Abstract
Mucor circinelloides and other members of Mucorales are filamentous fungi, widely used as model organisms in basic and applied studies. Although genetic manipulation methods have been described for some Mucoral fungi, construction of stable integrative transformants by homologous recombination has remained a great challenge in these organisms. In the present study, a plasmid free CRISPR-Cas9 system was firstly developed for the genetic modification of a Mucoral fungus. The described method offers a rapid but robust tool to obtain mitotically stable mutants of M. circinelloides via targeted integration of the desired DNA. It does not require plasmid construction and its expression in the recipient organism. Instead, it involves the direct introduction of the guide RNA and the Cas9 enzyme and, in case of homology directed repair (HDR), the template DNA into the recipient strain. Efficiency of the method for non-homologous end joining (NHEJ) and HDR was tested by disrupting two different genes, i.e. carB encoding phytoene dehydrogenase and hmgR2 encoding 3-hydroxy-3-methylglutaryl-CoA reductase, of M. circinelloides. Both NHEJ and HDR resulted in stable gene disruption mutants. While NHEJ caused extensive deletions upstream from the protospacer adjacent motif, HDR assured the integration of the deletion cassette at the targeted site.
Similar content being viewed by others
Introduction
Members of the order Mucorales constitute a remarkable group of filamentous fungi. Some of them have biotechnological importance as producers of extracellular hydrolytic enzymes (primarily of lipases and proteases), organic acids (e.g. lactic, fumaric and malic acids), alcohol and carotenoids1,2, others are used in starter cultures of oriental food fermentations3. Several species, among others Rhizopus oryzae and Lichtheimia corymbifera, can be agent of opportunistic human infections, known as mucormycoses, which are the second most frequent filamentous fungal deep mycoses after aspergilloses4,5. Besides, certain members of the order, such as Mucor circinelloides, Phycomyces blakesleeanus and R. oryzae, are widely used model organisms in genetic and molecular biological studies, among others on light sensing6, molecular regulation and signal processes7,8, sexual reproduction and differentiation9,10, morphogenesis7,11, carotenogenesis1,12 and pathogenicity13,14. Furthermore, M. circinelloides f. lusitanicus used in the present study has been involved in several studies addressed to the role of RNA interference in the fungal cell15, mechanism and role of morphological dimorphism7,16, production of enzymes, carotenoids and other metabolites1 and the genetic and molecular background of the pathogenicity of Mucoral fungi7,17,18.
Appropriate tools for genetic manipulation including efficient and reliable methods for genetic transformation are basic requirements of cell biological and molecular studies as well as of strain improvement by genetic and metabolic engineering. Although genetic transformation of different Mucoral fungi has been reported several times, targeted gene disruption, deletion and stable integration of the introduced DNA into the host genome have remained a great challenge in this fungal group (for detailed reviews in this topic, see refs1,19,20). Plasmid DNAs introduced into Mucoral fungi generally do not integrate into the genome even if they harbor long sequences homologous with the targeted sites and remain autonomously replicating elements frequently causing mitotic instability of the transformants21. Moreover, such plasmids often form high molecular weight concatenated structures, which make difficult the interpretation of the results of the transformation experiments19,20,21. The latter phenomenon is explained by the activity of the non-homologous end joining (NHEJ), which is the dominant mechanism of double-strand break repair over homologous recombination in these fungi21. Application of linear DNA fragments to force the integration into the genome may lead to ectopic integration and/or unwanted rearranges, formation of concatemers, excision and recircularization22,23. One of the reasons of the frequent application of M. circinelloides in molecular studies is the experience that this organism is more amenable for genetic manipulation than other Mucoral fungi and several transformation methods has been developed for this fungus24,25,26. However, gene disruption or integration by homologous recombination can hardly be carried out in this fungus too and stable integrative transformants has rarely been reported23. The difficulties to achieve homologous recombination hampers the construction of mutant libraries, characterization of gene functions, examination of the pathogenicity or performance of recombinant strain improvement studies.
The CRISPR-Cas9 system is a new and versatile genome editing system, which has already been used to target and disrupt genes in some filamentous fungi and yeast, for example in Aspergillus strains27,28, Trichoderma reseii 29, Saccharomyces cerevisiae 30,31,32, S. boulardii 33, Candida albicans 34, C. glabrata 35, Cryptococcus neoformans 36 and Penicillium chrysogenum 37. Different methods are available for the CRISPR-Cas9 based genome editing in filamentous fungi (for the state of the art achievements in this field, see the review of Shi et al.38). The in vivo expression strategies involve the construction of and transformation with one or two plasmids, which contain the cas9 gene and the sequences encoding the elements of the guide RNA (gRNA), i.e. the CRISPR-RNA (crRNA) and the trans-activating crRNA (tracrRNA), as well as the protospacer fragment and assure their expression. Although these approaches work well, application of expression vectors for CRISPR-Cas9 genome editing has some drawbacks. Besides that, construction of the corresponding plasmid expressing the cas9 and the appropriate gRNA can be time consuming and laborious, these systems allow the subsistence and replication of the plasmids after the genome edition event, which can narrow the applicability of the technique and the resulting mutants. For example, such mutants may harbor bacterial antibiotic resistance genes and other bacterial sequences used to select the plasmids during the construction, which is not allowed in many applications. Moreover, long time presence of plasmids and foreign DNA may cause off-target effect as well as unwanted reorganizations and degradations of the transferred DNA and the genome, which have been observed frequently in case of Mucoral fungi19,20,23.
