression vectors
The plasmid containing the IlVP gene and the plasmid pBI121 were cut with QuickCut SmaI and QuickCut ScaI, respectively, and incubated for 15 min at 30 °C. The two fragments were ligated in accordance with the DNA Ligation Kit Version 2.1 manual. A plant expression vector that included the CaMV 35S promoter, a NOS terminator and the IlVP gene was constructed. The recombinant plasmid was transformed into Escherichia coli strain DH5α. The cells were screened for kanamycin and rifampicin resistance. After purification of cells harbouring the recombinant plasmid, restriction enzyme digestion was performed.
Genetic transformation and identification of transgenic tobacco
Chemically competent cells of Agrobacterium tumefaciens strain EHA105 were prepared. After fusing the plant expression vector into EHA105 cells using the freeze–thaw method, tobacco strain ‘W38’ was transformed using the leaf-disc method. Tobacco leaf strips that showed expansion after placement on Murashige and Skoog (MS) culture medium lacking kanamycin were transfected by Agrobacterium for 7 min, blotted with sterile filter paper to remove excess liquid, and co-cultured for 2–3 days. The leaf strips were placed on kanamycin-containing differentiation medium (MS medium supplemented with 1 mg L−1 6-benzylaminopurine, 0.1 mg L−1 naphthaleneacetic acid, 50 mg L−1 kanamycin, and 500 mg L−1 carbenicillin). Callus was visible after 4 weeks of culturing on this medium. When generated shoots were 1–3 cm tall, they were transplanted onto MS medium supplemented with 50 mg L−1 kanamycin and 500 mg L−1 carbenicillin. Disinfected leaf strips were used as a control and were cultured on MS medium. When space was insufficient for growth, the generated shoots were transplanted into plastic culture pots containing a mixture of vermiculite and perlite (v/v, 3:1) and grown under a 16 h/8 h (light/dark) photoperiod and a light intensity of 600 µmol m−2 s−1 at 25 °C and 60% relative humidity. The plants were watered every 2 days with Hoagland’s solution. Total genomic DNA was extracted from the leaves of regenerated and WT plants in accordance with the Takara MiniBEST Plant Genomic DNA Extraction kit manual. PCR amplification was carried out with a TksGflex DNA Polymerase kit following the manufacturer’s instructions using plasmid DNA as a positive control, WT plant DNA as a negative control, and the following pair of specifically designed primers: F1 (5′-CATTGCTGGGATGGGTTC-3′) and R1 (5′-TCGTGGCTGCTCCTGTTC-3′). The PCR protocol consisted of pre-denaturation at 94 °C for 1 min, followed by 30 cycles of denaturation at 98 °C for 30 s, annealing at 55 °C for 15 s, and elongation at 68 °C for 1 min, followed by storage at 4 °C.
Southern Northern blot assays
Preparation of the Southern blot probe was carried out using a PCR DIG Probe Synthesis kit (Beijing Mylab Corporation). DNA samples (30 µg) were cut with DraI. The enzyme-digested product was purified and electrophoresed on a 1% agarose gel at 20 V. A capillary siphon was used for transfer of the purified product. After prehybridisation for 2 h, a radiolabeled probe was added and hybridised overnight.
DIG-PCR amplification was used to label probes for the Northern blot assay. After carrying out 1.1% formaldehyde denaturing agarose gel electrophoresis for 3 h at 50 V, a capillary siphon was used for transfer of the purified product. After prehybridisation for 2 h, the radiolabeled probe was added and hybridised overnight. Membrane washing and signal detection were conducted using a DIGD-210 Hybridization Detection II kit in accordance with the manufacturer’s instructions.
Assessment of salt tolerance of transgenic tobacco
Transgenic tobacco (T4 and T18) and WT plants grown under identical growth conditions were irrigated for 7 days using Hoagland’s solution containing 0, 50, 100, or 200 mM NaCl. Seven biological replicates were conducted, with three seedlings of each strain used per replicate. Root, stem, and leaf fresh weights, leaf relative water content, relative electrical conductivity, and Na+ and K+ concentrations were measured. After fresh weight measurements, roots, stems, and leaves were oven-dried at 80 °C to a constant weight, and the dry weight of each organ was recorded. The leaf relative water content and relative electrical conductivity were measured using Gao’s method43. Na+ and K+ concentrations in roots, stems, and leaves were measured using a flame emission spectrophotometer.
