Abstract
Streptococcus pneumoniae is a leading cause of morbidity and mortality globally. The Pilus-1 proteins, RrgA, RrgB and RrgC of S. pneumoniae have been previously assessed for their role in infection, invasive disease and as possible vaccine candidates. In this study we have investigated the glycan binding repertoire of all three Pilus-1 proteins, identifying that the tip adhesin RrgA has the broadest glycan recognition of the three proteins, binding to maltose/cellobiose, α/β linked galactose and blood group A and H antigens. RrgB only bound mannose, while RrgC bound a subset of glycans also recognized by RrgA. Adherence of S. pneumoniae TIGR4 to epithelial cells was tested using four of the oligosaccharides identified through the glycan array analysis as competitive inhibitors. The blood group H trisaccharide provided the best blocking of S. pneumoniae TIGR4 adherence. Adherence is the first step in disease, and host glycoconjugates are a common target for many adhesins. This study has identified Pilus-1 proteins as new lectins involved in the targeting of host glycosylation by S. pneumoniae.
Similar content being viewed by others
Introduction
Streptococcus pneumoniae is one of the leading causes of morbidity and mortality worldwide1,2,3. S. pneumoniae causes a range of diseases including pneumonia, meningitis, septicemia and otitis media, and produces a range of virulence factors including the toxin pneumolysin, pneumococcal surface protein A and pilus2,4,5,6. The current S. pneumoniae vaccines target the capsular polysaccharide, but cover only a subset of the 97 known capsular serotypes. Differences in serotype distribution between developed and developing countries, and serotype replacement in response to widespread use of the vaccines, are reducing the overall impact of the vaccines on the burden of pneumococcal disease1,3.
Expression of the pneumococcal Pilus-1, composed of three proteins RrgA, RrgB and RrgC, has been linked to pneumococcal meningitis in mouse infection models5,6,7. Pilus-1 was found to be required for the bacteria to breach the blood brain barrier5. The Pilus-1 protein complex consists of RrgB as the shaft protein, with RrgA as the tip adhesin and RrgC, which serves as a pilus anchor at the cell surface6,7,8. The S. pneumoniae Pilus-1 protein complex has been proposed as a novel vaccine target9,10. The RrgA and RrgB proteins were found to produce cross-protecting antibodies that led to blocking of adherence of S. pneumoniae to cells in culture and resulted in the opsonophagocytosis of S. pneumoniae 9,11. RrgA is also involved in regulation of the host immune response to S. pneumoniae by binding to both MAC-1 (complement receptor 3, CD11b/CD18)12 and Toll-like receptor 213. RrgA has also been shown to interact directly with cultured epithelial cells and extracellular matrix components including fibronectin and collagen10.
All of the known proteins that interact with RrgA are glycoproteins, indicating a potential role of the oligosaccharides in the interactions. In this study we aim to identify glycan targets of the Pilus-1 protein complex of S. pneumoniae.
Results
RrgA, RrgB and RrgC proteins bind glycans
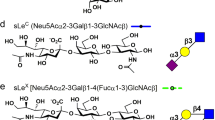
Glycan array analysis of the three Pilus-1 proteins from S. pneumoniae TIGR4 and RrgA from strain SPEC6B revealed differential glycan recognition between the four proteins tested. S. pneumoniae TIGR4 RrgB bound the least number of glycans, with only an α-mannobiose recognized (Table 1 and Dataset S1). RrgA from S. pneumoniae TIGR4 and RrgA from S. pneumoniae SPEC6B bound to a set of overlapping glycans including terminal galactose structures with both α and β linkages, glucose/maltose-related structures and blood group A (7 K and 392) and blood group H(O) (7 A) antigens (Table 1 and Dataset S1). No interactions with terminal galactose structures were observed when galactose was directly linked to glucose. S. pneumoniae SPEC6B RrgA also had binding to β-linked mannose and β-linked N-acetylglucosamine structures not bound by the TIGR4 RrgA. RrgC bound to structures that are the same or very similar to RrgA from TIGR4 with additional recognition of blood group B antigen (483), Lewis B (496) and hyaluronic acid (14I) (Table 1 and Dataset S1).