In a plasmid free strategy, a ribonucleoprotein (RNP) complex formed from the Cas9 enzyme and the in vitro transcribed gRNA is introduced into the cells to edit the targeted gene. Genome editing with RNP formation has been described in different human, animal and plant cell types and it was successfully used to modify the genome of P. chrysogenum 37,39,40,41,42,43,44. With this method the off-target effect is avoidable but the RNP can be degraded easily43.
Therefore, in this study, a CRISPR-Cas9 system without in vitro RNP formation and the using of plasmids has been developed and demonstrated as a tool for site-specific mutagenesis of M. circinelloides. The developed mutation strategy involves the introduction of only the gRNA, i.e. the duplex of the crRNA and the tracrRNA, into the fungal cells to guide the CRISPR-associated Cas9 nuclease and achieves RNA-guided double-strand breaks at the targeted DNA sequences. This plasmid free system has several advantages over the in vivo strategy; for example, no additional cloning steps are needed, the off-target effect can be avoided because of the transient exposure of Cas9 in the transformed cells and Cas9 can be active immediately after its uptake by the cells37.
To test this approach, two different genes of M. circinelloides, carB and hmgR2 encoding the enzymes phytoene dehydrogenase and 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) reductase, respectively, were disrupted and the mutants were characterized. HMG-CoA reductase is a key enzyme of the mevalonic acid pathway of terpene biosynthesis whereas phytoene dehydrogenase participates in the specific β-carotene biosynthesis, which can be considered as a side branch of the general mevalonic acid pathway.
Results
Disruption of the carB gene in M. circinelloides by NHEJ
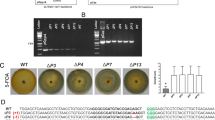
Protoplasts of the carotenoid producing M. circinelloides strains MS12 and CBS277.49 were co-transformed with the corresponding gRNA and the Cas9 nuclease. Selection of the transformants was based on the white color of their colonies. The original MS12 and CBS277.49 strains have yellow color due to their β-carotene content. Colonies, in which the targeted carB gene was disrupted, had white color since they were unable to synthesize β-carotene (Fig. 1A–C).
Colonies of Mucor circinelloides carB disruption mutants obtained by the CRISPR-Cas9 method. Panel A, selection of the transformed colonies obtained from the wild-type strain, CBS227.49 using NHEJ mutagenesis based on their white colony color. Panel B, colony color of the strain CBS227.49 and its carB′ disruption mutants (CBS227.49-carB′/1 and CBS227.49-carB′/2) created by NHEJ mutagenesis. Panel C, colony color of the strain MS12 and its carB disruption mutants created by NHEJ (MS12-carB′/1 and MS12-carB′/2) and HDR (MS12-carB + pyrG/1 and MS12-carB + pyrG/2) approaches.
To examine the effect of the amount of the gRNA and the Cas9 enzyme on the genome editing efficiency, different concentrations of these components ranging from 50 to 200 µM were added to the protoplasts (Table 1). If only 50 µM gRNA and 50 µM Cas9 were applied, no transformant colonies could be obtained. One hundred micromolar gRNA and Cas9, respectively, proved to be the most efficient to generate white colonies (i.e. to disrupt the carB gene) while the further increase of the gRNA and Cas9 amounts did not result in higher number of transformants. Thus, 100 µM of these components were used in all further experiments.
Using 100 µM gRNA and Cas9, transformation (i.e. disruption) frequencies were found to be 1.25 × 104 and 2 × 104 colonies per 105 protoplasts for MS12 and CBS277.49, respectively (Table 1), among which 9 and 13 colonies were isolated for the further analyses.
To prove the mutation, a region containing the targeted carB and the adjacent carRP (encoding another carotenogenic enzyme, the phytoene synthase/lycopene cyclase) genes was amplified by PCR from the DNA of the isolated transformants (positions of the primers are presented in Fig. 2A). Sequencing of the PCR products indicated that the mutation occurred at the targeted site in all of the tested transformants. However, extensive deletions (≥2.3 kb) with various lengths upstream from the protospacer, which involved a segment of the carB and almost the whole coding sequence of the carRP gene, were revealed in each case. Real-time quantitative reverse transcription PCR (qRT-PCR) analysis indicated the lack of carB transcripts in the transformants.
Genome editing strategy designed to disrupt the carB gene of Mucor circinelloides by the CRISPR-Cas9 method and positions of the primers used in the study (A) and PCR analysis of the transformants using the MccarB1 and MccarB2 primers (Table 2) (B). TGG indicates the PAM sequence; arrows show the orientations of the primers. Panel B: M, GeneRuler 1 kb DNA ruler (Thermo Scientific); 1, MS12 strain; 2, MS12-carB + pyrG/1 strain; 3, MS12- carB + pyrG/2 strain.
To test their mitotic stability, each transformant was passed several times onto selective and non-selective media. All of them proved to be mitotically stable retaining their white phenotype even after 20 cultivation cycles under non-selective growth conditions and the carB transcript could also not be detected by qRT-PCR in these colonies.
