Abstract
Osteoclasts can be differentiated from bone marrow-derived macrophages (BMDM). They play a key role in bone resorption. Identifying novel molecules that can regulate osteoclastogenesis has been an important issue. In this study, we found that FAM19A5, a neurokine or brain-specific chemokine, strongly stimulated mouse BMDM, resulting in chemotactic migration and inhibition of RANKL-induced osteoclastogenesis. Expression levels of osteoclast-related genes such as RANK, TRAF6, OSCAR, TRAP, Blimp1, c-fos, and NFATc1 were markedly decreased by FAM19A5. However, negative regulators of osteoclastogenesis such as MafB and IRF-8 were upregulated by FAM19A5. FAM19A5 also downregulated expression levels of RANKL-induced fusogenic genes such as OC-STAMP, DC-STAMP, and Atp6v0d2. FAM19A5-induced inhibitory effect on osteoclastogenesis was significantly reversed by a formyl peptide receptor (FPR) 2 antagonist WRW4 or by FPR2-deficiency, suggesting a crucial role of FPR2 in the regulation of osteoclastogenesis. Collectively, our results suggest that FAM19A5 and its target receptor FPR2 can act as novel endogenous ligand/receptor to negatively regulate osteoclastogenesis. They might be regarded as potential targets to control osteoclast formation and bone disorders.
Similar content being viewed by others
Introduction
Family with sequence similarity 19 (chemokine (C-C motif)-like) member A (FAM19A) encoded by TAFA genes (TAFA-1 to TAFA-5) are secretory proteins with cytokine-like properties1,2. Among these TAFA genes, TAFA mRNA expression levels are very high in the brain with very low levels in other organs1. TAFA4 has been regarded as a specific marker of C-low-threshold mechanoreceptors2. A previous study on the functional role of TAFA4 has demonstrated that genetic depletion of TAFA4 can cause severe mechanical and chemical hypersensitivity in response to injury2. Another previous report has shown that FAM19A4 encoded by TAFA4 can promote cellular migration and phagocytosis in macrophages3. In addition, FAM19A4 can directly bind to formyl peptide receptor (FPR) 1, its target receptor3. FPR1 is a well-known classical chemoattractant receptor for innate immune cells such as monocytes/macrophages and neutrophils3. However, the functional role or molecular target receptor of other FAM19A members, especially FAM19A5, has not been reported yet.
Osteoclasts are giant multinucleated cells with bone resorbing activity. They play essential roles in bone metabolism and homeostasis4,5. They can adhere to bone surface by interacting with extracellular matrix and degrade bone matrix6. Osteoclasts can be differentiated from monocyte/macrophage lineage7. Stimulation of macrophages with receptor activation of nuclear factor κB ligand (RANKL) in the presence of macrophage colony-stimulating factor (M-CSF) can elicit osteoclast differentiation8. During differentiation of osteoclasts, several osteoclast-associated genes including RANK, TRAF6, OSCAR, TRAP, and NFATc1 are upregulated4,5. Because osteoclasts have bone resorbing activity, several bone disorders including osteoporosis and rheumatoid arthritis are associated with enhanced osteoclast formation9. Considering the crucial role of osteoclasts in bone diseases, identifying molecules that can inhibit osteoclast differentiation is essential to control these diseases. In this study, we found that FAM19A5 stimulated mouse bone-marrow-derived macrophages (BMDMs) which could be differentiated into osteoclasts, leading to chemotactic migration of cells. We further investigated whether FAM19A5 could affect osteoclast formation from mouse BMDMs. Interestingly, we found that FAM19A5 strongly inhibited RANKL-induced osteoclastogenesis. Target receptor and signaling pathways involved in these processes are also examined in this study.
Results
FAM19A5 stimulates BMDM, leading to chemotactic migration via FPR2
It has been reported that FAM19A4 possesses cytokine-like property and stimulates macrophage chemotaxis3. In this study, we tested whether FAM19A5 could stimulate macrophage activity, especially chemotactic migration using Boyden chamber assay. FAM19A5 strongly stimulated chemotactic migration of BMDM, showing maximal activity at 10 μM (Fig. 1A). These results suggest that BMDMs are activated by FAM19A5. Chemokines and chemoattractant are known to stimulate macrophage chemotaxis through pertussis toxin (PTX)-sensitive G-protein(s)10. Our results showed that FAM19A5-induced BMDM chemotaxis was significantly blocked by PTX (Fig. 1B). As a control experiment, we found that WKYMVm (an agonist for FPR members)-stimulated BMDM chemotaxis was completely inhibited by PTX (Fig. 1B). These results suggest that FAM19A5 can stimulate BMDM chemotaxis via PTX-sensitive G-protein(s). Activation of BMDM by diverse extracellular stimuli can induce the activation of intracellular signaling kinases such as ERK and Akt11,12. Stimulation of BMDM with FAM19A5 also induced phosphorylation of ERK and Akt in a time-dependent manner, suggesting that FAM19A5 could stimulate ERK and Akt activities (Fig. 1C). FAM19A5-stimulated ERK phosphorylation was apparent at 2-30 min after stimulation. However, Akt phosphorylation was induced at 2–10 min. It then returned to its basal level after the stimulation (Fig. 1C). We then examined whether these ERK and Akt activities were required for FAM19A5-stimulated BMDM chemotaxis using specific inhibitors of kinases. FAM19A5-induced BMDM chemotaxis was almost completely inhibited by PD98059 (an ERK inhibitor), MK2206 (an Akt inhibitor), and LY294002 (a PI3K inhibitor) (Fig. 1D). These results suggest that FAM19A5-induced BMDM chemotaxis is mediated by ERK and Akt pathway.