Tonoplast vesicles isolation and Western blot
According to the method of Wang et al. (2000)44 with minor modifications, tonoplast enriched membrane vesicles were isolated. Briefly, about 100 mg leaves of WT and transgenic tobacco (T4 and T18) were selected under 200 mM NaCl treatment for 10 days, which were homogenized in extraction medium (pH 7.8) containing 250 mM mannitol, 1 mM DTT, 3 mM EGTA, 1% (w/v) PVP, 0.25 mM PMSF, 100 mM Tricine, 3 mM MgSO4. The homogenate was filtered using four layers of cheesecloth that was centrifuged at 12,000 × g for 15 min at 4 °C. Subsequently, these supernatant were centrifuged at 300,000 × g for 45 min and was suspended in suspension buffer (pH 7.5) with 250 mM mannitol, 2 mM DTT, 3 mM EGTA, 10 mM Hepes. The microsomal membrane vesicle suspension was loaded on a 1%/18% (w/w) Dextran T70 gradient in suspension buffer and then centrifuged at 100,000 × g for 2 h. The tonoplast-enriched membrane vesicle fraction located at the 1%/8% (w/w) Dextran T70 interface was carefully collected, diluted 4–5 fold with dilution buffer (pH 7.0) with 1 mM DTT, 3 mM MgSO4, 50 mM Hepes, 0.2 mM PMSF and then centrifuged at 300,000 × g for 45 min. For western blot, 100 μg tonoplast proteins were separated using 12% SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis), and was immunoblotted with antibody against H+-PPiase (Agrisera, Vännäs, SWEDEN). Blots were performed according to describe the methods of Sarafian et al. (1999)45 and Venancio et al. (2014)46.
Statistical analysis
Excel 2010 was used for data processing. SAS11.0 was used for variance and clustering analyses.
References
Zhu, J. K. Plant salt tolerance. Trends in Plant Sci. 6, 66–71 (2001).
Guo, Q. et al. Selective transport capacity for K+ over Na+ is linked to the expression levels of PtSOS1 in halophyte Puccinellia tenuiflora. Funct. Plant Biol. 39(12), 1047–1057 (2012).
Blumwald, E. & Poole, R. J. Na+/H+ antiporter in isolated tonoplast vesicles from storage tissue of Beta vulgaris. Plant Physiol. 78, 163–167 (1985).
Apse, M. P., Aharon, G. S., Snedden, W. A. & Blumwald, E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiporter in Arabidopsis. Science 285, 1256–1258 (1999).
Wei, A., He, C., Li, B., Li, N. & Zhang, J. The pyramid of transgenes TsVP and BetA effectively enhances the drought tolerance of maize plants. Plant Biotechnol. J. 9(2), 26–29 (2011).
Britten, C. J., Zhen, R. C., Kim, E. J. & Rea, P. A. Reconstitution of transport function of vacuolar H+-translocating inorganic pyrophosphatase. J. Biol. Chem. 267, 21850–21855 (1992).
Maeshima, M. Vacuolar H+-pyrophosphatase. Biochimica et Biophysica Acta 1465, 37–51 (2000).
Tanaka, Y., Chiba, K., Maeda, M. & Maeshima, M. Molecular cloning of cDNA for vacuolar membrane proton-translocating inorganic pyrophosphatase in Hordeum vulgare. Biochem. Bioph. Res. Co. 190, 1110–1114 (1993).
Kim, Y., Kim, E. J. & Rea, P. A. Isolation and characterization of cDNAs encoding the vacuolar H+-pyrophosphatase of Beta vulgaris. Plant Physiol. 106, 375–352 (1994).
Suzuki, Y. & Maeshima, M. Molecular cloning of vacuolar H+-pyrophosphatase and its expression during the development of pear fruit. Plant Cell Physiol. 40(8), 900–904 (1999).
Brini, F., Gaxiola, R. A., Berkowitz, G. A. & Masmoudi, K. Cloning and characterization of a wheat vacuolar cation/proton antiporter and pyrophosphatase proton pump. Plant Physiol. Bioch. 43, 347–354 (2005).