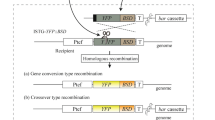
Integration of pyrG into the carB and the hmgR2 genes by homology directed repair (HDR)
Protoplasts of the M. circinelloides MS12 strain were co-transformed with the corresponding gRNA, the template DNA and the Cas9 nuclease. Template DNA served as the deletion cassette containing the pyrG gene as a selection marker and two fragments homologous to the target site to direct the HDR; pyrG encodes orotidine-5′-phosphate decarboxylase and complements the uracil auxotrophy of MS12. Figures 2A and 3A shows the genome editing strategy used in these experiments. Although the template DNAs harbored extensive homologous sequences, exclusive application of them (i.e. without the gRNA and Cas9) in different concentrations did not result in any transformants indicating the very low efficiency of homologous recombination in this fungus (Table 1). As the selection based on the complementation of uracil auxotrophy, control experiments using only the gRNA and the Cas9 without the appropriate template DNA also did not result in transformants (Table 1).
Genome editing strategy designed to disrupt the hmgR2 gene of Mucor circinelloides by the CRISPR-Cas9 method and positions of the primers used in the study (A) and PCR analysis of the transformants using the H2cDNS1 and H2cDNS8 primers (Table 2) (B). TGG indicates the PAM sequence; arrows show the orientations of the primers. Panel B: M, GeneRuler 1 kb DNA ruler (Thermo Scientific); 1, MS12 strain; 2, MS12-hmgR2 + pyrG strain.
When carB was targeted, transformation frequency was two colonies per 105 protoplasts (Table 1). Using the MccarB1 and MccarB2 primers (Table 2), a 3.2-kb fragment (Fig. 2B) was amplified by PCR from the DNA of the transformants designated as MS12-carB + pyrG/1 and MS12-carB + pyrG/2. In both cases, sequencing of the PCR products confirmed the disruption of the carB gene and the integration of the pyrG gene into the targeted site. Lack of transcripts was proven by qRT-PCR. Both transformants proved to be mitotically stable as their genotype indicated in Fig. 2 and, accordingly, their white phenotype (Fig. 1C) could be detected even after 20 cultivation cycles performed under non-selective conditions.
In case of the disruption of hmgR2, the transformation frequency was only one colony per 105 protoplasts (Table 1). Using H2cDNS1 and H2cDNS2 primers (Table 2), the expected 2.4-kb fragment containing the introduced pyrG (Fig. 3B) was amplified by PCR from the transformant designated as MS12-hmgR2 + pyrG. Sequencing also revealed that the CRISPR-Cas9-mediated HDR caused the expected modification (i.e. disruption of the hmgR2 by the integration of the pyrG) in the targeted site. No hmgR2 transcripts could be detected by qRT-PCR from this strain and it retained the integrated fragment and proved to be mitotically stable even after 20 cultivation cycles.
Characterization of the disruption mutants
Growth ability of the mutants was tested cultivating them on minimal medium (i.e. YNB) at different temperatures for four days in light and measuring the colony diameters daily. At the tested temperatures, significant differences in the growth of the MS12-hmgR2 + pyrG mutant and the original strains were not observed. However, all carB disruption mutants showed decreased colony size at 35 °C compared to the MS12 strain (Fig. 4).
Growth ability of the mutants obtained from the Mucor circinelloides MS12 strain at 35 °C. Strains were cultivated on YNB medium for four days under continuous light. Values followed by * and ** significantly differed from the corresponding value of the MS12 strain according to the two-sample t-test; p < 0.05 and p < 0.01, respectively.
All carB mutants failed to produce β-carotene and lycopene, while the carotenoid content of the hmgR2 mutant (457 ± 74 µg/g [dry weight]) did not change significantly compared to that of the original MS12 (476 ± 65 µg/g [dry weight]). In case of MS12-hmgR2 + pyrG, ergosterol content was also measured. Compared to that of the recipient MS12 strain (4.57 mg/g [dry weight]), it decreased (3.27 mg/g [dry weight]) significantly (p < 0.05) but not completely.
Growth of the different carB and hmgR2 disruption mutants and the MS12 strain under oxidative stress was also examined. Strains, in which carB was disrupted, showed no significant changes in their susceptibility to hydrogen peroxide (Supplementary Table S1). At the same time, the strain MS12-hmgR2 + pyrG displayed reduced growth in the presence of this compound (Supplementary Table S1).
Discussion
In this study, we transferred the Cas9 enzyme and the gRNA complex into the Mucoral fungus M. circinelloides, without in vitro RNP formation and the using of plasmids to disrupt the carB and the hmgR2 genes. The carB and the adjacent carRP genes are traditionally recommended for target sites of integration by homologous transformation as their disruption can be easily followed by the white phenotype of the transformant colonies24,45. Elements of the corresponding gRNA were simply synthesized by a commercial way and it was introduced into the recipient fungal strain together with the Cas9 enzyme using the polyethylene glycol (PEG)-mediated protoplast transformation technique, which is well established for these fungi1. This approach requiring only the in silico design of the protospacer sequence and the synthesis of the crRNA and the tracrRNA allows a fast but robust experimental setup and it is well known in genome editing of human and animal cells46.
