Abstract
Ceratophyllum demersum (common hornwort) is presently considered the worst invasive submerged aquatic macrophyte in New Zealand. We explored the global phylogeographic pattern of the species, based on chloroplast and nuclear DNA, in order to identify the origin of the invasive populations in New Zealand and to clarify if there were multiple introductions. The phylogeographic study identified geographically differentiated gene pools in North America, tropical Asia, Australia, and South Africa, likely native to these regions, and a recent dispersal event of a Eurasian-related haplotype to North America, New Zealand, Australia, and South Africa. At least two different invasive genotypes of this Eurasian-related haplotype have been found in New Zealand. One genotype is closely related to genotypes in Australia and South Africa, while we could not trace the closest relatives of the other genotype within our C. demersum sample set. Contrasting spectra of genetic distances in New Zealand and in a region within the native range (Denmark), suggest that the invasive population was founded by vegetative reproduction, seen as low genetic distances among genotypes. We also discovered the introduction of the same Eurasian-related haplotype in Australia and South Africa and that a cryptic invasion may be occurring in these continents.
Similar content being viewed by others
Introduction
Global trade and transport has dramatically increased the number of alien plant species and populations introduced to novel areas either intentionally or accidentally by human activity1,2. Islands often have unique and fragile ecosystems with ecological niches that have not been filled because of the distance from colonizing populations3,4. Furthermore, the geographical isolation of islands, such as New Zealand, makes them particularly vulnerable to invasions because of the lack of natural competitors and predators that control populations in their native ecosystems, i.e. introduced populations are released from the enemies of their native ranges according to the enemy release hypothesis5,6. The spread of alien species into New Zealand and the development of some of those into invasive nuisance plants threatening the indigenous flora have occurred several times in the past century and are likely to still occur7. Freshwater ecosystems are more sensitive to invasions than terrestrial habitats8, and in New Zealand several aquatic macrophyte species such as Ceratophyllum demersum L., Lagarosiphon major (Ridley) Moss and Egeria densa Planch. are spreading across freshwater ecosystems at a great cost to the local ecosystems and economy9.
In some instances, introduced genotypes may have conspecific native populations in the invaded range and be morphologically similar to the native genotypes but sufficiently different in ecology or physiology to become invasive and outcompete the conspecific native populations as well as other species. This type of invasion is called cryptic, as the new invaders are difficult to distinguish from native populations due to their morphological resemblance10. These invasions are particularly difficult to recognize before changes in ecosystems are observed10.
Ceratophyllum demersum L. (common hornwort) is presently considered the worst invasive submerged macrophyte in New Zealand as it grows vigorously, forming surface-reaching canopies, and replaces native plant communities3,11. It is also an economic burden as it clogs hydrostations and prevents fishing and recreational activities in several lakes due to production of thick biomass mats12. Ceratophyllum demersum achieved a cosmopolitan distribution about 2.5 million years before present13. Given the fossil record, the species is considered native to North America, Eurasia, Africa and Australia14,15,16. In New Zealand, C. demersum was recorded for the first time in 1961 in the Hawke’s Bay region on the east coast of the North Island. Since then it has spread to many lakes and streams across the North Island7. In 2002, C. demersum was also found in two locations in the South Island, where it has now been eradicated.
In this study, samples of C. demersum were collected from all over the world to investigate the phylogeography of the species and trace the origin of the invasive populations in New Zealand. We also assessed the role of genetic differentiation in the invasion in New Zealand, i.e. if the invasion is driven by a population with high genetic diversity, and thus has potential for rapid evolution, or by one or few genotypes with high phenotypic plasticity17. We used both chloroplast DNA sequences (cpDNA sequences) to trace long-distance dispersal events, and nuclear amplified fragment length polymorphisms (AFLPs) to resolve intra-haplotypic relationships (i.e. between individuals sharing chloroplast DNA), within distribution ranges.
Results
cpDNA sequences
In total, seven different haplotypes (Haplotype A-G, Fig. 1 and Supplementary Fig. S1) were identified by the cpDNA, within C. demersum (GenBank accession no. KJ093282- KJ093289 + KJ093291-KJ093300, Supplementary Table S1). The parsimony analysis (i.e. the simplest scientific explanation that fits the evidence) divided the haplotypes into two well-supported groups (Fig. 1 and Supplementary Fig. S1). One group included four haplotypes: the USA (US) (Haplotype A, GenBank accession no. KJ093282- KJ093284 and KJ093291- KJ093293, Cdemex 1 and rps16 respectively), Thailand (Th) (Haplotype C, GenBank accession no. KJ093287 and KJ093296), Australia (Aus) (Haplotype B, GenBank accession no. KJ093289 and KJ093300), and South Africa (SA) (Haplotype D, GenBank accession no. KJ093288 and KJ093299). The other group was represented by three closely related haplotypes (Haplotype E, F and G) differing in one substitution in the rps16 and one substitution in the Cdemex 1, and included most of our samples from the US, China (Ch), Aus, New Zealand (NZ), SA, and Europe (EU) independently from their geographic origin (Fig. 1). Of these three haplotypes, one haplotype (Haplotype G, GenBank accession no KJ093286 and KJ093295) was unique and found in one single sample collected in Italy (Trieste). The second haplotype (Haplotype E, GenBank accession no. KJ093285 Cdemex 1 and KJ093294 and KJ093297 rps16) was shared by all EU and Ch samples and showed variation at the microsatellite loci of the rps16 and Cdemex 1 regions. The third haplotype (Haplotype F, GenBank accession no. KJ093298, rps16) was shared by the samples from the US, NZ, Aus, and SA and did not show microsatellite variation in the two cpDNA regions studied. The phylogeographic structure remained unchanged when the microsatellite variation was omitted from the sequences (Supplementary Fig. S1). The microsatellite variation increased the geographic resolution within the first group and divided the US samples from those from Th, Aus, and SA samples (78% support for both groups), and suggested a closer relationship of the Th sample to the SA samples (83% support) rather than to the Aus samples (Fig. 1). Ceratophyllum demersum was separated from C. submersum in both trees with 100% support.