FAM19A5 stimulates BMDM chemotaxis via FPR2. (A) Mouse BMDMs were used for chemotaxis assay using multiwell chamber containing several concentrations (0, 0.1, 1, 2, 5, 10 μM) of FAM19A5 or 1 μM of WKYMVm for 2 h. (B) Mouse BMDMs were incubated in the absence or presence of 500 ng/ml PTX for 4 h and applied to the upper well of the multiwell chamber containing 2 μM of FAM19A5 or 1 μM of WKYMVm for 2 h. (C) Mouse BMDMs were stimulated with 2 μM of FAM19A5 for 0, 2, 5, 10, and 30 min. Total cell lysates were separated by SDS-PAGE. Levels of p-ERK and p-Akt were measured by Western blot analysis. Data are representative of three independent experiments (C). (D) Mouse BMDMs were incubated in the absence or presence of PD98059 (50 μM) for 60 min, LY294002 (50 μM) for 15 min, or MK-2206 (2 μM) for 20 min and applied to the upper well of the multiwell chamber containing 2 μM of FAM19A5 for 2 h. (E) Vector-, FPR1-, or FPR2- expressing RBL-2H3 cells were applied to the upper well of a multiwell chamber containing 2 μM of FAM19A5 for 4 h. The number of migrated cells was determined by counting under a light microscope (A,B,D,E). Data are presented as means ± SE (n = 3). Data are representative of at least three independent experiments. ns: not significant, *p < 0.05; **p < 0.01.
In a previous report, FAM19A4 has been demonstrated to be able to bind FPR1 and promote macrophage chemotaxis and phagocytosis3. In this study, we examined whether FAM19A5 could act on FPR family using vector-, FPR1-, or FPR2- expressing RBL-2H3 cells generated previously13. Stimulation of FPR2-expressing RBL-2H3 cells with FAM19A5 elicited chemotactic migration of these cells (Fig. 1E right). However, FAM19A5 failed to induce cellular migration of vector- or FPR1- expressing RBL-2H3 cells (Fig. 1E left). These results suggest that FAM19A5 can stimulate macrophage chemotaxis through FPR2, but not through FPR1. Collectively, we found that FAM19A5, a brain-specific chemokine, was a novel endogenous regulator of macrophage activity.
FAM19A5 inhibits osteoclast formation from mouse BMDMs
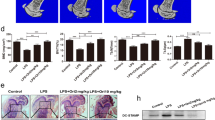
Our finding on the stimulatory effect of FAM19A5 on BMDM chemotactic migration led us to examine additional effects of FAM19A5 on BMDMs. BMDMs can be differentiated into osteoclasts which play an essential role in bone metabolism4,5,7. In this study, we examined whether FAM19A5 affected osteoclastogenesis using mouse BMDMs. Isolated mouse bone marrow cells were differentiated into BMDM by adding M-CSF. Incubation of BMDMs with M-CSF and RANKL-induced osteoclast formation based on tartrate-resistant acid phosphatase (TRAP) staining (Fig. 2A). Interestingly, addition of FAM19A5 strongly blocked osteoclast formation from BMDMs (Fig. 2A). Quantitation of osteoclast formation showed that FAM19A5 inhibited RANKL–induced osteoclast (TRAP + 3 < MNCs) formation by 73.23% (Fig. 2B left). Lipopolysaccharide (LPS), a well-known osteoclast formation inhibitory molecule, almost completely blocked RANKL-induced osteoclast formation (Fig. 2B left). FAM19A5 also inhibited TRAP + 5 < MNCs cell number by approximately 67.22% (Fig. 2B right). Several different concentrations of FAM19A5 were used to test their inhibitory effects on osteoclast formation. Addition of 0.1–2 μM of FAM19A5 markedly blocked osteoclast formation (Fig. 2C). Since FAM19A5 is a recombinant protein expressed in E. coli, we investigated if the osteoclast formation inhibitory effect of FAM19A5 could be induced by LPS endotoxin using polymyxin B, an endotoxin inhibitor. FAM19A5-induced inhibitory effects on osteoclast formation was not affected by polymyxin B (Fig. 2D). However, LPS-induced osteoclast formation inhibitory effect was blocked by polymyxin B (Fig. 2D). We also investigated whether the inhibitory effect of FAM19A5 on osteoclast formation was mediated by cell death during osteoclastogenesis. Addition of FAM19A5 failed to affect cell viability under osteoclastogenesis condition (Fig. 2E). These results suggest that FAM19A5 can block osteoclast formation from BMDMs without affecting cell viability.