Surface Plasmon Resonance analysis of RrgA and glycans identified by array analysis
To validate the glycan array results and to determine the dissociation equilibrium constant (KD) of the interactions, surface plasmon resonance was performed between free oligosaccharides and captured RrgA proteins (Table 2). The highest affinity (smallest KD) interaction was between RrgA proteins from both S. pneumoniae TIGR4 and SPEC6B and the H trisaccharide type 3/4 (Table 2).
Inhibition of adherence of RrgA-expressing S. pneumoniae TIGR4 using free oligosaccharides
S. pneumoniae TIGR4 and S. pneumoniae TIGR4ΔrrgA were tested for adherence differences using A549 human lung carcinoma and Detroit 562 pharyngeal carcinoma cell lines. These cell lines are representative of the two most important sites for colonization/infection of humans by S. pneumoniae. S. pneumoniae TIGR4ΔrrgA was significantly less adherent to both cell lines, with RrgA contributing to around 50% of the adherence of S. pneumoniae TIGR4 to A549 cells and 70% of the adherence to Detroit 562 cells (Table 3).
The Detroit 562 and A549 cell lines were examined for the presence of RrgA target glycans using lectin array analysis (see Dataset S2). Lectins recognizing terminal β-linked galactose were observed for both cells. The A549 cells were recognized by the lectins AAA and RSL, both of which recognize structures containing α1-2 linked fucose, while the Detroit 562 cells did not (Dataset S2). Lectins with α1-2 linked fucose specificity recognize blood group antigens, particularly the H (O) blood group antigens.
Adherence of S. pneumoniae TIGR4 to A549 cells was then re-examined using four of the oligosaccharides identified through the glycan array analysis as competitive inhibitors. The blood group H type 3/4 trisaccharide provided the best blocking of S. pneumoniae TIGR4 adherence, with 67.7% inhibition (Table 4; P < 0.001). In addition, maltose, cellobiose and blood group A tetrasaccharide all reduced adherence by around 56% (Table 4; P < 0.001 in all cases). Interestingly, the residual A549 adherence observed for TIGR4ΔrrgA (approximately 50% relative to wild type TIGR4) could also be competitively inhibited by a further 30–46% by cellobiose and blood group A tetrasaccharide and H trisaccharide (Table 4; P < 0.001).
Discussion
Adherence to host tissues is the first step in disease and host glycoconjugates are a common target for numerous bacterial adhesins14,15,16, including pili/fimbriae17,18. Pilus-1 of S. pneumoniae has been shown to interact with Toll-like receptor 2 and MAC-112,13 and epithelial cells and ECM components10, all of which are glycosylated.
The glycan array analysis of RrgA from S. pneumoniae TIGR4 and SPEC6B identified a cluster of oligosaccharide binding that was consistent between the two proteins. These proteins have previously been shown to bind to both cells and ECM components equally, indicating recognition of uncapped β-linked galactose. Binding of S. pneumoniae RrgA to β-linked galactose and blood group A glycan is consistent with interactions between S. pneumoniae and a wide variety of cell surfaces in the human host.
The array binding was investigated further using surface plasmon resonance analysis. Binding to blood group antigens, β-galactose, maltose, cellobiose and GlcNAc were confirmed through SPR analysis, with the highest affinity interaction observed occurring between RrgA from TIGR4 with the H-type 3/4 oligosaccharide (Table 2). As observed in the array experiment, no concentration dependent interaction was observed using SPR for terminal galactose with underlying glucose (lactose; Table 2). The SPR analysis found high affinity binding between RrgA and Lewis X (KD < 500 nM; Table 2).