cpDNA regions (Cdemex 1 and rps16) consensus tree from jackknife analysis with 37% deletion (including cpDNA microsatellite loci). Black bold versus non-bold and dashed line indicate three different haplotypes differing in one base substitution within the global unresolved groups with 85% support. Haplotypes A-G are indicated in brackets after the sampleID. Different colours indicate variation of microsatellite loci within the European haplotype (incl. China, 154Ch and New Zealand, 109NZ).
AFLPs
The AFLP median-joining network divided the samples into three major groups: (1) C. submersum, (2) EU, and (3) the rest of the world, being the US, tropical Asia (Th and Ch), Aus, SA, and NZ in our sample set (Fig. 2). In agreement with the cpDNA sequences, the US and some Aus samples appeared as two well-resolved branches within the third group. The samples from NZ split into different branches with unresolved connections mostly to the Aus and SA nodes. No phylogeographic structure was detected within the EU group, or among the Danish (DK) samples (dark green nodes in Fig. 2). Two C. demersum EU samples appeared to belong to the C. submersum branch.
Median-joining network of AFLP data. The different colours indicate different geographic regions or different taxa. Ceratophyllum submersum (orange) and C. demersum: Europe (light green, Denmark: dark green), Asia (light blue), Australia (dark blue), New Zealand (purple), South Africa (dark red), and USA (red).
The spectra of genetic distances showed a similar discontinuous pattern with distinct groups of genetic distances in NZ and in the native range in DK (Fig. 3). The DK spectrum of pairwise genetic distances, which we used as a reference spectrum for interbreeding and non-interbreeding populations, split the genetic distances into two groups (Fig. 3a). The first group (range 0–30) showed a modal distribution and included pairwise genetic distances within C. demersum and within C. submersum. The second group (range 32–49) included pairwise genetic distances between C. demersum and C. submersum. Three samples of C. demersum showed similar genetic distances both to the other samples of C. demersum and to C. submersum. These may be regarded as an intermediate group of genetic distances (range 21–30) between the two species. One of these three samples was 201DK, which the network placed along the C. submersum branch, while the other two samples were included in the EU group (Fig. 2). The potential group of intermediate genetic distances between the two species prompted suspicion of morphologically undetectable interspecific hybrids might occasionally occur in C. demersum populations. Gene diversity (h) was 0.161 within the Danish C. demersum population and it decreased to 0.132 when the possible hybrids were omitted from the analysis (Table 1).
The spectrum of pairwise genetic distances within the NZ populations suggested two reproductively isolated C. demersum gene pools, given the bimodal and discontinuous distribution of genetic distances (Fig. 3b). The second peak included the genetic distances between the sample 132NZ (from Lake Tarawera) and the rest of the NZ samples, and revealed at least two discontinuously related genotypes and pointed to the possibility of two introductions. Compared with the spectrum of the DK populations, the NZ samples showed shorter pairwise genetic distances and a decreasing curve of genetic distances, whereas the genetic distances with sample 132NZ were of the same order of magnitude as between C. demersum and its suspected hybrids with C. submersum in DK. Gene diversity (h) was 0.110 in the NZ population and decreased to 0.069 when sample 132NZ was removed (Table 1). In an attempt to rule out the possibility of two morphologically similar Ceratophyllum species in NZ, we aligned our cpDNA sequences with those of C. echinatum and C. submersum in GenBank. Given the close AFLP relationship of the NZ samples to the US samples sharing the same haplotype, we assumed that C. echinatum might have contributed alleles to the gene pool of the US-Asian-Aus-NZ-SA branch of the network (Fig. 2). Ceratophyllum echinatum is morphologically similar to C. demersum and C. submersum. However, none of the NZ sequences matched the sequences of either C. echinatum or C. submersum in GenBank (matK and ITS).
Nei’s unbiased AFLP genetic identities showed that the most similar gene pools in our sample set to that of the NZ invasive population are in SA, Aus, and in Ch (for sample 172NZ), based on the pairwise genetic identities (Table 2). However, discontinuous pairwise genetic distances within these regions indicated two distantly related gene pools in SA and Aus (Table 3). The NZ samples had the shortest genetic distances to samples in the south of SA and in the east coast of Aus, and such samples in SA and Aus had shorter genetic distances to each other than to the conspecific genotypes in SA and Aus (Table 3). AFLP gene diversity (h) increased to 0.072 for the population of the NZ samples and its shortest genetically distant genotypes in SA and Aus (Table 1).