Targeted gene disruption can be achieved by the error-prone NHEJ-mediated mutagenesis generating insertions or deletions (indels) at the site of the strand break caused by the Cas9 enzyme. In this way, carB of M. circinelloides could be successfully disrupted with considerable frequency and the resulting mutants were found to be stable. However, more than 2.3 kb long deletions were detected upstream from the protospacer adjacent motif (PAM) sequence in the resulting mutants. These deletions also affected the adjacent carRP gene, which is in the same carotenogenic cluster as carB and encodes the phytoene synthase/lycopene cyclase enzyme. Indels induced by NHEJ have most frequently been found to be a few tens of nucleotides long in fungi27,28. Thus, the presence of such extensive deletions is unusual and makes difficult the molecular analysis of the NHEJ-generated disruption mutants in Mucor. Moreover, it may lead to unwanted consequences, such as disruption of the adjacent gene or genes. Therefore, the application of a template DNA to induce HDR is preferred for the CRISPR-Cas9 mutagenesis of M. circinelloides.
Double-strand DNA break can increase the frequency of DNA repair by HDR as it was proven in S. cerevisiae and C. glabrata 30,35. To test and compare the efficiency of this method, we targeted two different genes (i.e. carB and hmgR2) transferring an appropriate template DNA (functioning as the deletion cassette) together with the gRNA and the Cas9 into the recipient strain. Transformation frequency was relatively low in both cases. However, it should be emphasized that several previous attempts using the classical genetic transformation methods to integrate the same template DNAs into the Mucor genome had remained unsuccessful.
HDR resulted in stable transformants, in which gene disruption occurred via the integration of the selection marker at the appropriate sites, indicating the usefulness of this methodology to obtain targeted gene disruption and/or integration in Mucor. Stability of the mutants was proven; reorganizations or degradations of the integrated DNA were not found.
Some features of the disrupted mutants were characterized. Apart from the yellow color, morphology of the mutants did not differ from those of the original strains. This lack of morphological consequences of the disruption of carotenoid biosynthesis was also observed in other organisms, such as Fusarium fujikuroi, Neurospora crassa, Xanthophyllomyces dendrorhous or P. blakesleeanus 47. At the same time, mutants showed reduced growth at 35 °C suggesting that carotenoid content has effect on the survival at higher temperatures. Previously, it was also found that carotenoid composition and amount may have a protective effect on M. circinelloides at higher temperatures25,48. In contrast with the fact that β-carotene and carotenoids are effective antioxidants and they are thought to be protective against oxidative stress in fungi47, susceptibility of the carB mutants to hydrogen peroxide did not change compared to that of the original strains suggesting that carotenoid content has not a significant role in the defense against oxidative stress in Mucor, at least against the tested oxidizing agent.
Due to the disruption of hmgR2, ergosterol content decreased while the carotenoid content did not change. M. circinelloides has three hmgR genes, i.e. hmgR1, hmgR2 and hmgR3, among which hmgR2 was previously found to be expressed predominantly during the whole life cycle of the fungus48. Manipulation of hmgR genes has been successfully used to change the carotenoid content in different organisms49,50,51. Previously, overexpression of hmgR2 and hmgR3 enhanced the carotenoid production of M. circinelloides 48. The result that disruption of hmgR2 did not affect the carotenoid content reinforce the overlapping functions of the two genes and the suggestion that hmgR3 has the major role in the carotenoid biosynthesis.
HMG-CoA reductase activity is thought to affect adaptation to certain environmental challenges, such as osmotic changes, different temperatures or oxidative stress52,53,54, via its role in the biosynthesis of the membrane component, ergosterol and other terpenoids, such as prenyl groups of certain proteins and ubiquinon55. Disruption of hmgR2 gene did not result in reduced or altered the fungal growth at the different temperatures. Previously, transcript abundance of hmgR2 found to be independent from the different cultivation temperatures48. Although ergosterol content decreased after disruption of hmgR2, it remained at a considerable level, which also reinforce the overlapping function of the other hmgR genes. The strain MS12-hmgR2 + pyrG displayed reduced growth in the presence of hydrogen peroxide. This feature can be in connection with the decreased ergosterol content. In S. cerevisiae, low ergosterol level caused sensitivity to paraquat-induced oxidative stress56,57. In fact, membrane permeability plays important role in the adaptation to oxidative stress and the altered sterol production may result in higher sensitivity to inducers of oxidative stress58,59,60.
This is the first report of the successful application of the CRISPR-Cas9 system for genetic modification of a Mucoral fungus. Moreover, an approach using directly the guide RNA and the template DNA together with the Cas9 enzyme without the need of plasmid construction was suggested and tested. This technique allows the robust and stable modification (i.e. gene deletion, disruption and/or integration) of M. circinelloides via the HDR mechanism. Elements of the genome editing system can be easily introduced into the fungus by a simple genetic transformation method, such as the well-developed PEG-mediated transformation. As the described method offers a rapid and reliable genome modification tool to construct mutant strains, it can enhance the various basic and applied studies involving Mucoral strains as model organisms.
Materials and Methods
Strains, media and growth conditions
Mucor circinelloides f. lusitanicus CBS 277.49 (wild type) and MS12 (leuA − and pyrG −) strains, were used in this study. The latter strain is auxotrophic for leucine and uracil but wild-type for the carotenoid biosynthesis48. For nucleic acid and carotenoid extractions, 106 sporangiospores were plated onto solid minimal medium (YNB; 10 g glucose, 0.5 g yeast nitrogen base without amino acids (Difco), 1.5 g (NH4)2SO4, 1.5 g sodium glutamate and 20 g agar per liter) supplemented with leucine and/or uracil (0.5 mg/ml) if required. Fungal cultures were grown for 4 days under continuous light at 25 °C. To examine the effect of the temperature on the growth of the mutants, fungal strains were cultivated on solid YNB at 20, 25, 30 and 35 °C and the number of the plated spores was 104. To test the mitotic stability of the transformants, malt extract agar (MEA; 10 g glucose, 5 g yeast extract, 10 g malt extract and 20 g agar per liter) was used as a complete, i.e. non-selective, medium.