The significance of these interactions in terms of interaction with host surfaces was further confirmed by competitive inhibition of TIGR4 adherence to A549 cells by exogenous maltose, cellobiose, blood group A type 3/4 and blood group H type 3/4 oligosaccharides. Interestingly, although RrgA is thought to be the most important component of the Pilus-1 structure in terms of mediating adherence, the residual A549 adherence exhibited by TIGR4ΔrrgA could be inhibited further, albeit to a slightly lesser extent, by the three exogenous oligosaccharides that were most effective against TIGR4. There are multiple S. pneumoniae surface proteins that have been shown to have a role in adherence to host epithelial cells, most notably CbpA, which is known to bind to cell surface glycoconjugates19, indicating that the residual binding observed in the TIGR4ΔrrgA is likely to be another adhesin.
The direct interaction between RrgA and Lewis X was not seen on the array, but Lewis X was a terminal group on several of the larger fucosylated structures identified (Table 1; 8 J and 8 K). This may indicate that the binding of Lewis X requires a specific presentation that is available in solution, but not available on the array except as a part of a larger structure. Lewis X is a member of the Lewis system histo-blood group antigens, which were originally identified on RBCs where they are acquired from the plasma as glycolipids20 and is expressed on most cell types but is over-expressed by human tumor cells from various sites21.
S. pneumoniae is a host-adapted pathogen that infects humans so it is unexpected to observe the binding of the Rrg proteins to many non-human and non-mammalian glycans. Rrg proteins recognised maltose and cellulose saccharides, repeating units of glucose typically produced by plants, algae and some bacteria22. This interaction may allow S. pneumoniae to interact with other bacterial species either directly or to the matrix of bacteria in biofilm communities.
RrgC and RrgA also bound to the non-human terminal α1-3-linked galactose structures. RrgC bound four of the 11 terminal α1-3-linked galactose structures present on the array, while RrgA only bound 1-2 terminal α1-3-linked galactose structures on the array (Table 1). This binding does not correlate with the host adapted nature of S. pneumoniae with α1-3-linked galactose widely expressed in non-human mammals but not in humans. The expression of α1-3-linked galactose is only observed in humans that produce blood group B antigens, structures not recognised on the array by the Rrg proteins.
The Pilus-1 complex of S. pneumoniae is known to be an important mediator of adherence of S. pneumoniae to host epithelial cells. This binding appears to be glycan related, as RrgA was found to have lectin activity, recognising a range of common host cell surface glycans. All three proteins that make up the Pilus-1 complex recognise glycans as recombinant soluble proteins. Thus, we have identified three new lectins produced by S. pneumoniae, with the pilius tip protein RrgA directly involved in interactions between pathogen and host.
Materials and Methods
Cloning, expression and purification of Rrg proteins
Genes were cloned into pET protein expression vectors and proteins were expressed and purified as previously described10.
Glycan Array
Glycan array slides were printed using SuperEpoxy 3 activated substrates as previously described23 using an ArrayIt Spotbot Extreme 3 contact printer with solid metal pins. The Glycan array binding experiments were performed and analysed as previously described16. Briefly 1 μg of protein in 1xPBS containing 1 mM MgCl2 and 1 mM CaCl2 was pre-complexed with mouse anti-His antibody (Cell signaling), Alexa555 rabbit anti-mouse IgG and Alexa555 goat anti-rabbit IgG and allowed to bind to a pre-blocked (1% bovine serum albumin in PBS) for 15 minutes. Slides were washed three times for 2 minutes in 1xPBS, dried by centrifugation and scanned and analysed using the Scan Array Express software package (Perkin Elmer) and Microsoft Excel for statistical analysis (Student’s unpaired t-test of fluorescence of background spots vs fluorescence of glycan printed spots).