The relationship of sample 172NZ from the most northern sampling locality in NZ with the Ch sample (Table 2) appeared weaker than the relationship of this sample with the SA and Aus samples (Table 3). We could not trace any close relatives of sample 132NZ collected in Lake Tarawera within our sample set.
Discussion
Phylogeography of Ceratophyllum demersum
The parsimony analyses of cpDNA sequences combined with AFLPs suggest the worldwide phylogeographic structure of C. demersum to consist of two layers (Fig. 1). The first layer is a relic of an ancient disjunct distribution marked by the well-resolved branches in the consensus trees of the US, Th, Aus and SA samples (Haplotype A–D), and the second layer is unstructured which is likely due to a recent dispersal event of two closely related haplotypes. One of these haplotypes is dominant in the modern Eurasian populations (Haplotype E) and the other has dispersed across North America, NZ, Aus and SA (Haplotype F). The median-joining network of nuclear AFLPs (Fig. 2) has a similar structure of well resolved and unresolved branches which further support the occurrence of ancient, likely native populations in the US and Aus (well resolved branches), and of undifferentiated, intercontinentally-wide spread populations of closely related genotypes, occurring in North America, NZ, Aus and SA (unresolved branches), likely resulting from recent dispersal. In the US and Aus, where the recently introduced genotypes are sympatric with the ancient native populations, the network suggests that gene exchange has occurred, as some genotypes of the introduced haplotypes are placed in the branches of the native haplotypes. Long-distance dispersal by birds is regarded as a viable explanation for widely disjunct aquatic plant distributions13. However, human-mediated dispersal, such as the aquarium trade, is also a well-documented source of exotic propagules in the recent invasion history of many aquatic species18 and may have facilitated recent and repeated dispersal events among continents, and contact among previously isolated populations as suggested by the present study.
C. demersum in New Zealand
Ceratophyllum demersum is a popular ornamental plant for aquaria and was sold in NZ until it was banned in 19827. In NZ, it shows typical invasive traits such as high growth rates and creation of monocultures19,20. In our study, at least two introduced genotypes of C. demersum were found in NZ. One of these was found in several lakes in NZ. The nuclear DNA of this frequent genotype is closely related to that of genotypes in SA and Aus sharing the same Eurasian-related haplotype (Haplotype F) (Fig. 2). The other genotype was found only once in our study, at Lake Tarawera in the North Island, and we could identify neither its origin nor its closest relationships within our sample set, despite it being the same haplotype as the rest of the NZ samples (Haplotype F). The species is spreading effectively in New Zealand with aid from fishing boats and boat trailers9, but also naturally via downstream transport of vegetative fragments7. Vegetative reproduction is an advantage for introduced species in a novel environment as it enables the species to establish and spread instantly after introduction and this has been found to be positively correlated with invasion success21. Field observations suggest that C. demersum is reproducing primarily by fragmentation in New Zealand7 and seeds have never been observed in the populations (Dr J Clayton, pers. comm.). This is supported by our findings, as the spectrum of genetic distances (Fig. 3b) indicates that the two genotypes found in NZ do not interbreed, and that the population of the most frequent of the two genotypes has been founded by clonal reproduction. The short pairwise genetic distances among the samples of the frequent genotype (mode = 2, Fig. 3) and the decreasing distribution of genetic distances fit the clonal mode of reproduction22. In the case of sexually reproducing and outcrossing organisms, a normal distribution of genetic distances is expected22, that is, a pattern more similar to the one we detected in DK with a mode of genetic distances around 10. Nevertheless, a larger sample of populations from NZ should be analysed to conclusively address vegetative vs. sexual reproduction and interbreeding within and between the two distantly related genotypes because the study of Les in Wisconsin, USA, showed that sexual recombination can occasionally occur in C. demersum populations, despite genetic diversity may be low23.
C. demersum invasion in NZ is driven by high phenotypic plasticity
Invasive populations are often found to have lower genetic diversity than the populations in their native ranges due to genetic bottlenecks and founder effect24,25. This diminishes the width of environmental conditions under which the introduced populations may thrive, but this can be counteracted by a high level of plasticity in the introduced populations2,26. Populations harbouring inherently high phenotypic plasticity can hypothetically inhabit a diverse range of new localities without undergoing genetic adaptation through natural selection. Phenotypic plasticity has therefore been proposed as a factor contributing to the development of invasiveness in plants17,27,28,29,30,31,32,33.
In two earlier studies, we explored the phenotypic plasticity of a population in New Zealand compared to a population from within the native range (DK). From the results in the present study we can conclude, that the previously studied population from NZ belongs to the most frequent genotype found in New Zealand. The NZ population was shown to have a higher level of phenotypic plasticity in photosynthesis and relative growth rate in response to temperature and nitrogen concentration in the water compared to the native population34,35. The low levels of genetic diversity in C. demersum in New Zealand detected in this study may therefore be overcome by high inherent phenotypic plasticity in the present genotypes, as also shown for other aquatic macrophytes32,36.