Molecular techniques, gRNA and construction of the deletion cassette
Genomic DNA and total RNA were isolated using the ZR Fungal/Bacterial DNA MiniPrep (Zymo Research) and the Direct-zol RNA MiniPrep (Zymo Research) kits, respectively, according to the instructions of the manufacturer. PCR products were isolated and concentrated using Zymoclean Large Fragment DNS Recovery Kit (Zymo Research) and DNA Clean & Concentrator-5 (Zymo Research). To design the oligonucleotide sequences used in the study sequence data available in the M. circinelloides CBS277.49v2.0 genome database (DoE Joint Genome Institute; http://genome.jgi-psf.org/Mucci2/Mucci2.home.html 61) were used. The protospacer sequences designed to target the DNA cleavage in the carB and hmgR2 gene (CBS277.49v2.0 genome database ID: Mucci1.e_gw1.1.244.1) was the following: 5′-gagcgcattcaagaccacct-3′and 5′-ctctgatatcgtacgcccct-3′ which corresponds to the fragment between the nucleotide positions 220 and 239 downstream from the start codon of carB gene and 2117 and 2137 downstream from the start codon of hmgR2. Using these sequences, the Alt-R CRISPR crRNA and Alt-R CRISPR-Cas9 tracrRNA molecules were purchased from IDT. To form the crRNA:tracrRNA duplexes (gRNAs), the Nuclease-Free Duplex Buffer (IDT) was used according the instructions of the manufacturer. Deletion cassette also functioning as a template DNA for HDR was constructed by PCR using the Phusion Flash High-Fidelity PCR Master Mix (Thermo Scientific). In case of carB gene, at first, two fragments 992 and 986 nucleotides upstream and downstream from the protospacer sequence, respectively, and the M. circinelloides pyrG gene (CBS277.49v2.0 genome database ID: Mucci1.e_gw1.3.865.1) together with its promoter and terminator sequences were amplified using the primers listed in Table 2. The amplified fragments were fused in a subsequent PCR using the nested primers carBcrispfuz7 and carBcrispfuz8 (Table 2); the ratio of the fragments in the reaction was 1:1:1. The structure of the 3.7-kb fusion product constituting the deletion cassette and location of the primers are presented in Fig. 2A. To disrupt hmgR2 two fragment 1817 and 1698 nucleotides upstream and downstream from the protospacer sequence were fused also with pyrG in a subsequent PCR using nested primers h2p7 and h2p8 (Table 2).
qRT-PCR analysis
Transcript level of the carB and hmgR2 genes were analyzed by using the real-time qPCR technique. Reverse transcription reactions were carried out with the Maxima H Minus First Strand cDNA Synthesis Kit (Thermo Scientific) using random hexamer and oligo(dT)18 primers, following the instructions of the manufacturer. The qPCR experiments were performed in a CFX96 real-time PCR detection system (Bio-Rad) using the Maxima SYBR Green qPCR Master Mix (Thermo Scientific) and the primers presented in Table 2. The relative quantification of the copy number and the gene expression was achieved with the 2−ΔΔCt method using62 the actin gene (scaffold_07: 2052804-2054242) of M. circinelloides as a reference. Experiments were performed in biological and technical triplicates.
Transformation
Deletion cassette, guide RNA and Cas9 enzyme (Alt-R S.p. Cas9 Nuclease, IDT) were introduced together into the M. circinelloides MS12 strain by PEG-mediated protoplast transformation according to van Heeswijck and Roncero63. Protoplasts were prepared as described earlier64. In a transformation reaction, 5, 10 or 15 µg template DNA (deletion cassette), 50, 100 or 200 µM gRNA and 50, 100 or 200 µM Cas9 nuclease were added to the protoplasts. If the double-strand break was expected to be repaired by NHEJ instead of HDR, only the gRNA and the Cas9 were added to the protoplasts of the MS12 or the CBS CBS277.49 strain in the abovementioned amounts. Transformants were selected on solid YNB medium by the white color of the transformant colonies due to the disruption of the carB gene and/or the complementation of the uracil auxotrophy of the MS12 strain due to the expression of the pyrG gene. As a control, the transformation procedure was also carried out on the protoplasts without adding any transforming agents (i.e. the template DNA and the elements of the CRISPR-Cas9 system) in all cases. None of these control experiments resulted in transformant colonies.
Hydrogen peroxide susceptibility tests
Sensitivity of the fungal strains to menadione and H2O2 was examined in a 96-well microtiter plate assay. H2O2 were dissolved in liquid YNB media to prepare stock solutions. Final concentrations of H2O2 in the wells ranged from 0 to 0.5 mM; chemicals were diluted with liquid YNB medium. Inocula were prepared also in liquid YNB and the final amount of the sporangiospores in the wells was set to 104. Plates were incubated for 48 h at 25 °C and the optical density of the fungal cultures was measured at 620 nm using a Jupiter HD plate reader (ASYS Hitech). The uninoculated medium was used as the background for the calibration and fungal growth in the chemical-free medium was considered as 100%; all experiments were performed in triplicates.