FAM19A5 blocks RANKL-induced osteoclastogenesis. (A,B) Mouse BMDMs were stimulated with FAM19A5 (2 μM) or LPS in the presence of M-CSF (30 ng/ml) and RANKL (100 ng/ml) for 4 days. All cells were stained with TRAP staining solution. TRAP+ 3 < MNCs (B, left) and TRAP+ 5 < MNCs (B, right) were counted. (C) Mouse BMDMs were stimulated with various concentrations of FAM19A5 (0, 0.01, 0.1, 1, and 2 μM) in the presence of M-CSF (30 ng/ml) and RANKL (100 ng/ml) for 4 days. (D) Mouse BMDMs were pre-incubated with polymyxin B (10 μM) prior to FAM19A5 (2 μM) and LPS (1 μg/ml) treatment in the presence of M-CSF (30 ng/ml) and RANKL (100 ng/ml) for 4 days. (E) Mouse BMDMs were stimulated with 2 μM FAM19A5 (left) or various concentrations of FAM19A5 (0, 0.01, 0.1, 1, and 2 μM) (right) in the presence of M-CSF (30 ng/ml) and RANKL (100 ng/ml) for 4 days. After adding CCK-8 assay solution, the OD was measured at 450 nm. Data in these panels are representative of three independent experiments (A). Data are presented as means ± SE (n = 3). Data are representative of at least three independent experiments (B–E). ns: not significant, *p < 0.05; **p < 0.01.
FAM19A5 regulates the expression of RANKL-induced osteoclast-related genes
Since osteoclasts are specialized cells differentiated from BMDMs, differentiated osteoclast are known to express several osteoclast-related genes14,15. Previous studies have reported that osteoclasts express RANK, TRAF6, OSCAR, TRAP, and NFATc1 16,17. In this study, we also found that differentiation of osteoclast from BMDMs by RANKL upregulated the expression of these osteoclast-related genes by qPCR analysis (Fig. 3A). RANKL-induced upregulation of these osteoclast-related genes was markedly inhibited by FAM19A5 (Fig. 3A). Protein expression of RANK, TRAF6, c-fos, and NFATc1 was also induced by RANKL. However, such effect of RANKL was markedly blocked by FAM19A5 (Fig. 3B). MafB and IRF-8 are well-known negative regulators of osteoclast formation18,19. Stimulation of BMDMs with RANKL elicited downregulated expression of MafB and IRF8 (Fig. 3C). However, addition of FAM19A5 blocked the downregulation of these two negative regulators of osteoclast formation (Fig. 3C).
FAM19A5 regulates RANKL-induced gene expression during osteoclastogenesis. (A,C,D) Mouse BMDMs were stimulated with FAM19A5 (2 μM) in the presence of M-CSF (30 ng/ml) and RANKL (100 ng/ml) for 3 days. Cells were harvested for RNA preparation. qPCR was performed using specific primers for RANK, TRAF6, OSCAR, TRAP, Blimp1, c-FOS, NFATc1, MafB, IRF-8, DC-STAMP, OC-STAMP, Atp6v0d2, and GAPDH. (B) Cells were harvested and Western blot analysis was conducted using anti-RANK, anti-TRAF6, anti-c-fos, anti-NFATc1, and β-actin antibodies. Data are presented as means ± SE (n = 3). Data are representative of three independent experiments (A,C,D). *p < 0.05; **p < 0.01; ***p < 0.001. Results shown are representative of three independent experiments (B).
During osteoclast differentiation, osteoclast precursors need to be fused together to generate immature and mature osteoclasts20. The fusion process is regulated by several genes, including dendritic cell-specific transmembrane protein (DC-STAMP), osteoclast stimulatory transmembrane protein (OC-STAMP), and Atp6v0d2 21,22. In this study, we also observed that expression levels of DC-STAMP, OC-STAMP, and Atp6v0d2 were markedly increased in the presence of M-CSF plus RANKL in BMDMs by qPCR analysis (Fig. 3D). We then tested the effect of FAM19A5 on the expression of these fusogenic genes. The addition of FAM19A5 significantly decreased expression levels of these osteoclast fusogenic genes (Fig. 3D). Collectively, these results suggest that FAM19A5 can regulate the expression of osteoclast differentiation and fusion-associated genes, leading to inhibition of osteoclastogenesis.