More Ceratophyllum species in New Zealand?
The genus Ceratophyllum includes at least six species of which only C. demersum is cosmopolitan, whereas the other species have restricted distribution ranges37. The other Ceratophyllum species are allopatric among each other, but sympatric with C. demersum, suggesting that hybridization opportunities might have occurred for C. demersum 37. Furthermore, hybridization can stimulate the evolution of invasiveness38 and has occurred frequently in similar invasive aquatic plants, such as the invasive watermilfoils Myriophyllum spicatum x sibiricum 39 and Myriophyllum heterophyllum x hippuroides 19 in North America. There is no evidence of hybrids from our study. However, the high genetic distances between the two NZ genotypes comparable to those between the C. demersum and C. submersum populations in the native range rise a suspicion that hybrids may exist (Fig. 3). Such distances may be due to the occurrence of two Ceratophyllum taxa or an introgressed taxon in our sample set. Only a study of nuclear DNA variation can confirm or rule out the hypothesis of two taxa.
Possible cryptic invasion in SA and Aus
The introduction of conspecific genotypes may hide cryptic invasions in regions where the species is native10,40. In our study the populations in SA and Aus have high AFLP genetic identity to the genotype that is most frequent in the invasive population in NZ (Table 3, Fig. 2). The SA and Aus genotypes also share the same haplotype and a vigorous growth and similar invasive behaviour (J Coetzee and Dr T Bickel, pers. comm.). However, in SA and Aus, C. demersum is not classified as “invasive”, as this term by definition is restricted to non-native, introduced species41. In the sampled regions in SA and Aus there are also native populations and the introduction of a new haplotype might have been unobserved. C. demersum is still sold as an aquarium plant in SA and Aus, which may have been the source of a recent introduction in these countries. Strikingly, the genetic structure of the Eurasian haplotype (Haplotype F) occurring in the US, NZ, Aus and SA is almost identical to that of the invasive haplotype M of Phragmites australis (Cav.) Trin ex Steudel (common reed) in North America42, which can serve as a reference of a recent cryptic invasion whose genetic dynamics have been exhaustively studied and documented10,42,43,44,45,46. A case of convergent evolution in NZ, Aus and SA cannot be ruled out; however, it would be very unlikely that the same mutations (in the chloroplast and in the nuclear DNA) had occurred independently in three disjunct ranges. In the case of independent mutations we would also expect diverging alleles among the populations of the three continents due to allopatric evolution, and an increase in gene diversity when the genotypes of the three populations are considered together. In our study, the genotypes in NZ, Aus and SA shared the same cpDNA sequences and diverged in 2 to 11 AFLP fragments, gene diversity did not increase significantly when we merged the Aus, SA and NZ genotypes and was about half of that found in the Danish population (Table 3). This is in contrast with local speciation processes and supports the hypothesis of a cryptic invasion.
Materials and Methods
Plant material
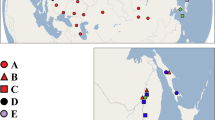
Shoots of C. demersum were collected by colleagues and botanists in geographically widespread locations covering as much of the distribution range of the species as possible, and include both native and introduced populations (Fig. 4). A total of 65 samples from different locations were included in the analyses. We also collected samples of C. submersum in Denmark and in the Czech Republic to use as an outgroup for our phylogeography. We checked taxonomic leaf traits of the samples received in order to confirm the species identification made by the collectors. C. demersum has dichotomous leaves forked 1–2 times, into 2–4 leaf segments and C. submersum leaves are most often forked 2–3 times, producing 3–8 leaf segments47.
Sampling locations for C. demersum: Europe (light green), Denmark (dark green), Asia (light blue), Australia (dark blue), New Zealand (purple), South Africa (dark red), and USA (red), and for C. submersum (orange). The samples are placed according to coordinates determined by the collectors. Individual sample numbers have been added where they were relevant. The maps were produced using the computer program ArcGIS 10.5 R-ESRI (www.esri.com).
DNA Extractions
DNA was extracted from apical parts of shoots dried with silica gel using the E.Z.N.A.TM SP Plant DNA Kit (Omega Bio-Tek Inc., Norcross, Georgia, USA). The protocol was modified for dried samples as suggested by Lambertini et al.42 to include grinding of the sample in liquid nitrogen and a small amount of autoclaved sand prior to addition of extraction buffer. In addition, incubation at 65 °C was prolonged to 60 min and incubation on ice was 30 min.