Analysis of the carotenoid and ergosterol content
Carotenoid extraction was performed as described earlier65. For HPLC analysis of the extracts, dried samples were re-dissolved in 100 μL tetrahydrofuran supplemented with butylated hydroxytoluene (100 μg/mL) and 2 μL was subjected onto a Prodigy ODS-3 (4.6 × 150, ODS 3 μm) column (Phenomenex). Separation was carried out using a modular Shimadzu low-pressure gradient HPLC system as described previously23. A β-carotene lycopene standards (Sigma) were used for identification and quantification. Analysis and extraction of ergosterol were performed as described earlier48.
Data availability
The authors declare that all data supporting the findings of this study are available within the article and its Supplementary Information file, or are available from the corresponding author upon request.
References
Papp, T. et al. Improvement of industrially relevant biological activities in Mucoromycotina fungi, Gene Expression Systems in Fungi: Advancements and Applications (eds Schmoll, M. and Dattenböck, C.) 97–118 (Springer, 2016).
Voigt, K. et al. Genetic and metabolic aspects of primary and secondary metabolism of the Zygomycetes, The Mycota; III. Biochemistry and Molecular Biology (ed. Hoffmeister, D.) 361–385 (Springer, 2016).
Babu, P. D., Bhakyaraj, R. & Vidhyalakshmi, R. A low cost nutritious food “tempeh”- a review. World J. Dairy Food Sci. 4, 22–27 (2009).
Katragkou, A., Walsh, T. J. & Roilides, E. Why is mucormycosis more difficult to cure than more common mycoses? Clin. Microbiol. Infect. 20, 74–81 (2014).
Riley, T. T., Muzny, C. A., Swiatlo, E. & Legendre, D. P. Breaking the Mold: A review of Mucormycosis and current pharmacological treatment options. Ann. Pharmacother. 50, 747–757 (2016).
Corrochano, L. M. & Garre, V. Photobiology in the Zygomycota: multiple photoreceptor genes for complex responses to light. Fungal Genet. Biol. 47, 893–899 (2010).
Lee, S. C. et al. Calcineurin orchestrates dimorphic transitions, antifungal drug responses and host-pathogen interactions of the pathogenic mucoralean fungus. Mucor circinelloides. Mol. Microbiol. 97, 844–865 (2015).
Ruiz-Vázquez, R. M., Nicolás, F. E., Torres-Martínez, S. & Garre, V. Chapter two-distinct RNAi pathways in the regulation of physiology and development in the fungus Mucor circinelloides. Adv. Genet. 91, 55–102 (2015).
Camino, L. P., Idnurm, A. & Cerdá-Olmedo, E. Diversity, ecology, and evolution in Phycomyces. Fungal Biol. 119, 1007–1021 (2015).
Wöstemeyer, J. et al. Pheromone action in the fungal groups Chytridiomycetes and Zygomycetes and in the Oophytes, The Mycota (ed. Wendland, J.) 203–234 (Springer, 2016).
Wang, L. & Lin, X. Morphogenesis in fungal pathogenicity: shape, size, and surface. PLoS Pathog. 8, e1003027, https://doi.org/10.1371/journal.ppat.1003027 (2012).
Zhang, Y. et al. A new regulatory mechanism controlling carotenogenesis in the fungus Mucor circinelloides as a target to generate β-carotene over-producing strains by genetic engineering. Microb. Cell Fact. 15, 99, https://doi.org/10.1186/s12934-016-0493-8 (2016).
Ibrahim, A. S. et al. The high affinity iron permease is a key virulence factor required for Rhizopus oryzae pathogenesis. Mol. Microbiol. 77, 587–604 (2010).
Gebremariam, T. et al. CotH3 mediates fungal invasion of host cells during mucormycosis. J. Clin. Invest. 124, 237–250 (2014).
Nicolás, F. E. et al. The RNAi machinery controls distinct responses to environmental signals in the basal fungus Mucor circinelloides. BMC Genomics 16, 237, https://doi.org/10.1186/s12864-015-1443-2 (2015).
Valle-Maldonado, M. I. et al. Selection of reference genes for quantitative real time RT-PCR during dimorphism in the zygomycete Mucor circinelloides. Mol. Biol. Rep. 42, 705–711 (2015).
Lee, S. C., Li, A., Calo, S. & Heitman, J. Calcineurin plays key roles in the dimorphic transition and virulence of the human pathogenic zygomycete Mucor circinelloides. PLoS Pathog. 9, e1003625, https://doi.org/10.1371/journal.ppat.1003625 (2013).
Li, C. H. et al. Sporangiospore size dimorphism is linked to virulence of Mucor circinelloides. PLoS Pathog. 7, e1002086, https://doi.org/10.1371/journal.ppat.1002086 (2011).
Papp, T., Csernetics, Á., Nyilasi, I., Ábrók, M. & Vágvölgyi, C. Genetic transformation of Zygomycetes fungi, Progress In Mycology (eds Rai, M. K. & Kövics, G. J.) 75–94 (Springer, 2010).
Ibrahim, A.S. & Skory, C.D. Genetic manipulation of Zygomycetes in Medical mycology: cellular and molecular techniques. (ed. Kavanagh, K.) 305–326 (John Wiley & Sons, 2007).