FPR2 mediates FAM19A5-induced inhibitory effects on osteoclast formation
Since we found that FAM19A5 could stimulate BMDM chemotaxis via FPR2, we investigated whether FPR2 played a role in FAM19A5-induced inhibition of osteoclast formation. Cellular signaling of FPR2 is mediated by PTX-sensitive G-proteins23. Therefore, we tested the effect of PTX on FAM19A5-induced inhibitory effect on osteoclast formation. Pre-incubation of BMDMs with PTX (which inactivates G proteins) prior to the addition of FAM19A5 reversed FAM19A5-induced inhibitory effect on osteoclast formation (Fig. 4A). These results suggest that FAM19A5-induced osteoclastogenesis inhibitory effect is mediated by PTX-sensitive G proteins. We then investigated the role of FPR2 on FAM19A5′s inhibitory effect on osteoclast formation using WRWWWW (WRW4)24, an FPR2 antagonist. FAM19A5-induced inhibitory effect on osteoclast formation was significantly reversed by WRW4 (Fig. 4B). However, LPS-induced inhibitory effect on osteoclast formation was not affected by WRW4 (Fig. 4B). In addition to FPR2, FPR1 is another member of FPR that plays a crucial role in the regulation of leukocyte chemotactic migration25. We also tested the role of FPR1 on FAM19A5-induced inhibitory effect on osteoclast formation. Pre-incubation of BMDMs with cyclosporin H, an FPR1 antagonist, prior to addition of FAM19A5 failed to affect FAM19A5-induced inhibitory effect on osteoclast formation (Fig. 4C). These results suggest that FAM19A5-induced osteoclast formation inhibition is mediated by FPR2, but not by FPR1.
Inhibitory effects of FAM19A5 on RANKL-induced osteoclast formation are mediated by FPR2. (A–C) Mouse BMDMs were pre-incubated with PTX (100 ng/ml), WRW4 (60 μM) or cyclosporin H (10 μM) prior to FAM19A5 (2 μM) treatment in the presence of M-CSF (30 ng/ml) and RANKL (100 ng/ml) for 4 days. All cells were stained with TRAP staining solution. Osteoclasts were considered for TRAP+ 3 < MNCs. Data are presented as means ± SE (n = 3). Data are representative of at least two independent experiments. ns: not significant, **p < 0.01.
FPR2-deficiency blocks FAM19A5-induced macrophage activation but reverses inhibitory effect of FAM19A5 on osteoclast formation
To support our notion that FPR2 could mediate FAM19A5-induced macrophage activation and osteoclast differentiation inhibition, we generated FPR2 knockout (FPR2−/−) mice by TALEN-mediated gene targeting (Fig. 5A). FPR2 gene knockout was confirmed by T7E1 assay (Fig. 5B). Since we found that FAM19A5 stimulated BMDM chemotaxis through ERK and Akt pathway, we investigated whether FPR2-deficiency affected FAM19A5-induced BMDM chemotaxis and ERK/Akt phosphorylation using BMDM from FPR2+/+ or FPR2−/− mice. As expected, we observed that FAM19A5-stimulated BMDM chemotaxis and ERK/Akt phosphorylation were strongly blocked by FPR2 deficiency (Fig. 5C,D). We also tested the effect of FPR2 deficiency on FAM19A5-induced inhibitory effect on osteoclast formation. FAM19A5′s inhibitory effect on RANKL-induced osteoclast differentiation was significantly reversed by FPR2 deficiency (Fig. 5E). Taken together, these results strongly indicate that FPR2 plays an essential role in FAM19A5-induced macrophage chemotaxis, ERK/Akt phosphorylation, and FAM19A5′s inhibition on osteoclastogenesis.
FPR2-deficiency blocks FAM19A5-induced BMDM chemotaxis and FAM19A5′s inhibitory effects on osteoclastogenesis. (A) TALEN recognition site for the generation of FPR2 knockout mice. (B) T7E1 assay of WT, heterozygotes, and homozygotes of FPR2 knockout mice. (C) BMDMs generated from FPR2+/+ or FPR2−/− mice were subjected to chemotaxis assay using several concentrations (0, 0.01, 0.1, 1, and 2 μM) of FAM19A5. The number of migrated cells was determined by counting under a light microscope. (D) Mouse BMDMs generated from FPR2+/+ or FPR2−/− mice were stimulated with 2 μM of FAM19A5 for 0, 2, 5, 10, and 30 min. Total cell lysates were separated by SDS-PAGE. Levels of p-ERK and p-Akt were measured by Western blot analysis. (E) Mouse BMDMs were stimulated with FAM19A5 (2 μM) in the presence of M-CSF (30 ng/ml) and RANKL (100 ng/ml) for 4 days. All cells were stained with TRAP staining solution. The number of cells with TRAP+ 3 < MNCs was counted. Data in these panels are representative of three independent experiments (D). Data are presented as means ± SE (n = 3). Data are representative of three independent experiments (C,E). *p < 0.05; **p < 0.01.
Discussion
Although several previous reports have shown that FAM family members are involved in some biological functions, including the regulation of leukocyte adhesion, migration3,26, phagocytosis3, vascular inflammation27, and microglia/macrophage polarization28, studies on the functional role of FAM19A5 are limited. In this study, we demonstrated that FAM19A5, an endogenous brain-specific chemokine, strongly inhibited RANKL-induced osteoclast generation, suggesting a novel inhibitory functional role of FAM19A5 in osteoclastogenesis. Since osteoclasts play essential roles in bone metabolism by mediating bone resorption, resulting in bone disorders such as osteoporosis and rheumatoid arthritis, our findings provide a novel insight on the pathophysiological role of FAM19A5 in bone metabolism and bone-related disorders.