AFLP
The AFLP genotyping was performed using a slightly modified protocol according to Vos et al.48. Several primer combinations were initially tested on a subset of samples including the most geographically diverse samples to identify primer combinations that produced an adequate number of well separated, clearly polymorphic and reproducible fragments. The full set of samples were analysed using two sets of primers, based on the number of well separated, clear polymorphic fragments: (1) E-CAGcy (5′-GACTGCGTACCAATTCCAG-3′) + M-CAG (5′-GATGAGTCCTGAGTAACAG-3′) and (2) E-CGTcy (5′-GACTGCGTACCAATTCCGT-3′) + M-AGG (5′-GATGAGTCCTGAGTAAAGG-3′). Primers were 6-FAM labelled. Restriction digestion and adapter ligation were performed simultaneous in a 20 µL reaction on approx. 20 ng of genomic DNA, using 1.25 U of each of the restriction enzymes EcoRI and MseI and 0.3 U T4 DNA ligase to ligate 1.25 pmol EcoRI and 12.5 pmol MseI double-stranded nucleotide adapters. We incubated the restriction and ligation reaction in a thermal cycler (Gradient Cycler, Bio-rad Laboratories, Hercules, California, USA) at 37 °C for 4 h followed by a 0.1 °C/s decrease to 16 °C for 2 h and finally 10 min at 70 °C. The restricted DNA was diluted four times prior to preamplification. One-nucleotide selective primers were used in the preamplification, performed with the program: 20 cycles of 94 °C for 30 s, 56 °C for 60 s, 72 °C for 60 s. The preamplification product was diluted 10 times prior to selective amplification. For the selective amplification the program was: 94 °C for 30 s, 65 °C for 30 s (decreased by 0.7 °C/cycle for the subsequent 12 cycles) and 72 °C for 60 s, followed by 23 cycles at 94 °C for 30 s, 56 °C for 30 s and 72 °C for 60 s. The PCR product from the selective amplification was diluted 20 times prior to electrophoresis. The AFLPs were run in an ABI sequencer with an internal ladder in each sample containing 16 fragments ranging in size from 35 bp to 500 bp.
DNA sequencing
We downloaded all C. demersum sequences from GenBank and tested variation in 13 chloroplast regions: trnQ-rps1649, trnT-trnL50, trnL-trnF50, matK51, rbcL gene52,53, rbcL–psaI54, rpl1655, rpl3249 and the two nuclear internal transcribed spacers of nuclear ribosomal DNA (ITS) 56 in a subset of intercontinental samples. Given the low intraspecific variation in the published and tested sequences, we also screened variable regions within the chloroplast genome sequenced by Moore et al.57 (accession no. EF614270.1, GI: 148508422) and designed specific primers in two non-coding regions rich in microsatellites (Cdemex 1 and 2).
Based on the variation in the subset of samples, we chose to include two regions in the analysis, which showed both substitutions and variation at the microsatellite loci. We chose the rps16 gene encoding the S16′ protein, which is a component of the 40 S ribosomal subunit and is amplified by forward primer (rps16F) 5′-GTGGTAGAAAGCAACGTGCGACTT-3′ and reverse primer (rps16R) 5′-TCGGGATCGAACATCAATTGCAAC-3′58 and a microsatellite rich region of the cpDNA amplified by the specifically designed forward primer (Cdemex 1 F) 5′-CTATTGACCCCAAGTTCCCT-3′ and reverse primer (Cdemex 1 R) 5′-CAGGCCGTATCGACGGAAAG-3′.
The mastermix for one amplification reaction included 10 ng DNA, 5 µl Red Taq DNA Polymerase 2X Mastermix (VWR – Bie & Berntsen, Herlev, Denmark), 5 pmol of forward and reverse primer, sterile water was added to reach a final volume of 10 µl. PCR amplification was run with an initial step of 3 min at 95 °C, followed by 40 cycles of 95 °C for 30 s, 52 °C for 40 s, 72 °C for 1 min and terminated by a final step of 72 °C for 7 min. Subsequently, 3 µl of PCR products were run on a 1% agarose gel with EtBr for approx. 1 hour (120 V) to check amplification quality and quantity. Sequencing with both forward and reverse primers was performed at GATC Biotech AG, Köln, Germany. All different haplotypes obtained by the cpDNA sequencing were deposited in GenBank.
Data analysis
AFLPs
Genious R6 ver. 6.0.6 (Biomatters Ldt., Auckland, New Zealand) was used to analyse the AFLP data. The internal ladder peaks were aligned and the resulting aligned chromatograms were scored manually for the presence/absence of peaks of equal size.
For the first primer combination we scored fragments in the range between 70 to 300 bp in size. For the second primer combination we scored fragments between 40 and 400 bp. In total we scored 104 polymorphic fragments including both C. demersum and C. submersum. The DNA was extracted a second time from samples showing diverging chromatograms. The subset of samples used for primer screening was amplified multiple times to ensure reproducibility of the scored peaks. We removed genetically identical AFLP profile samples which were collected from the same locations. We entered question marks for uncertain peaks, either because of low intensity or uncertain size.
cpDNA sequences
The sequences were aligned with the program Geneious R6 ver. 6.0.6. To avoid overestimation of the number of mutations associated with mononucleotide repeats we coded repeat motifs (microsatellites) as multistate characters59. Hereby, differences due to the number of repeats were counted as one character, independently of their number. Furthermore, indels were coded with two states (presence = 1, absence = 0). The final matrix for the two sequences combined contained 1518 characters. There were 9 substitutions and 5 variable microsatellite loci in total among the C. demersum samples. Because of possible DNA slippage at microsatellite loci, the data set was analysed with and without microsatellite repeat motifs, and haplotypes were defined only by substitutions.