Surface Plasmon Resonance analysis
The interactions between the Rrg proteins and test glycans were analysed using surface plasmon resonance (SPR) using a Biacore T100 system as described Shewell et al.16 with the following modifications. Proteins were immobilised onto a CM5 chip at pH 4.5, flow rate of 10 µL/min for 420 seconds with an ethanolamine blank flow cell as a control. Glycans were tested between 160 nM and 100 µM. All data was double reference subtracted.
Mutagenesis of rrgA in S. pneumoniae TIGR4
The rrgA gene in TIGR4 was deleted in-frame and replaced with an erythromycin resistance cassette (erm) by direct transformation with a linear DNA fragment constructed by overlap-extension PCR, essentially as previously described24. This involved using primer pairs rrgAF/rrgAermR and rrgAermF/rrgAR to amplify 5′ and 3′ regions flanking rrgA from TIGR4 template DNA, and J214/J215 to amplify erm from pVA838 (Table 5). Primers included overlapping sequences such that the three PCR products could be fused in a second round of PCR using primer pair rrgAF/rrgAR Erythromycin-resistant transformants were screened by PCR for deletion of rrgA, confirmed by DNA sequencing and designated TIGR4ΔrrgA.
Competitive inhibition of adherence of S. pneumoniae with free glycans
Adherence assays on A549 (human type II pneumocyte) and Detroit 562 (human nasopharyngeal carcinoma) cells were carried out as previously described25. A549 and Detroit 562 cells were grown in Dulbecco’s Modified Eagle Medium (DMEM; Gibco), or a 1:1 mix of DMEM and Ham’s F-12 Nutrient Mixture (Gibco), respectively, supplemented in both cases with 5% fetal bovine serum, 2 mM L-glutamine, 50 IU of penicillin and 50 μg/ml streptomycin. Confluent monolayers in 24-well plates were washed with PBS and infected with pneumococci (approximately 5 × 105 CFU per well) in a 1:1 mixture of the respective culture medium (without antibiotics) and C + Y medium, pH 7.423. Plates were centrifuged at 500 × g for 5 min, and then incubated at 37 °C in 5% CO2 for 2.5 h. Monolayers were washed 3 times in PBS, and adherent bacteria were released by treatment with 100 μl trypsin/EDTA, followed by 400 μl 0.025% Triton X-100. Lysates were serially diluted and plated on blood agar to enumerate adherent bacteria. Total adherence (mean ± SEM for 4-9 replicates) was expressed as a percentage of that for TIGR4 (or TIGR4ΔrrgA where appropriate) without additives. For inhibition assays, free oligosaccharides were used at a final concentration of 200 μM (>100 × KD) throughout the adherence assay. Data were analysed using Student’s un-paired t-test (two-tailed).
Lectin array analysis of Detroit 562 and A549 cells
The two cells lines used for adherence assays, A549 and Detroit 562, were analysed for cell surface glycans using lectin arrays. Lectin arrays were printed using an ArrayJet Argus Marathon Inkjet Bio-Printing System on Arrayit SME3 substrates. The lectins are immobilized to the epoxy activated substrate through non-specific amine coupling through free amines on the lectin proteins and the epoxide groups on the glass. Arrays were neutralized and performed as previously described26, with the exception that Bodipy 558/568 succinimidyl ester (Thermo Scientific) was used in place of CFDA-SE. Briefly, cells were collected and washed in 1xPBS prior to being labelled for 30 minutes at 37 °C with Bodipy 558/568 succinimidyl ester. Cells were washed three times with PBS and resuspended in PBS containing 1 mM MgCl2 and 1 mM CaCl2 at 107 cells/mL and 300 µL was applied to the array in a 125 µL frame without a coverslip. Cells were allowed to interact with the lectins on the array for 30 minutes and then the slides were washed three times with PBS, fixed in 4% formaldehyde and dried by centrifugation. Slides were scanned on an Innopsys InnoScan 1100AL to acquire the data of which lectins bound to the cells and analysed using Innopsys Mapix data acquisition and analysis software and Microsoft Excel for statistical analysis (Student’s unpaired t-test of fluorescence of background spots vs fluorescence of lectin printed spots).
