Abstract
Organelle acquisitions via endosymbioses with prokaryotes were milestones in the evolution of eukaryotes. Still, quite a few uncertainties have remained for the evolution in the early stage of organellogenesis. In this respect, rhopalodiacean diatoms and their obligate cyanobacterial endosymbionts, called spheroid bodies, are emerging as new models for the study of organellogenesis. The genome for the spheroid body of Epithemia turgida, a rhopalodiacean diatom, has unveiled its unique metabolic nature lacking the photosynthetic ability. Nevertheless, the genome sequence of a spheroid body from a single lineage may not be sufficient to depict the evolution of these cyanobacterium-derived intracellular structures as a whole. Here, we report on the complete genome for the spheroid body of Rhopalodia gibberula, a lineage distinct from E. turgida, of which genome has been fully determined. Overall, features in genome structure and metabolic capacity, including a lack of photosynthetic ability, were highly conserved between the two spheroid bodies. However, our comparative genomic analyses revealed that the genome of the R. gibberula spheroid body exhibits a lower non-synonymous substitution rate and a slower progression of pseudogenisation than those of E. turgida, suggesting that a certain degree of diversity exists amongst the genomes of obligate endosymbionts in unicellular eukaryotes.
Similar content being viewed by others
Introduction
Photosynthesis and aerobic respiration were introduced to eukaryotes through the acquisition of plastids and mitochondria, respectively. The two metabolic abilities were primarily innovated among prokaryotes and incorporated into the eukaryotic cellular system via endosymbioses of a photosynthetic bacterium and a bacterium generating energy under the presence of oxygen. Thus, the evolutionary process that transformed an endosymbiotic bacterium into a host-controlled organelle (organellogenesis) is considered one of the critical phenomena in understanding the origin and diversification of modern eukaryotes. Although much effort has been taken to elucidate how organellogenesis proceeded by studying the extant eukaryotes harbouring mitochondria and/or plastids, many uncertainties remained for the early stages of organellogenesis. As both mitochondria and plastids have already been established as organelles, the systems for maintenance and functions of the two organelles in modern eukaryotes most likely retained little information regarding the early process of organellogenesis. In this respect, unicellular eukaryotic cells hosting bacterial intracellular symbionts have been paid attention as new model systems to study organellogenesis1,2. These symbionts were established more recently than mitochondria or plastids in eukaryotic evolution, and have been anticipated to be more informative for studying organellogenesis.
Diatoms belonging to the family Rhopalodiaceae have been nominated as one of the new models for studying organellogenesis2,3,4. The Rhopalodiaceae is a group of pennate diatoms that includes three genera, namely Rhopalodia, Epithemia, and Protokeelia 5. The genera Rhopalodia and Epithemia are known to possess cyanobacterial endosymbionts called “spheroid bodies” in addition to plastids and mitochondria2,6,7. Unlike other diatoms, Rhopalodia and Epithemia species containing the spheroid bodies can grow in media with little or no nitrogen source7, and the nitrogen fixation capacities in R. gibba and E. turgida were experimentally confirmed3,8. Consequently, it is now widely accepted that the spheroid bodies (cyanobacterial endosymbionts) in rhopalodiacean diatoms operate nitrogen fixation and supply nitrogen compounds to the host diatoms8. Indeed, molecular phylogenies confirmed a close relationship between the spheroid bodies and nitrogen-fixing cyanobacteria belonging to the genus Cyanothece 6,8.
The spheroid bodies inside cells of rhopalodiacean diatoms are separated from the cytoplasm by an envelope, which is thought to consist of two menbranes based on previous studies7,8,9. Although the spheroid bodies still retain the structural characteristic derived from their cyanobacterial ancestor such as thylakoid membranes, the intracellular structure bears neither prominent pigmentation nor chlorophyll autofluorescence10. The spheroid bodies are obligate endosymbionts, as these structures have never been cultivated outside of the host cells8 and are passed to the daughter cells through binary cell division10,11. Furthermore, both host (diatom) and symbiont (cyanobacteria) phylogenies suggested that the endosymbiosis of a nitrogen-fixing cyanobacterium, which later gave rise to the spheroid body, was established in the common ancestor of species in genera Rhopalodia and Epithemia, and inherited throughout the host speciation6. By taking into account the information from fossil records, the acquisition of the spheroid body could be traced back to the middle Miocene epoch, approximately 12 Mya6.