The resulting AFLP and cpDNA sequencing results were analysed separately. The cpDNA sequences were analysed with PAUP, ver. 4.0b10 (Phylogenetic Analysis Using Parsimony, Swofford). The data were subject to a jackknife analysis with 37% character deletion, “full heuristic search” and 1000 replicates60 and the trees were rooted with five samples of C. submersum as a functional outgroup. The consensus tree obtained from the cpDNA sequence matrix was compared with that obtained by the same matrix of cpDNA sequence data, but omitting the microsatellite variation.
The AFLP binary matrix was analysed in a median-joining network with the program Network ver. 4.6.1.1. Phylogenetic Network Constructions (Fluxus Technologies Ltd., Clare, Suffolk, UK) which combines parsimony with minimum spanning tree. The data from the AFLPs were also used to calculate pairwise Nei’s unbiased genetic identities61 among geographic regions with GenAlEx ver. 6.562 and Euclidean pairwise genetic distances (i.e pairwise no. of changes). Euclidean genetic distances were used to construct the spectrum of genetic distances and assess reproduction mode of the NZ population. A normal distribution of genetic distance frequencies indicates sexual reproduction, whereas a decreasing curve of genetic distances peaking close to zero is more likely due to clonal reproduction and somatic mutations22, and/or artefactual AFLP polymorphisms. We compared the NZ spectrum of genetic distance with that of a native population in Denmark (DK). We included C. demersum and C. submersum populations in the DK spectrum in order to have a reference of genetic distance patterns within the interbreeding population of C. demersum and between the non-interbreeding populations of C. demersum and C. submersum.
We also calculated AFLP gene diversity (h) (or expected heterozygosity) within the interbreeding Danish population and the introduced population in NZ, and within haplotypes and populations at different geographic scales, in order to identify populations of closely related genotypes.
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Bossdorf, O. et al. Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia 144, 1–11, https://doi.org/10.1007/s00442-005-0070-z (2005).
Roman, J. & Darling, J. A. Paradox lost: genetic diversity and the success of aquatic invasions. Trends Ecol. Evol. 22, 454–464 (2007).
Wells, R. D. S., de Winton, M. D. & Clayton, J. S. Successive macrophyte invasions within the submerged flora of Lake Tarawera, Central North Island, New Zealand. New Zeal. J. Mar. Fresh. 31, 449–459, https://doi.org/10.1080/00288330.1997.9516778 (1997).
Denslow, J. S. Weeds in Paradise: Thoughts on the Invasibility of Tropical Islands. Ann. Mo. Bot. Gard. 90, 119–127, https://doi.org/10.2307/3298531 (2003).
Thomas, M. B. & Reid, A. M. Are exotic natural enemies an effective way of controlling invasive plants? Trends Ecol. Evol. 22, 447–453 (2007).
Keane, R. M. & Crawley, M. J. Exotic plant invasions and the enemy release hypothesis. Trends Ecol. Evol. 17, 164–170, https://doi.org/10.1016/S0169-5347(02)02499-0 (2002).
de Winton, M. D., Champion, P. D., Clayton, J. S. & Wells, R. D. S. Spread and status of seven submerged pest plants in New Zealand lakes. New Zeal. J. Mar. Fresh. 43, 547–561 (2009).
Shea, K. & Chesson, P. Community ecology theory as a framework for biological invasions. Trends Ecol. Evol. 17, 170–176, https://doi.org/10.1016/S0169-5347(02)02495-3 (2002).
Compton, T. J., De Winton, M., Leathwick, J. R. & Wadhwa, S. Predicting spread of invasive macrophytes in New Zealand lakes using indirect measures of human accessibility. Freshwater Biol. 57, 938–948, https://doi.org/10.1111/j.1365-2427.2012.02754.x (2012).
Saltonstall, K. Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. P. Natl Acad. Sci. USA 99, 2445–2449 (2002).
de Winton, M. D., Clayton, J. S. & Edwards, T. Incorporating invasive weeds into a plant indicator method (LakeSPI) to assess lake ecological condition. Hydrobiologia 691, 47–58 (2012).
Pieterse, A. H. In Aquatic weeds: the ecology and management of nuisance aquatic vegetation (eds A. H. Pieterse & K. J. Murphy) Ch. 1, 3–16 (Oxford University Press, 1990).
Les, D. H., Crawford, D. J., Kimball, R. T., Moody, M. L. & Landolt, E. Biogeography of discontinuously distributed hydrophytes: A molecular appraisal of intercontinental disjunctions. Int. J. Plant Sci. 164, 917–932 (2003).
Herendeen, P. S., Les, D. H. & Dilcher, D. L. Fossil Ceratophyllum (Ceratophyllaceae) from the Tertiary of North America. Am. J. Bot. 77, 7–16 (1990).
Dilcher, D. L. & Wang, H. An early cretaceous fruit with affinities to Ceratophyllaceae. Am. J. Bot. 96, 2256–2269 (2009).
Short, P. S. In Flora of the Darwin Region Northern Territory botanical bulletin no. 37 (eds P. S. Short & I. D. Cowie) 1–2 (Northern Territory Herbarium, Department of Natural Resources, Environment, the Arts and Sport, 2011).