References
Alderson, M. R. Status of research and development of pediatric vaccines for Streptococcus pneumoniae. Vaccine 34, 2959–2961, https://doi.org/10.1016/j.vaccine.2016.03.107 (2016).
Feldman, C. & Anderson, R. Epidemiology, virulence factors and management of the pneumococcus. F1000Research 5, 2320, https://doi.org/10.12688/f1000research.9283.1 (2016).
Rodgers, G. L. & Klugman, K. P. The future of pneumococcal disease prevention. Vaccine 29(Suppl 3), C43–48, https://doi.org/10.1016/j.vaccine.2011.07.047 (2011).
Barnett, T. C. et al. Streptococcal toxins: role in pathogenesis and disease. Cellular microbiology 17, 1721–1741, https://doi.org/10.1111/cmi.12531 (2015).
Iovino, F. et al. Pneumococcal meningitis is promoted by single cocci expressing pilus adhesin RrgA. The Journal of clinical investigation 126, 2821–2826, https://doi.org/10.1172/JCI84705 (2016).
Nelson, A. L. et al. RrgA is a pilus-associated adhesin in Streptococcus pneumoniae. Molecular microbiology 66, 329–340, https://doi.org/10.1111/j.1365-2958.2007.05908.x (2007).
LeMieux, J., Hava, D. L., Basset, A. & Camilli, A. RrgA and RrgB are components of a multisubunit pilus encoded by the Streptococcus pneumoniae rlrA pathogenicity islet. Infection and immunity 74, 2453–2456, https://doi.org/10.1128/IAI.74.4.2453-2456.2006 (2006).
Hilleringmann, M. et al. Molecular architecture of Streptococcus pneumoniae TIGR4 pili. The EMBO journal 28, 3921–3930, https://doi.org/10.1038/emboj.2009.360 (2009).
Ahmed, M. S. et al. Immune responses to pneumococcal pilus RrgA and RrgB antigens and their relationship with pneumococcal carriage in humans. The Journal of infection 68, 562–571, https://doi.org/10.1016/j.jinf.2014.01.013 (2014).
Moschioni, M. et al. The two variants of the Streptococcus pneumoniae pilus 1 RrgA adhesin retain the same function and elicit cross-protection in vivo. Infection and immunity 78, 5033–5042, https://doi.org/10.1128/IAI.00601-10 (2010).
Amerighi, F. et al. Identification of a monoclonal antibody against pneumococcal pilus 1 ancillary protein impairing bacterial adhesion to human epithelial cells. The Journal of infectious diseases 213, 516–522, https://doi.org/10.1093/infdis/jiv461 (2016).
Orrskog, S. et al. Pilus adhesin RrgA interacts with complement receptor 3, thereby affecting macrophage function and systemic pneumococcal disease. mBio 4, e00535–00512, https://doi.org/10.1128/mBio.00535-12 (2012).
Basset, A. et al. Toll-like receptor (TLR) 2 mediates inflammatory responses to oligomerized RrgA pneumococcal pilus type 1 protein. The Journal of biological chemistry 288, 2665–2675, https://doi.org/10.1074/jbc.M112.398875 (2013).
Day, C. J. et al. Glycan:glycan interactions: High affinity biomolecular interactions that can mediate binding of pathogenic bacteria to host cells. Proceedings of the National Academy of Sciences of the United States of America 112, E7266–7275, https://doi.org/10.1073/pnas.1421082112 (2015).
Lehmann, F., Tiralongo, E. & Tiralongo, J. Sialic acid-specific lectins: occurrence, specificity and function. Cellular and molecular life sciences: CMLS 63, 1331–1354, https://doi.org/10.1007/s00018-005-5589-y (2006).