Regarding FAM19A5′s inhibitory effects against osteoclast formation, we demonstrated that FPR2, its target receptor, mediated the process. FAM19A5-inhibited osteoclastogenesis was blocked by WRW4, an FPR2 antagonist (Fig. 4B). Moreover, the inhibitory effect of FAM19A5 on RANKL-induced osteoclastogenesis was not apparent in BMDMs generated from FPR2-deficient mice (Fig. 5E). These results strongly indicate that FAM19A5 inhibits RANKL-induced osteoclast differentiation by stimulating FPR2. Several previous reports have reported that FPR2 play an essential role in the regulation of macrophage/monocyte differentiation into other cells such as foam cells29,30,31, dendritic cells32, and macrophage polarization33. We have previously demonstrated that FPR2 can mediate cellular differentiation such as foam cell formation induced by oxLDL or serum amyloid A (SAA)29,30. FPR2 can also mediate immune-modulating peptide WKYMVm-induced dendritic cell maturation inhibition32. Li et al. have demonstrated that FPR2 agonists SAA and LL-37 can elicit M2-like macrophage polarization33. This process is mediated by FPR233. However, in this study, we report that FPR2 is involved in the regulation of osteoclastogenesis. The role of FPR2 in FAM19A5-induced cellular response was also observed in this study because FAM19A5 stimulated macrophage chemotactic migration which was blocked by PTX, a Gi-protein inhibitor. Moreover, we demonstrated that FAM19A5 stimulated chemotactic migration of FPR2-expressing RBL-2H3 cells, but not that of vector-expressing RBL-2H3 cells (Fig. 1), indicating that FPR2 could mediate FAM19A5-induced chemotactic migration. FAM19A5-stimulated BMDM chemotaxis and ERK/Akt phosphorylation were also markedly attenuated by FPR2 deficiency (Fig. 5C,D). These results strongly support the notion that FPR2 mediates FAM19A5-induced biological functions such as inhibition of osteoclastogenesis.
Through studying the mechanism of action involved in the inhibitory effect of FAM19A5 on RANKL-induced osteoclastogenesis, we demonstrated that FAM19A5 could modulate gene expression induced by RANKL. RANKL-induced osteoclast-associated genes (RANK, TRAF6, OSCAR, TRAP, Blimp1, c-fos, and NFATc1) were downregulated by FAM19A5. However, negative regulator genes of osteoclastogenesis were upregulated by FAM19A5 (Fig. 3A–C). Osteoclast fusogenic genes were strongly decreased by FAM19A5 (Fig. 3D). Since FAM19A5-induced inhibitory effect on osteoclastogenesis was mediated by FPR2, these results suggested that there might be cross-talks between FAM19A5-stimulated FPR2 signaling and RANKL-induced RANK signaling in macrophages. FAM19A5-induced FPR2 signaling might block RANK signaling, resulting in inhibition of osteoclast-associated genes expression and increased negative regulator of osteoclast genes. In a previous report, we have demonstrated that SAA can block osteoclast formation by stimulating c-fms shedding34. Therefore, we also examined the effect of FAM19A5 on c-fms shedding and found that stimulation of BMDMs with FAM19A5 did not induce c-fms shedding (data not shown). These results suggest that FAM19A5 blocks osteoclast differentiation by mechanism different from SAA-induced one, at least partly.
Although FAM19A5 was known to be a brain-specific chemokine, we found that FAM19A5 negatively regulated osteoclast differentiation from macrophages. It has been previously reported that other FAM19 members such as FAM19A3 and FAM19A4 can regulate macrophage activity3,28. Considering our findings and others’ findings, FAM19 members might be involved in the regulation of peripheral immune cell activity, especially macrophages. Because studies on expression of FAM19A5 are limited, it is currently unclear whether the expression level of FAM19A5 will change during specific pathological progress. A recent proteomics study has reported that FAM19A5 level is differentially expressed in cholangiocarcinoma compared to that in benign biliary tract diseases27. In this study, we could not detect any expression of FAM19A5 in BMDM or osteoclast through RT-PCR analysis (data not shown). However, since BMDM express target receptor for FAM19A5, FPR2, these cells can be activated by FAM19A5 to mediate biological function. Future studies are needed to determine the expression of FAM19A5 in specific disease conditions to further understand the biological role of FAM19A5 in diseases. Our findings provide important insights to the pathophysiological role of FAM19A5 in disease states.
In conclusion, we found that stimulation of BMDMs with FAM19A5, a member of brain-specific chemokine, strongly blocked RANKL-induced osteoclast differentiation. We demonstrated that FAM19A5-induced inhibition of osteoclastogenesis was mediated by FPR2. Taken together, our results provide novel insights on the functional role of FAM19A5 and its target receptor FPR2 as crucial candidate for modulating osteoclast formation and bone disorders.