We have reported the complete genome sequence of a spheroid body in one of the rhopalodiacean diatoms, Epithemia turgida 3. The complete spheroid body genome sequence revealed an apparent reductive nature of the genome with regard to both the size and gene repertoire. Strikingly, the spheroid body genome in E. turgida appeared to lack most of the genes involved in photosynthesis, indicating that an autotrophic lifestyle is impossible for the cyanobacterium-derived intracellular structure. In contrast, the genes related to nitrogen fixation were well conserved in the spheroid body genome, consistent with the proposed function of the intracellular structure in the diatom cell. The first complete spheroid body genome sequence illuminated recent metabolic adaptations to an intracellular lifestyle as well as genome reductions driven by metabolic adaptations. However, except the spheroid body genome of E. turgida, only a partial genomic data from the spheroid body of Rhopalodia gibba was available12. The knowledge learned from the single complete spheroid body genome may not be sufficient to depict the evolutionary process worked on the endosymbiont and its genome during transition from a cyanobacterial endosymbiont to a host-controlled intracellular structure. To explore the genetic and metabolic divergences among the spheroid bodies and the dynamics in the reductive process shaped the extant spheroid body genomes, here we report the complete genome sequence for the spheroid body of Rhopalodia gibberula, which is genetically and morphologically distinct from E. turgida. The detailed comparison between the E. turgida and R. gibberula spheroid body genomes provided new insights into the genome evolution of the obligate cyanobacterial endosymbiont in rhopalodiacean diatoms.
Results and Discussion
Overall difference between the two spheroid body genomes
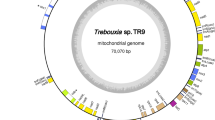
A culture strain of Rhopalodia gibberula was established from a sample collected in a fresh water pond at Tsukuba city, Japan. DNA extracted from a spheroid body-enriched cellular fraction of R. gibberula was amplified and analysed with Illumina MiSeq. After assembling short reads from the sequencing and gap-filling by PCR, we successfully obtained a single circular chromosome for the spheroid body of R. gibberula (RgSB). The complete RgSB genome was found to be ~3.02 Mbp in size (Fig. 1a, Table 1), which is over 200 Kbp larger than the previously reported genome of the spheroid body of E. turgida (EtSB)3, but smaller than the genomes of free-living cyanobacteria closely related to the spheroid bodies, e.g., Cyanothece sp. PCC 8801 (4.68 Mbp13) and Cyanothece sp. ATCC 51142 (5.36 Mbp14). The number of rRNA gene clusters and tRNA genes were identical between the RgSB and EtSB genomes (2 and 39, respectively; Table 1). The GC content was almost identical between the two spheroid body genomes (33–34%; Table 1). Although the size of the RgSB genome is larger than that of the EtSB genome by >200 Kbp, the two genomes appeared to be similar to each other as a whole and no large sequence blocks unique to the RgSB genome were found (Fig. 1b). The spheroid body genomes have been rearranged during the divergence of rhopalodiacean diatoms, as inversions and/or translocations were detected between the two spheroid body genomes (Fig. 1b). 1,671 and 1,720 open reading frames (ORFs) were predicted in the RgSB and EtSB genomes (Tables S1 and S2), respectively, and 60.3 and 54.5% of the predicted ORFs (i.e. 1,007 and 937 ORFs) in the RgSB and EtSB genomes, respectively, were assigned into any of functional categories in the Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology (KO). We found that the majority of KO IDs from the RgSB and EtSB genomes were shared between the two genomes (844 out of 909 and 849, respectively; Fig. 1c). However, the number of unique KO IDs was found to be different between the two genomes. The RgSB genome had 65 KO IDs that were absent in the RgSB genome, while only five KO IDs were unique to the EtSB genome (Fig. 1c). Increment of insertion sequence (IS) is believed to play a key role in the genome reduction15. We found 22 and 18 IS elements in the RgSB and EtSB genomes, respectively. None of the IS elements found in the two spheroid body genomes are likely transposable, as their internal ORFs for transposases are fragmented or severely truncated.