Richardson, D. M. & Pysek, P. Plant invasions: Merging the concepts of species invasiveness and community invasibility. Prog. Phys. Geog. 30, 409–431 (2006).
Lockwood, J. L., Hoopes, M. F. & Marchetti, M. P. Invasion Ecology. 1 edn, (Blackwell Publishing Ltd., 2007).
Spencer, W. & Bowes, G. In Aquatic weeds: the ecology and management of nuisance aquatic vegetation (eds A. H. Pieterse & K. J. Murphy) Ch. 4, 39–73 (Oxford University Press, 1990).
Wells, R. D. S., Bannon, H. J. & Hicks, B. J. Control of macrophytes by grass carp (Ctenopharyngodon idella) in a Waikato drain, New Zealand. New Zeal. J. Mar. Fresh. 37, 85–93 (2003).
Kolar, C. S. & Lodge, D. M. Progress in invasion biology: Predicting invaders. Trends Ecol. Evol. 16, 199–204 (2001).
Rozenfeld, A. F. et al. Spectrum of genetic diversity and networks of clonal organisms. J. Roy. Soc. Interface 4, 1093–1102 (2007).
Les, D. H. Genetic diversity in the monoecious hydrophile Ceratophyllum (Ceratochyllaceae). Am. J. Bot. 78, 1070–1082 (1991).
Ward, S. M., Gaskin, J. F. & Wilson, L. M. Ecological genetics of plant invasion: What do we know? Invas. Plant Sci. Mana. 1, 98–109 (2008).
Amsellem, L., Noyer, J. L., Le Bourgeois, T. & Hossaert-Mckey, M. Comparison of genetic diversity of the invasive weed Rubus alceifolius Poir. (Rosaceae) in its native range and in areas of introduction, using amplified fragment length polymorphism (AFLP) markers. Mol. Ecol. 9, 443–455 (2000).
Geng, Y.-P. et al. Phenotypic plasticity rather than locally adapted ecotypes allows the invasive alligator weed to colonize a wide range of habitats. Biol. Invasions 9, 245–256 (2007).
Sultan, S. E. Phenotypic plasticity for plant development, function and life history. Trends Plant Sci. 5, 537–542 (2000).
Sakai, A. K. et al. The population biology of invasive species. Annual Review of Ecological Systems 32, 305–332 (2001).
Richards, C. L., Bossdorf, O., Muth, N. Z., Gurevitch, J. & Pigliucci, M. Jack of all trades, master of some? On the role of phenotypic plasticity in plant invasions. Ecol. Lett. 9, 981–993, https://doi.org/10.1111/j.1461-0248.2006.00950.x (2006).
Hulme, P. E. Phenotypic plasticity and plant invasions: is it all Jack? Funct. Ecol. 22, 3–7 (2008).
Ren, M. X. & Zhang, Q. G. The relative generality of plant invasion mechanisms and predicting future invasive plants. Weed Res. 49, 449–460 (2009).
Riis, T. et al. Invasion strategies in clonal aquatic plants: are phenotypic differences caused by phenotypic plasticity or local adaptation? Ann. Bot.-London 106, 813–822 (2010).
Davidson, A. M., Jennions, M. & Nicotra, A. B. Do invasive species show higher phenotypic plasticity than native species and, if so, is it adaptive? A meta-analysis. Ecol. Lett. 14, 419–431 (2011).
Hyldgaard, B. & Brix, H. Intraspecies differences in phenotypic differences: Invasive versus non-invasive populations of Ceratophyllum demersum. Aquat. Bot. 97, 49–56 (2012).
Hyldgaard, B., Sorrell, B., Olesen, B., Riis, T. & Brix, H. Geographically distinct Ceratophyllum demersum populations differ in growth, photosynthetic responses and phenotypic plasticity to nitrogen availability. Funct. Plant Biol. 39, 774–783 (2012).
Zhang, Y. Y., Zhang, D. Y. & Barrett, S. C. H. Genetic uniformity characterizes the invasive spread of water hyacinth (Eichhornia crassipes), a clonal aquatic plant. Mol. Ecol. 19, 1774–1786 (2010).
Les, D. H. The evolution of achene morphology in Ceratophyllum (Ceratophyllaceae), IV. Summary of proposed relationships and evolutionary trends. Syst. Bot. 14, 254–262 (1989).
Ellstrand, N. C. & Schierenbeck, K. A. Hybridization as a stimulus for the evolution of invasiveness in plants? P. Natl Acad. Sci. USA 97, 7043–7050 (2000).
Sturtevant, A. P. et al. Molecular characterization of Eurasian watermilfoil, Northern milfoil, and the invasive interspecific hybrid in Michigan lakes. J. Aquat. Plant Manage. 47, 128–135 (2009).
Pysek, P. et al. Hitting the right target: taxonomic challenges for, and of, plant invasions. AoB Plants 5 (2013).
Richardson, D. M. et al. Naturalization and invasion of alien plants: concepts and definitions. Divers. Distrib. 6, 93–107 (2000).
Lambertini, C. et al. A phylogeographic study of the cosmopolitan genus Phragmites (Poaceae) based on AFLPs. Plant Syst. Evol. 258, 161–182 (2006).