Materials and Methods
Materials
Human recombinant FAM19A5 was purchased from Biovendor (Brno, Czech Republic). Murine recombinant M-CSF and murine RANKL were purchased from Peprotech (Rocky Hill, NJ, USA). LPS was purchased from Sigma-Aldrich (St. Louis, MO, USA). WKYMVm and WRW4 were synthesized from Anygen (Gwangju, Korea). Polymyxin B, PTX, and PD98059 were purchased from Calbiochem (San Diego, CA, USA). LY294002 was obtained from BIOMOL Research Laboratories, Inc. (Plymouth Meeting, PA, USA). MK-2206 was purchased from Selleck Chemicals (Houston, TX, USA). Phospho-Akt (Ser473), Akt, phospho-ERK, ERK, RANK, TRAF6, and β-actin antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). c-fos and NFATc1 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Mouse BMDM generation
All animal experiments were performed in accordance with the Korea Food and Drug Administration guidelines. Protocols were approved by the Animal Care and Use Committee, Sungkyunkwan University. Mouse BMDMs were generated according to a previous report35. Briefly, bone marrow cells were isolated by flushing femurs and tibias of wild-type C57BL/6 mice of five to seven weeks of age with ice-cold PBS. Bone marrow progenitor cells were cultured in α-MEM supplemented with 10% fetal bovine serum (FBS) for one day. After harvesting non-adherent cells, they were cultured for three days with α-MEM supplemented with 10% FBS and 30 ng/ml M-CSF.
Chemotaxis assay
Chemotaxis assays were performed using multiwell chambers (Neuroprobe Inc., Gaithersburg, MD, USA) according to a previous report36. Briefly, generated mouse BMDMs were suspended in opti-MEM medium at a concentration of 1 × 106 cells/ml and 25 μl was placed onto the upper well of a chamber separated by a 5-μm polyhydrocarbon filter from a FAM19A5-containing lower well. Polycarbonate filters (8-μm pore size) precoated with 50 μg/ml rat type I collagen (Collaborative Research, Bedford, MA, USA) in DMEM medium were used for chemotaxis assay using vector-, FPR1-, or FPR2- expressing RBL-2H3 cells. After incubation at 37°C for 2 h (4 h for vector-, FPR1-, or FPR2-expressing RBL-2H3 cells), non-migrated cells were removed by scraping. Cells that migrated across the filter were dehydrated, fixed, and stained with hematoxylin (Sigma-Aldrich, St. Louis, MO, USA). Stained cells in each well were counted under a light microscope.
Osteoclast differentiation and TRAP staining
Mouse BMDMs (1 × 104 cells/well) were differentiated into osteoclasts using M-CSF (30 ng/ml) and RANKL (100 ng/ml) according to a previous report37. Differentiated osteoclasts were stained for TRAP using acid phosphatase leukocyte diagnostic kit (Sigma-Aldrich, St. Louis, MO, USA) as described previously34. Stained cells in five randomly chosen fields (200 × magnification) were then counted for each well.
Cell viability assay
Cell viability assay was conducted using cell counting kit-8 solution (Dojindo, Rockville, MD, USA) as described previously38.
Quantitative polymerase chain reaction (qPCR) analysis
qPCR was designed based on consensus sequences of each alignment and performed using Rotor-gene Q (2plex on PC) instrument (QIAGEN; Hilden, Germany) with SYBR Green qPCR Mix (Biofact). The following primers were used for qPCR: RANK-forward, 5′-AGAAGACGGTGCTGGAGTCT-3′; RNAK-reverse, 5′-TAGGAGCAGTGAACCAGTCG-3′; TRAF6-forward, 5′-GCCCAGGCTGTTCATAATGT-3′; TRAF6-reverse, 5′-TCGCCCACGTACATACTCTG-3′; OSCAR-forward, 5′-CTGCTGGATACGGATCAGCTCCCCAGA-3′; OSCAR-reverse, 5′-CCAAGGAGCCAGAACCTTCGAAACT-3′; TRAP-forward, 5′-CAGTTGGCAGCAGCCAAGGAGGAC-3′; TRAP-reverse, 5′-TCCGRGCTCGGCGATGGACCAGA-3′; Blimp1-forward, 5′-TGCTTATCCCAGCACCCC-3′; Blimp1-reverse, 5′- CTTCAGGTTGGAGAGCTGACC -3′; c-fos-forward, 5′-AGAGCGGGAATGGTGAAGAC-3′; c-fos-reverse, 5′-GCTGCATAGAAGGAACCGGA-3′; NFATc1-forward, 5′- CAACGCCCTGACCACCGATAG -3′; NFATc1-reverse, 5′-GGGAAGTCAGAAGTGGGTGGA-3′; MafB-forward, 5′-AGTGTGGAGGACCGCTTCTCT-3′; MafB-reverse, 5′-CAGAAAGAACTCAGGAGAGGAGG-3′; IRF-8-forward, 5′AGACGAGGTTACGCTGTGC-3′; IRF-8-reverse, 5′- TCGGGGACAATTCGGTAAACT -3′. Data were normalized against the expression of GAPDH as an internal control.
Western blot assay
Cell extracts were prepared in lysis buffer containing 20 mM HEPES (pH7.2), 10% glycerol, 150 mM NaCl, 1% Triton X-100, 50 mM NaF, 1 mM Na3VO4, 10 μg/ml leupeptin, 10 μg/ml aprotinin, and 1 mM PMSF. Western blot analysis was conducted as described previously39.