The spheroid body genome of Rhopalodia gibberula and comparison with the spheroid body genome of Epithemia turgida. (a) Map of the circular chromosome of the spheroid body of the diatom Rhopalodia gibberula (RgSB). Dark and light green bars in the outermost and second outermost circles show the positions of protein-coding genes on the plus and minus strands, respectively. The two circles also contain rRNA gene clusters shown by red bars. The bars on the second innermost circle indicate pseudogenes, and the innermost circle shows the GC content (window size: 5,000 bp). (b) Whole-genome comparison between the RgSB and Epithemia turgida spheroid body (EtSB). The RgSB genome sequence was aligned based on the EtSB genome sequence. Syntenic regions between the two genomes are colour-coded. The regions coloured in purple, dark green, light green and dark blue were found to be inverted in the RgSB genome relative to the EtSB genome. (c) Comparison of the unique KEGG orthology (KO) ID repertoire of the RgSB and EtSB genomes. The numbers in parentheses indicate the total number of unique KO IDs in each genome.
Besides the circular chromosome of the RgSB, we also obtained a ~6 Kbp contig (Dataset S1) carrying ORFs identified in previously sequenced cyanobacterial genomes. The fragment was closely related to the ~5.7 Kbp contig found in the EtSB genome sequencing3, suggesting that the fragment is conserved between the two distinct rhopalodiacean species. These fragments may correspond to plasmids of the spheroid bodies, albeit we are still unsure of their cellular localization at this point.
Pseudogenisation is less advanced in the RgSB genome than the EtSB genome
The complete genome sequence of the EtSB demonstrated that the symbiont has lost its photosynthetic ability entirely as a consequence of adaptations to an endosymbiotic lifestyle3. Most genes for components of photosystems I and II as well as other proteins essential for photosynthesis appeared to be absent in the EtSB genome. Consistent with the non-photosynthetic nature of the EtSB, entire genes for the chlorophyll a (Chl-a) biosynthetic pathway were found to be pseudogenised or undetected in the genome. The Calvin cycle also appeared to be incomplete because both rbcL and rbcS encoding the large and small subunits, respectively, of ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) were pseudogenised. The putative metabolic functions retained in the RgSB, which were reconstructed from the genome sequence generated in this study, were essentially identical to those retained in the EtSB. For instance, we failed to find any functional genes for photosystem I or II in the RgSB genome, indicating that the symbiont lacks photosynthetic ability. Nevertheless, we noticed that genes, which became dispensable for an endosymbiotic lifestyle, were degraded more rapidly in the EtSB genome than the RgSB genome. The most prominent examples are the biosynthetic pathways for Chl-a and vitamin B12, which are described below in detail.
Chl-a and vitamin B12 possess tetrapyrrole rings as their backbones, and these biosynthetic pathways branch from the heme biosynthetic pathway (Fig. 2a). We identified all of the 10 genes that composes the Chl-a biosynthetic pathway in the RgSB genome. Consistent with the non-photosynthetic nature of the EtSB, all of these genes were predicted to be dysfunctional in the EtSB genome3. In contrast, 9 out of the 10 genes (chlB, chlD, chlE, chlG, chlH, chlI, chlL, chlM, and chlN) in the RgSB genome appeared to bear no apparent signs of dysfunctionalisation (Fig. 2b, Table S3). A single pseudogene in the Chl-a biosynthetic pathway in the RgSB genome encoded cyanobacterial 3,8-divinyl chlorophyllide reductase (bciB)16, of which the ORF was disrupted by a frame shift and in-frame stop codons (Fig. 2b). These data suggest that the RgSB cannot produce Chl-a due to the lack of a functional bciB, but other genes involved in the corresponding pathway carry intact ORFs. Our analysis on the non-synonymous substitution rates (dN values) revealed dN values of genes for the Chl-a biosynthesis were found to be significantly greater than the values from other genes in the RgSB genome (Figure S1). This observation suggests that the functional constraint on the Chl-a biosynthetic pathway as a whole has been loosened, implying that the genes involved in this pathway have already been pseudogenised albeit without apparent signs of dysfunctionalisation in their primary structure. In the case of the vitamin B12 biosynthetic pathway, 17 out of the 18 genes were identified in the RgSB genome (Table S3). Note that cobR, which encodes cobyrinic acid a,c-diamide reductase, was excluded from the discussion below, as this gene was not identified in the two spheroid body genomes or their free-living relatives such as Cyanothece spp. The EtSB cannot synthesize vitamin B12, as 14 of the 17 genes were found to be pseudogenized or undetected in the genome (Fig. 2b, Table S3). On the other hand, none of the same set of genes appeared to bear any obvious signs of dysfunctionalisation in the RgSB genome (Fig. 2b), implying that the RgSB can synthesize the vitamin B12 coenzyme by itself. In support of this idea, the dN values calculated from genes for the vitamin B12 biosynthesis showed no significant elevation when compared to the values of genes for Chl-a biosynthesis (Figure S1).