Shewell, L. K. et al. The cholesterol-dependent cytolysins pneumolysin and streptolysin O require binding to red blood cell glycans for hemolytic activity. Proceedings of the National Academy of Sciences of the United States of America 111, E5312–5320, https://doi.org/10.1073/pnas.1412703111 (2014).
Abraham, S. N., Sun, D., Dale, J. B. & Beachey, E. H. Conservation of the D-mannose-adhesion protein among type 1 fimbriated members of the family Enterobacteriaceae. Nature 336, 682–684, https://doi.org/10.1038/336682a0 (1988).
Wurpel, D. J. et al. F9 fimbriae of uropathogenic Escherichia coli are expressed at low temperature and recognise Galbeta1-3GlcNAc-containing glycans. PloS one 9, e93177, https://doi.org/10.1371/journal.pone.0093177 (2014).
Rosenow, C. et al. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Molecular microbiology 25, 819–829 (1997).
Marcus, D. M. & Cass, L. E. Glycosphingolipids with Lewis blood group activity: uptake by human erythrocytes. Science 164, 553–555 (1969).
Yuriev, E., Farrugia, W., Scott, A. M. & Ramsland, P. A. Three-dimensional structures of carbohydrate determinants of Lewis system antigens: implications for effective antibody targeting of cancer. Immunol Cell Biol 83, 709–717, https://doi.org/10.1111/j.1440-1711.2005.01374.x (2005).
Lee, Y. C. Isolation and characterization of lipopolysaccharides containing 6-O-methyl-D-glucose from Mycobacterium species. The Journal of biological chemistry 241, 1899–1908 (1966).
Waespy, M. et al. Carbohydrate recognition specificity of trans-sialidase lectin domain from Trypanosoma congolense. PLoS neglected tropical diseases 9, e0004120, https://doi.org/10.1371/journal.pntd.0004120 (2015).
Iannelli, F. & Pozzi, G. Method for introducing specific and unmarked mutations into the chromosome of Streptococcus pneumoniae. Molecular biotechnology 26, 81–86, https://doi.org/10.1385/MB:26:1:81 (2004).
Harvey, R. M. et al. The variable region of pneumococcal pathogenicity island 1 is responsible for unusually high virulence of a serotype 1 isolate. Infection and immunity 84, 822–832, https://doi.org/10.1128/IAI.01454-15 (2016).
Arndt, N. X., Tiralongo, J., Madge, P. D., von Itzstein, M. & Day, C. J. Differential carbohydrate binding and cell surface glycosylation of human cancer cell lines. Journal of cellular biochemistry 112, 2230–2240, https://doi.org/10.1002/jcb.23139 (2011).
Acknowledgements
This work was supported by a Smart Futures Fund Research Partnerships Program Grant (MPJ) and National Health and Medical Research Council (NHMRC) Program Grant 1071659 (to JCP and MPJ), Russian Science Foundation grant # 14-50-00131 (NB). JCP is a NHMRC Senior Principal Research Fellow. KLS is a NHMRC Career Development Fellow.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: L.E.H., A.W.P., J.C.P., J.T., M.P.J., C.J.D., K.L.S. Performed the experiments: S.S., V.M., A.W.P., L.E.H., C.J.D., N.B., R.M.H. Analyzed the data: L.E.H., A.W.P., C.J.D., J.C.P., M.P.J. Wrote the paper: C.J.D., A.W.P., J.C.P., M.P.J. Reviewed and approved the final version of the manuscript: C.J.D., A.W.P., R.M.H., L.E.H., K.L.S., J.T., N.B., S.S., V.M., J.C.P., M.P.J.
Corresponding authors
Ethics declarations
Competing Interests
This work was cosponsored by Novartis Vaccines, now acquired by the GSK group of companies.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Day, C.J., Paton, A.W., Harvey, R.M. et al. Lectin activity of the pneumococcal pilin proteins. Sci Rep 7, 17784 (2017). https://doi.org/10.1038/s41598-017-17850-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-17850-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.