FPR2 knockout mouse generation, maintenance, and genotyping
FPR2 knockout mice were generated as described previously40. Briefly, TALENs specific for FPR2 were microinjected into fertilized embryos with C57BL/6 N background. Mutations on FPR2 were screened by T7E1 assay. Mutant mouse line with 163 bp deletion was selected and confirmed by Sanger sequencing (Cosmo Genetech). Mice were maintained on a normal diet (PicoLab® Rodent Diet 20, Orientbio) under a 12 h light/dark cycle. Genotypes of mice were determined by PCR with following primers; 5′-TCAGGTGCAGACAAAATGGA-3′ (forward) and 5′-TTTGAGCCCAGGATCCAAAGT-3′ (reverse).
Statistical analysis
Results were analyzed with GraphPad prism software (GraphPad Software, Inc., La Jolla, CA, USA). Statistical analysis was performed using one-way analysis of variance followed by Student’s t-test. All results are expressed as mean ± SE. A p value < 0.05 was considered statistically significant.
Data availability statement
No datasets were generated or analysed during the current study.
References
Tom Tang, Y. et al. TAFA: A Novel Secreted Family with Conserved Cysteine Residues and Restricted Expression in the Brain. Genomics 83, 727–734 (2004).
Delfini, M. C. et al. TAFA4, a chemokine-like protein, modulates injury-induced mechanical and chemical pain hypersensitivity in mice. Cell Rep. 5, 378–388 (2013).
Wang, W. et al. FAM19A4 is a novel cytokine ligand of formyl peptide receptor 1 (FPR1) and is able to promote the migration and phagocytosis of macrophages. Cell. Mol. Immunol. 12, 615–624 (2015).
Karsenty, G. & Wagner, E. F. Reaching a genetic and molecular understanding of skeletal development. Dev. Cell. 2, 389–406 (2002).
Miyamoto, T. Regulation of Osteoclast Differentiation and Bone Homeostasis. Anti-Aging Med. 9, 161–164 (2012).
Xiong, J. et al. Matrix-embedded cells control osteoclast formation. Nat. Med. 17, 1235–1241 (2011).
Boyle, W. J., Simonet, W. S. & Lacey, D. L. Osteoclast differentiation and activation. Nature 423, 337–342 (2003).
Wada, T., Nakashima, T., Hiroshi, N. & Penninger, J. M. RANKL-RANK signaling in osteoclastogenesis and bone disease. Trends Mol. Med. 12, 17–25 (2006).
Adamopoulos, I. E. & Mellins, E. D. Alternative pathways of osteoclastogenesis in inflammatory arthritis. Nat. Rev. Rheumatol. 11, 189–194 (2015).
Migeotte, I., Communi, D. & Parmentier, M. Formyl peptide receptors: A promiscuous subfamily of G protein-coupled receptors controlling immune responses. Cytokine Growth Factor Rev. 17, 501–519 (2006).
Edward, T. R. et al. ERK signaling is essential for macrophage development. PLoS One 10, e0140064 (2015).
Verqadi, E. et al. Akt signaling pathway in macrophage activation and M1/M2 polarization. J. Immunol. 198, 1006–1014 (2017).
Rong, H. et al. The synthetic peptide Trp-Lys-Tyr-Met-Val-D-Met is a potent chemotactic agonist for mouse formyl peptide receptor. J. Immunol. 165, 4598–4605 (2000).
Feng, X. RANKing intracellular signaling in osteoclasts. IUBMB life 57, 389–395 (2005).
Nakagawa, N. et al. RANK is the essential signaling receptor for osteoclast differentiation factor in osteoclastogenesis. Biochem. Biophys. Res. Commun. 253, 395–400 (1998).
Lamothe, B. et al. TRAF6 ubiquitin ligase is essential for RANKL signaling and osteoclast differentiation. Biochem. Biophys. Res. Commun. 359, 1044–1049 (2007).
Koga, T. et al. Costimulatory signals mediated by the ITAM motif cooperate with RANKL for bone homeostasis. Nature 428, 758–763 (2004).
Kim, K. et al. MafB negatively regulates RANKL-mediated osteoclast differentiation. Blood 109, 3253–3259 (2007).
Zhao, B. et al. Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nat. Med. 15, 1066–1071 (2009).
Ishii, M. & Saeki, Y. Osteoclast cell fusion: Mechanisms and molecules. Mod. Rheumatol. 18, 220–227 (2008).
Yang, M. et al. Osteoclast stimulatory transmembrane protein(OC-STAMP), a novel protein induced by RANKL that promotes osteoclast differentiation. J. Cell Physiol. 215, 497–505 (2009).
Yagi, M. et al. DC-STAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells. J. Exp. Med. 202, 345–351 (2005).
Ye, R. D. et al. International Union of Basic and Clinical Pharmacology. LXXIII. Nomenclature for the formyl peptide receptor (FPR) family. Pharmacol. Rev. 61, 119–161 (2009).
Bae, Y. S. et al. Identification of peptides that antagonize formyl peptide receptor-like 1-mediated signaling. J. Immunol. 173, 607–14 (2004).