Chlorophyll a and vitamin B12 biosynthetic pathways in the spheroid bodies of Rhopalodia gibberula (RgSB) and Epithemia turgida (EtSB). (a) Pathways for chlorophyll a and vitamin B12 biosynthesis. Each arrow indicates a single reaction. Gene names corresponding to each reaction are displayed. The step indicated by a dashed grey arrow is catalysed by cobyrinic acid a,c-diamide reductase (encoded by cobR) in diverse photosynthetic organisms, but this gene was not detected in the genome of the RgSB, EtSB, or their close free-living relatives (i.e. Cyanothece spp.). (b) Status of genes for the chlorophyll a and vitamin B12 biosynthetic pathways in the RgSB and EtSB genomes. A filled box indicates the presence of an intact gene, whereas a box with a X mark and a blank box designate the presence of a pseudogene and the absence of a gene, respectively.
Vitamin B12 synthesized in the RgSB can be utilized for vitamin B12-dependent enzymes. We found metH encoding a vitamin B12-dependent enzyme, methionine synthase (5-methyltetrahydrofolate-homocysteine methyltransferase), which catalyzes the final reaction of the methionine biosynthesis, in the RgSB genome (locus ID: RGRSB_0352). This enzyme is most likely functional in the RgSB, as (i) its amino acid sequence bears a high identity to the orthologue in Cyanothece sp. PCC 8801, a free-living relative of the spheroid bodies (Figure S2), and (ii) the dN value for metH was not significantly high as it was not detected as an outlier (the Grubbs’ test, P < 0.005). Interestingly, we noticed the possibility of methionine synthase being also functional in the EtSB, which cannot synthesize vitamin B12 by itself. metH appeared to be retained intact in the EtSB genome (locus ID: ETSB_0394), and its amino acid sequence keeps high identities to both orthologues in theRgSB and free-living cyanobacteria (Figure S2). If methionine synthase is genuinely functional in the EtSB, the intracellular structure need to uptake external vitamin B12. Diverse bacteria including Cyanothece spp. uptake vitamin B12 by BtuCD-F transport system, one of ABC transporters17. Nevertheless, no gene for the BtuCD-F transport system was found in the EtSB (or RgSB) genome. Thus, the EtSB uptakes external vitamin B12 by either of the two systems described below—1) an as-yet-unidentified transporter system equipped originally with the spheroid bodies or 2) the transporter supplied from the host (diatom) cell. If the latter is the case, it is attractive to speculate that the genes for the BtuCD-F transport system were relocated from the endosymbiont genome to the host genome (i.e., endosymbiotic gene transfer or EGT18), and the gene products are now targeted to the envelope of the EtSB. Alternatively, the host supplies a transporter system of non-cyanobacterial origin to the EtSB to uptake vitamin B12. Future studies on vitamin B12 transport in E. turgida and its spheroid body may provide keys to judge whether EGT occurred in the rhopalodiacean diatom system. To tackle the above issue, both genome data of the host diatoms and proteomic data of the spheroid body envelope are indispensable.