Gao, F. et al. Cloning of an H+-PPase gene from Thellungiella halophile and its heterologous expression to improve tobacco salt tolerance. J. Exp. Bot. 57(12), 3259–3270 (2006).
Gan, X. Y., Gong, L., Shi, L. & Song, Y. X. Cloning and sequence analysis of a cDNA encoding vacuolar H + -PPase gene from Haloxylon ammodendron. Acta Agriculturae Boreali-occidentalis Sinica 23(11), 198–203 (2014).
Bao, A. K. et al. Vacuole membrane H+-PPase and salt tolerance of plants. Plant Physiol. Co. 42(4), 777–783 (2006).
Colombo, R. & Cerana, R. Enhanced activity of tonoplast pyrophosphatase in NaCl-grown cells of Daucus carota. J. Plant Physiol. 142, 226–229 (1993).
Ballesteros, F., Donaire, J. P. & Belver, A. Effects of salt stress on H+-ATPase and H+-PPase activities of tonoplast-enriched vesicles isolated from sunflower roots. Plant Physiol. 97, 259–268 (1996).
Guo, S. L. et al. Molecular cloning and characterization of a vacuolar H+-pyrophosphatase gene, SsVP, from the halophyte Suaeda salsa and its overexpression increases salt and drought tolerance of Arabidopsis. Plant Mol. Biol. 60, 41–50 (2006).
Vera-Estrella, R., Barkla, B. J., Garcia-Ramirez, L. & Pantoja, O. Salt stress in Thellungiella halophila activates Na+ transport mechanisms required for salinity tolerance. Plant Physiol. 139, 1507–1517 (2005).
Matsumoto, H. & Chung, G. C. Increase in proton-transport activity of tonoplast vesicles as an adaptive response of barley roots to NaCl stress. Plant Cell Physiol. 29(7), 1133–1140 (1988).
Barkla, B. J. & Pantoja, O. Physiology of ion transport across the tonoplast of higher plants. Ann. Rev. Plant Physiol. Mol. Bio. 47, 159–184 (1996).
Chen, M. J. & Jia, S. X. China Forage Plant. 1440–1441 (China Agriculture Press, 2002).
Shi, X. X., Mao, P. C., Zhang, G. F. & Meng, L. The comparison of drought resistance on 15 Chinese Iris germpalsm at seedling stage. Acta Agrestia Sinica. 15(4), 352–358 (2007).
Guo, Q. et al. Antioxidative systems, metal ion homeostasis and cadmium distribution in Iris lactea exposed to cadmium stress. Ecotox. Environ. Safe. 139, 50–55 (2017).
Mao, P. C., Tian, X. X. & Meng, L. Evaluation of salt tolerance for 16 Iris lactea var. chinensis accessions at seedling stage. Pratacultural Sci. 30(1), 35–43 (2013).
Guo, J. Y., Li, S. S., Guo, Q., Meng, L. & Yang, X. P. Cloning and sequence analyzing of the IlVP gene fragment in Iris lactea. Genomics and Appl. Biol. 34(12), 2667–2673 (2015).
Meng, L., Xiao, K. & Zhao, M. L. Technological system of tissue culture and rapid propagation of Iris lactea Pall. var. chinensis (Fisch.) Koidz. Bull. Bot. Res. 29(2), 193–197 (2009).
Gaxiola, R. A. et al. Genetic manipulation of a “Vacuolar” H+-PPase: from salt tolerance to yield enhancement under phosphorus-deficient soils. Plant Physiol. 159, 3–11 (2012).
Blumwald, E. Tonoplast vesicles for the study of ion transport in plant vacuoles. Acta Physiol. Plant. 9, 731–734 (1987).
Maeshima, M. Tonoplast transporters: organization and function. Ann. Rev. Plant Physiol. Mol. Bio. 52, 469–497 (2001).
Lü, S. Y. et al. cDNA cloning of vacuolar H+-pyrophosphatase and its expression in Hordeum brevisubulatum (Trin.) Link in response to salt stress. Agr. Sci. China. 4(4), 247–251 (2005).
Yao, M. H. et al. Overexpression of the halophyte Kalidium foliatum H+-Pyrophosphatase gene confers salt and drought tolerance in Arabidopsis thaliana. Mol. Biol. Rep. 39, 7989–7996 (2012).