Meussen, B. J., de Graaf, L. H., Sanders, J. P. & Weusthuis, R. A. Metabolic engineering of Rhizopus oryzae for the production of platform chemicals. Appl. Microbiol. Biotechnol. 94, 875–886 (2012).
Michielse, C. B. et al. Development of a system for integrative and stable transformation of the zygomycete Rhizopus oryzae by Agrobacterium-mediated DNA transfer. Mol. Gen. Genomics 271, 499–510 (2004).
Papp, T. et al. Canthaxanthin production with modified Mucor circinelloides strains. Appl. Microbiol. Biotechnol. 97, 4937–4950 (2013).
Gutiérrez, A., López-García, S. & Garre, V. High reliability transformation of the basal fungus Mucor circinelloides by electroporation. J. Microbiol. Methods 84, 442–446 (2011).
Papp, T., Csernetics, Á., Nyilasi, I., Vágvölgyi, C. & Iturriaga, E. A. Integration of a bacterial β-carotene ketolase gene into the Mucor circinelloides genome by the Agrobacterium tumefaciens-mediated transformation method, Microbial Carotenoids from Fungi: Methods and Protocols, Methods in Molecular Biology (ed. Barredo, J. L.) 123–132 (Humana Press, 2012).
Torres-Martínez, S. et al. Molecular tools for analysis in the zygomycete Mucor circinelloides, Microbial Carotenoids from Fungi: Methods and Protocols, Methods in Molecular Biology (ed. Barredo, J.) 85-107 (Humana Press, 2012).
Nødvig, C. S., Nielsen, J. B., Kogle, M. E. & Mortensen, U. H. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS One 10, e0133085, https://doi.org/10.1371/journal.pone.0133085 (2015).
Fuller, K. K., Chen, S., Loros, J. J. & Dunlap, J. C. Development of the CRISPR/Cas9 system for targeted gene disruption in Aspergillus fumigatus. Eukaryot. Cell 14, 1073–1080 (2015).
Liu, R., Chen, L., Jiang, Y., Zhou, Z. & Zou, G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discovery 1, 15007, https://doi.org/10.1038/celldisc.2015.7 (2015).
DiCarlo, J. E. et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas system. Nucleic Acid Res. 41, 4336–4343 (2013).
Zhang, G. C. et al. Construction of a quadruple auxotrophic mutant of an industrial polypoid Saccharomyces cerevisiae strain by using RNA-guided Cas9 nuclease. Appl. Environ. Microbiol. 80, 7694–7670 (2014).
Mans, R. et al. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of a multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 15, fov004, https://doi.org/10.1093/femsyr/fov004 (2015).
Liu, J. J. et al. Metabolic engineering of probiotic Saccharomyces boulardii. Appl. Environ. Microbiol. 82, 2280–2287 (2016).
Min, K., Ichikawa, Y., Woolford, C. A. & Mitchell, A. P. Candida albicans gene deletion with transient CRISPR-Cas9 system. mSphere 1, e00130–16, https://doi.org/10.1128/mSphere.00130-16 (2016).
Enkler, L., Richer, D., Marchand, A. L., Ferrandon, D. & Jossinet, F. Genome engineering in the yeast pathogen Candida glabrata using the CRISPR-Cas9 system. Sci. Rep. 6, 35766, https://doi.org/10.1038/srep35766 (2016).
Arras, S. D. et al. Targeted genome editing via CRISPR in the pathogen Cryptococcus neoformans. PLoS One 11, e0164322, https://doi.org/10.1371/journal.pone.0164322 (2016).
Pohl, C., Kiel, J. A. K. W., Driessen, A. J. M., Bovenberg, R. A. L. & Nygård, Y. CRISPR/Cas9 based genome editing of Penicillium chrysogenum. ACS Synth. Biol. 5, 754–764 (2016).
Shi, T.-Q. et al. CRISPR/Cas9-based genome editing of the filamentous fungi: the state of the art. Appl. Microbiol. Biotechnol. 101, 7435–7443 (2017).
Cho, S. W., Lee, J., Caroll, D., Kim, J. S. & Lee, J. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics 195, 1177–1180 (2013).
Sung, Y., Kim, J., Kim, H., Lee, J. & Jeon, J. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonuclease. Genome Res. 24, 125–131 (2014).
Zuris, J. A. et al. Cationic lipidmediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 33, 73–80 (2014).
Ramakrishna, S. et al. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 24, 1020–1027 (2014).
Kim, S., Kim, D., Cho, S. W., Kim, J. & Kim, J. S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019 (2014).
Lin, S., Staahl, B. R., Alla, R. K. & Doudna, J. A. Enhanced homology directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3, e04766, https://doi.org/10.7554/eLife.04766 (2014).
Garre, V., Barredo, J. L. & Iturriaga, E. A. Transformation of Mucor circinelloides f. lusitanicus protoplasts. Genetic Transformation Systems in Fungi (ed. van den Berg, M.A.) 49–59 (Springer, 2015).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nature Protocols 8, 2281–2308 (2013).
Avalos, J. & Limón, M. C. Biological roles of fungal carotenoids. Curr. Genet. 61, 309–324 (2015).
Nagy, G. et al. Transcription analysis of the three HMG-CoA reductase genes of Mucor circinelloides. BMC Microbiol. 14, 93, https://doi.org/10.1186/1471-2180-14-93 (2014).