Lambertini, C. et al. Tracing the origin of Gulf Coast Phragmites (Poaceae): A story of long-distance dispersal and hybridization. Am. J. Bot. 99, 538–551 (2012).
Kettenring, K. M. & Mock, K. E. Genetic diversity, reproductive mode, and dispersal differ between the cryptic invader, Phragmites australis, and its native conspecific. Biol. Invasions 14, 2489–2504, https://doi.org/10.1007/s10530-012-0246-5 (2012).
Meyerson, L. A. & Cronin, J. T. Evidence for multiple introductions of Phragmites australis to North America: Detection of a new non-native haplotype. Biol. Invasions 15, 2605–2608, https://doi.org/10.1007/s10530-013-0491-2 (2013).
Lambertini, C. Heteroplasmy due to chloroplast paternal leakage: another insight into Phragmites haplotypic diversity in North America. Biol. Invasions 18, 2443–2455, https://doi.org/10.1007/s10530-016-1193-3 (2016).
Lowden, R. M. Studies on the submerged genus Ceratophyllum L. in the neotropics. Aquat. Bot. 4, 127–142 (1978).
Vos, P. et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23, 4407–4414 (1995).
Shaw, J., Lickey, E. B., Schilling, E. E. & Small, R. L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am. J. Bot. 94, 275–288 (2007).
Taberlet, P., Gielly, L., Pautou, G. & Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 17, 1105–1109 (1991).
Cuénoud, P. et al. Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am. J. Bot. 89, 132–144 (2002).
Les, D. H., Garvin, D. K. & Wimpee, C. F. A phylogeny of ancient genus Ceratophyllum (Ceratophyllaceae) derived from DNA sequence data. Am. J. Bot. 79, 151 (1992).
Savolainen, V. et al. Phylogenetics of flowering plants based on combined analysis of plastid atpB and rbcL gene sequences. Syst. Biol. 49, 306–362 (2000).
Saltonstall, K. A set of primers for amplification of noncoding regions of chloroplast DNA in the grasses. Mol. Ecol. Notes 1, 76–78 (2001).
Zhang, W. Phylogeny of the grass family (Poaceae) from rpl16 intron sequence data. Mol. Phylogenet. Evol. 15, 135–146 (2000).
White, T. J., Bruns, T., Lee, S. & Taylor, J. W. In PCR Protocols: A guide to methods and applications (eds M. A. Innis, D. H. Gelfand, J. J. Sninsky & T. J. White) Ch. 38, 315–322 (Academic Press, Inc., 1990).
Moore, M. J., Bell, C. D., Soltis, P. S. & Soltis, D. E. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. P. Natl Acad. Sci. USA 104, 19363–19368 (2007).
Oxelman, B., Lidén, M. & Berglund, D. Chloroplast rps16 intron phylogeny of the tribe Sileneae (Caryophyllaceae). Plant Syst. Evol. 206, 393–410 (1997).
Doyle, J. J. Gene trees and species trees: molecular systematics as one-character taxonomy. Syst. Bot. 17, 144–163 (1992).
Farris, J. S., Albert, V. A., Källersjö, M., Lipscomb, D. & Kluge, A. G. Parsimony jackknifing outperforms neighbor-joining. Cladistics 12, 99–124 (1996).
Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89, 583–590 (1978).
Peakall, R. & Smouse, P. E. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research - an update. Bioinformatics 28, 2537–2539 (2012).
Acknowledgements
Brian K. Sorrell provided valuable comments to the manuscript. Peder K. Bøcher helped creating the maps in Figure 4. Camilla Håkansson is thanked for her invaluable help in the DNA lab. We thank Mats Gustafsson, Paola Balboni, Lubomir Adamec, Nicola Bressi, Arunothai Jampeetong, Guilia Cavalli, Tenna Riis, Deborah Hofstra, John Clayton, Marianne Krag Petersen, Judit Padisák, Jane Roberts, Francisco Comín, Jason Nicol, Damien Cook, Stig Madsen, Poul Evald, Einar Flensted-Jensen, Katka Diáková, Tobias Bickel, Philip Weyl, and Julie Coetzee for collecting samples for this study.
Author information
Authors and Affiliations
Contributions
B.H. designed the study, performed the data analyses, made figures and tables and wrote the first draft of the manuscript with guidance from C.L. and H.B. as part of her PhD studies. B.H. and technician Camilla Håkansson performed the lab work. B.H. and C.L. jointly discussed the data and text and improved the various drafts of the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hyldgaard, B., Lambertini, C. & Brix, H. Phylogeography reveals a potential cryptic invasion in the Southern Hemisphere of Ceratophyllum demersum, New Zealand’s worst invasive macrophyte. Sci Rep 7, 16569 (2017). https://doi.org/10.1038/s41598-017-16712-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-16712-8
This article is cited by
-
Towards global dominance of invasive alien plants in freshwater ecosystems: the dawn of the Exocene?
Hydrobiologia (2021)
-
Why are tall-statured energy grasses of polyploid species complexes potentially invasive? A review of their genetic variation patterns and evolutionary plasticity
Biological Invasions (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.