Dorward, D. A. et al. The role of formylated peptides and formyl peptide receptor 1 in governing neutrophil function during acute inflammation. Am. J. Pathol. 185, 1172–1184 (2015).
Peng, X. et al. Identification of FAM3D as a novel endogenous chemotaxis agonist for the FPRs (formyl peptide receptors). J. Cell Sci. 129, 1831–1842 (2016).
Janvilisri, T., Leelawat, K., Roytrakul, S., Paemanee, A. & Tohtong, R. Novel Serum Biomarkers to Differentiate Cholangiocarcinoma from Benign Biliary Tract Diseases Using a Proteomic Approach. Dis. Markers. https://doi.org/10.1155/2015/105358 (2015). dx.doi.org/.
Shao, Y., Deng, T., Zhang, T., Li, P. & Wang, Y. FAM19A3, a novel secreted protein, modulates the microglia/macrophage polarization dynamics and ameliorates cerebral ischemia. FEBS Lett. 589, 467–475 (2015).
Lee, H. Y., Oh, E., Kim, S. D., Seo, J. K. & Bae, Y. S. Oxidized low-density lipoprotein-induced foam cell formation is mediated by formyl peptide receptor 2. Biochem. Biophys. Res. Commun. 443, 1003–1007 (2014).
Lee, H. Y., Kim, S. D., Baek, S. H., Choi, J. H. & Bae, Y. S. Role of formyl peptide receptor 2 on the serum amyloid A-induced macrophage foam cell formation. Biochem. Biophys. Res. Commun. 433, 255–259 (2013).
Lee, H. Y., Lee, M. & Bae, Y. S. Formyl Peptide Receptors in Cellular Differentiation and Inflammatory Diseases. J. Cell. Biochem. 118, 1300–1307 (2017).
Kang, H. K. et al. The Synthetic Peptide Trp-Lys-Tyr-Met-Val-D-Met Inhibits Human Monocyte-Derived Dendritic Cell Maturation via Formyl Peptide Receptor and Formyl Peptide Receptor-Like 2. J. Immunol. 175, 685–692 (2005).
Li, Y. et al. Pleiotropic regulation of macrophage polarization and tumorigenesis by formyl peptide receptor-2. Oncogene 30, 3887–3899 (2011).
Oh, E. et al. Serum amyloid A inhibits RANKL-induced osteoclast formation. Exp. Mol. Med. 47, e194, https://doi.org/10.1038/emm.2015.83 (2015).
Kim, K. et al. MicroRNA-26a Regulates RANKL-Induced Osteoclast Formation. Mol Cells. 38, 75–80 (2015).
Bae, Y. S. et al. Identification of novel chemoattractant peptides for human leukocytes. Blood 97, 2854–2862 (2001).
Gupta, N. et al. IL-3 inhibits human osteoclastogenesis and bone resorption through downregulation of c-Fms and diverts the cells to dendritic cell lineage. J. Immunol. 185, 2261–2272 (2010).
Li, N. et al. Low concentration of quercetin antagonizes the cytotoxic effects of anti-neoplastic drugs in ovarian cancer. PLoS One 9, e100314, https://doi.org/10.1371/journal.pone.0100314 (2014).
Lee, H. Y. et al. Sphingosylphosphorylcholine stimulates CCL2 production from human umbilical vein endothelial cells. J. Immunol. 186, 4347–4353 (2011).
Sung, Y. H. et al. Knockout mice created by TALEN-mediated gene targeting. Nat. Biotechnol. 31, 23–24 (2013).
Acknowledgements
This study was supported by grant of the National Research Foundation of Korea [2015R1A2A1A10054567 (YB)].
Author information
Authors and Affiliations
Contributions
M.P. acquired, analyzed, interpreted data, and designed study; H.K., H.L., and Y.B. analyzed, interpreted data, and designed study; M.L., B.P., E.C., and J.S. supplied materials for this study and interpreted data; M.P. and Y.B. drafted manuscript; All authors reviewed the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Park, M.Y., Kim, H.S., Lee, M. et al. FAM19A5, a brain-specific chemokine, inhibits RANKL-induced osteoclast formation through formyl peptide receptor 2. Sci Rep 7, 15575 (2017). https://doi.org/10.1038/s41598-017-15586-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-15586-0
This article is cited by
-
Circulating levels of FAM19A5 are inversely associated with subclinical atherosclerosis in non-alcoholic fatty liver disease
BMC Endocrine Disorders (2021)
-
The effect of the WKYMVm peptide on promoting mBMSC secretion of exosomes to induce M2 macrophage polarization through the FPR2 pathway
Journal of Orthopaedic Surgery and Research (2021)
-
FAM19A5l Affects Mustard Oil-Induced Peripheral Nociception in Zebrafish
Molecular Neurobiology (2021)
-
HNGF6A Inhibits Oxidative Stress-Induced MC3T3-E1 Cell Apoptosis and Osteoblast Phenotype Inhibition by Targeting Circ_0001843/miR-214 Pathway
Calcified Tissue International (2020)
-
VU0155069 inhibits inflammasome activation independent of phospholipase D1 activity
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.