Wang, G. Y. & Keasling, J. D. Amplification of HMG-CoA reductase production enhances carotenoid accumulation in Neurospora crass. Metab. Eng. 4, 193–201 (2002).
Shimada, H. et al. Increased carotenoid production by the food yeast Candida utilis through metabolic engineering of the isoprenoid pathway. Appl. Environ. Microbiol. 64, 2676–2680 (1998).
Yan, G. L., Wen, K. R. & Duan, C. Q. Enhanvement of β-carotene production by over-expression of HMG-CoA reductase coupled with addition of ergosterol biosynthesis inhibitors in recombinant Saccharomyces cerevisiae. Curr. Microbiol. 64, 159–163 (2012).
Vaupotič, T., Veranic, P., Petrovič, U., Gunde-Cimerman, N. & Plemenitaš, A. HMG-CoA reductase regulated by environmental salinity and activity is essential for halotolerance in halophilic fungi. Stud. Mycol. 61, 61–66 (2008).
Bidls, K. A., Hanson, T. E., Howell, K. & Nannen, J. HMG-CoA reductase is regulated by salinity at the level of transcription in Haloferax volcanii. Extremophiles 11, 49–55 (2007).
Rep, M., Krantz, M., Thevelein, J. M. & Hohmann, S. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p6Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J. Biol. Chem. 275, 8290–8300 (2000).
Kuzina, V. & Cerdá-Olmedo, E. Ubiquinone and carotene production in the Mucorales Blakeslea and Phycomyces. Appl. Microbiol. Biotechnol. 76, 991–999 (2007).
Marisco, G., Saito, S. T., Ganda, I. S., Brendel, M. & Pungartnik, C. Low ergosterol content in yeastadh1 mutant enhances chitin maldistribution and sensitivity to paraquat-induced oxidative stress. Yeast 28, 263–373 (2011).
Vincent, J. H., Anthony, G. B., Anthony, D. O., Peter, J. R. & Ian, W. D. Yeast genome-wide expression analysis identifies a strong ergosterol and oxidative stress response during the initial stages of an industrial large fermentation. Appl. Environ. Microbiol. 67, 4777–4787 (2003).
Higgins, V. J., Beckhouse, A. G., Oliver, A. D., Rogers, P. J. & Dawes, I. W. Yeast genome-wide expression analysis identifies a strong ergosterol and oxidative stress response during the initial stages of an industrial lager fermentation. Appl. Environ. Microbiol 69, 4777–4787 (2003).
Branco, M. R., Marinho, H. S., Cyrne, L. & Antunes, F. Decrease of H2O2 plasma membrane permeability during adaptation to H2O2 in Saccharomyces cerevisiae. J. Biol. Chem. 279, 6501–6506 (2004).
Thorpe, G. W., Fong, C. S., Alic, N., Higgins, V. J. & Dawes, I. W. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc. Natl. Acad. Sci. USA 101, 6564–6569 (2004).
Corrochano, L. M. et al. Expansion of signal transduction pathways in fungi by extensive genome duplication. Curr. Biol. 26, 1577–1584 (2016).
Livak, K. J. & Schmitgen, T. D. Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408 (2001).
van Heeswijk, R. & Roncero, M. I. G. High frequency transformation of Mucor with recombinant plasmidDNA. Carlsberg Res. Commun. 49, 691–702 (1984).
Nagy, Á., Vágvölgyi, C., Balla, É. & Ferenczy, L. Electrophoretic karyotype of Mucor circinelloides. Curr. Genet. 26, 45–48 (1994).
Papp, T. et al. Heterologous expression of astaxanthin biosynthesis genes in Mucor circinelloides. Appl. Microbiol. Biotechnol. 69, 526–531 (2006).
Acknowledgements
This study was supported by the “Momentum” Grant of the Hungarian Academy of Sciences (LP2016-8/2016) and the project GINOP-2.3.2-15-2016-00035. Á.Cs. is supported by the Hungarian Academy of Sciences Postdoctoral Research Programme (Postdoc-16, 45.0.080).
Author information
Authors and Affiliations
Contributions
G.N. and T.P. designed the approach and the experiments, participated in the experimental work, managed the study and wrote the manuscript. C.S., Á.C., A.G.V. and E.J.T. participated in the experimental work. C.V. participated in the data analysis, evaluation of the results and drafting the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nagy, G., Szebenyi, C., Csernetics, Á. et al. Development of a plasmid free CRISPR-Cas9 system for the genetic modification of Mucor circinelloides . Sci Rep 7, 16800 (2017). https://doi.org/10.1038/s41598-017-17118-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-17118-2
This article is cited by
-
Development of versatile and efficient genetic tools for the marine-derived fungus Aspergillus terreus RA2905
Current Genetics (2022)
-
Transcriptome sequencing and global analysis of blue light-responsive genes provide clues for high carotenoid yields in Blakeslea trispora
International Microbiology (2022)
-
Applications of CRISPR/Cas gene-editing technology in yeast and fungi
Archives of Microbiology (2022)
-
Exploring nano-enabled CRISPR-Cas-powered strategies for efficient diagnostics and treatment of infectious diseases
Journal of Nanostructure in Chemistry (2022)
-
Ten decadal advances in fungal biology leading towards human well-being
Fungal Diversity (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.