Blumwald, E. & Gelli, A. Secondary inorganic ion transport in plant vacuoles. Adv. Bot. Res. 25, 401–407 (1997).
Wang, Y. Q., Jin, S. X., Min, L. & He, X. The Sophora alopecuroid H+-PPase gene SaVP1 confers multiple abiotic stress tolerance in Arabidopsis. Plant Mol. Biol. Rep. 33, 923–930 (2015).
Gaxiola, R. A. et al. Drought-and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc. Natl. Acad. Sci. USA 98, 11444–11449 (2001).
Gaxiola, R. A. et al. The Arabidopsis thaliana proton transporters, AtNHXl and AVP1 can function in cation detoxification in yeast. Proc. Natl. Acad. Sci. USA 96, l480–1485 (1999).
Bao, A. K. et al. Over expression of the Arabidopsis H+-PPase enhanced resistance to salt and drought stress in transgenic alfalfa (Medicago sativa L.) Plant Sci. 176, 232–240 (2009).
Song, H., Nan, Z. B., Cai, X. N., Zhong, X. X. & Gu, H. R. Clone and sequence analysis of 5′ Race of PvVP1 in Paspalum vaginatum. Acta Prataculturae Sinica. 23(5), 168–174 (2014).
Li, Y. Y., Guo, J. R., Yang, M. F. & Wang, B. S. Effects of KCl and NaCl treatment on growth and water metabolism of halophyte Suaeda salsa seedlings. J. Plant Physiol. Mol. Bio. 29, 576–580 (2003).
Fukuda, A. & Tanaka, Y. Effects of ABA, auxin, and gibberellin on the expression of genes for vacuolar H+-inorganic pyrophosphatase, H+-ATPase subunit A, an Na+/H+ antiporter in barley. Plant Physiol. Bio. 44, 351–358 (2006).
Bao, A. K. et al. Co-expression of tonoplast Cation/H+ antiporter and H+-pyrophosphatase from xerophyte Zygophyllum xanthoxylum improves alfalfa plant growth under salinity, drought and field conditions. Plant Biotechnol. J. 14, 964–975 (2016).
Zhao, F. Y., Zhang, X. J., Li, P. H., Zhao, Y. X. & Zhang, H. Co-expression of the Suaeda salsa SsNHX1 and Arabidopsis AVP1 confer greater salt tolerance to transgenic rice than the single. SsNHX1. Mol. Breeding 17, 341–353 (2006).
Yang, H. et al. Enhanced phosphorus nutrition in monocots and dicots over-expressing a phosphorus-responsive type I H+-pyrophosphatase. Plant Biotechnol. J. 5, 735–745 (2007).
Gao, J. F. Plant Physiology Experiment Guide. 14–15 (Higher Education Press, 2006).
Wang, B. S., Ratajczak, R. & Zhang, J. H. Activity, amount and subunit composition of vacuolar-type H+-ATPase and H+-PPase in wheat roots under severe NaCI stress. J Plant Physiol. 157, 109–116 (2000).
Retamal, C. A., Thiebaut, P. & Alves, E. W. Protein purification from polyacrylamide gels by sonication extraction. Anal. Biochem. 268, 15–20 (1999).
Venancio, J. B. et al. A vacuolar H+-pyrophosphatase differential activation and energy coupling integrate the responses of weeds and crops to drought stress. Biochim. Biophys. Acta 1840, 1987–1992 (2014).
Acknowledgements
This research was financially supported by the Beijing Natural Science Foundation (grant no. 6142007) and the Scientific Innovation Ability Construction Project (grant no. KJCX20140103) of the Beijing Academy of Agriculture and Forestry Sciences.
Author information
Authors and Affiliations
Contributions
L.M. conceived and designed the experiments. S.L., J.G., Q.G., P.M. and X.T. prepared for sample and performed the experiments. L.M. analysed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Meng, L., Li, S., Guo, J. et al. Molecular cloning and functional characterisation of an H+-pyrophosphatase from Iris lactea . Sci Rep 7, 17779 (2017). https://doi.org/10.1038/s41598-017-18032-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-18032-3
This article is cited by
-
Effect of salt stress on the genes expression of the vacuolar H+ -pyrophosphatase and Na+/H+ antiporter in Rubia tinctorum
Molecular Biology Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.