Difference in non-synonymous substitution rate between the two spheroid body genomes
In the previous section, we focused on two metabolic pathways with differential tempo of pseudogenisation between the RgSB and EtSB genomes. In this section, we explore the evolutionary mechanism underlying the difference observed between the two spheroid body genomes by calculating and comparing the evolutionary rates of the genes shared between the two genomes. 1,450 orthologous genes were extracted from the two spheroid body genomes, and the number of non-synonymous substitutions from the most recent common ancestor was estimated. The differences in dN values (ΔdN), which were calculated by subtracting the EtSB values from the corresponding RgSB values, were found to be almost normally distributed, but the entire distribution was skewed toward the negative side (Fig. 3). We conducted the Wilcoxon signed-rank test to examine the null hypothesis, which assumes that the dN values were not significantly different between the paired RgSB and EtSB genes (i.e., ΔdN = 0). The null hypothesis was rejected at the 0.1% level (P < 0.001), suggesting that the EtSB genes tended to have evolved at higher non-synonymous substitution rates than the RgSB genes. The difference in dN value between the RgSB and EtSB genomes explains well the difference in tempo of pseudogenisation between the two genomes (Fig. 2b). In the ancestral spheroid body genome, a large number of genes should have become dispensable along with the transition from a photosynthetic/free-living to a non-photosynthetic/endosymbiotic lifestyle. Nevertheless, the fates of the dispensable genes most likely varied between the descendent genomes evolving with different evolutionary rates. The chance to receive apparent signs of dysfunctionalisation (e.g., nonsense substitutions) was most likely higher in the EtSB genome than that of RgSB genome, leading to the difference in the Chl-a and vitamin B12 biosynthetic pathways observed between the two genomes (see the previous section).
Overall difference in non-synonymous substitution rates between the Rhopalodia gibberula spheroid body (RgSB) and Epithemia turgida spheroid body (EtSB) genomes. For each orthologous pair of the EtSB and RgSB genes, non-synonymous substitution rates (dN values) were estimated, and then the difference between the two corresponding dN values (ΔdN) were calculated. To calculate ΔdN, the dN values of a EtSB gene was subtracted from the value of the corresponding gene in the RgSB genome. The mean of the total ΔdN values was -0.0101 (indicated by a solid line). Outliers were not included in this histogram.
Conclusion
Obligate bacterial endosymbionts in unicellular eukaryotes are regarded as models to retrace the evolutionary transition from a free-living bacterium to an organelle integrated into the host eukaryotic cell. In this regard, the spheroid bodies in rhopalodiacean diatoms are expected to provide unique evolutionary insights into how the intracellular structures specialized for nitrogen fixation emerged through endosymbiosis. The current study on the two spheroid body genomes demonstrated that a certain degree of diversity exists among the endosymbiont genomes in the diatoms. Such genomic diversity has also been reported from cyanobacterium-derived structures/endosymbionts recently integrated into eukaryotic cells: (i) cyanobacterium-derived structures (chromatophores) in testate amoebae Paulinella spp.19,20,21, and (ii) nitrogen-fixing obligate cyanobacterial endosymbionts (UCYN-A cyanobacteria) associated with a particular group of haptophytes22,23. We need to further assess the genomic diversity observed in the aforementioned endosymbionts carefully to separate the genomic changes, which were critical for obligate endosymbiotic lifestyles and organellogenesis, from random changes that accumulated in the endosymbiont genomes.
Materials and Methods
DNA preparation from the spheroid bodies of Rhopalodia gibberula cells
Rhopalodia gibberula cells were found in a sample collected from a pond in Namiki park, Tsukuba, Ibaraki, Japan (36°03.56 North, 140°08.32 East). A single cell was isolated by micropipetting and cultured clonally in nitrogen-depleted medium (CSi-N6). The cultured diatom cells were mildly disrupted by vortexing with glass beads (~1 mm diameter) for five minutes, and then the spheroid bodies were separated roughly from other intracellular particles by discontinuous density Percoll gradient centrifugation as previously described in Nakayama et al.3. After the centrifugation, the absence of the host DNA-containing organelles (i.e., diatom nuclei, mitochondria, and plastids) in the spheroid body-enriched fraction was confirmed under a light microscope. The spheroid body-enriched fraction was subjected to whole-genome amplification using the REPLI-g mini kit (Qiagen). The whole-genome amplicon was de-branched with S1 nuclease to reduce chimeric sequences during the amplification reaction.
Genome sequencing and assembly
Amplified DNA was subjected to a library construction using a TruSeq Nano DNA sample preparation kit (Illumina) with 550 bp inserts, as manufacturer’s recommendations. The library analyzed on an Illumina MiSeq platform (300 bp, paired-end), yielding ~51 million short reads. The first 5 and last 50 bases of each read as well as reads with low sequencing quality were removed using FASTQ_Trimmer and FASTQ_Quality_Filter, respectively, which are both included in the FASTX_Toolkit program package (ver. 0.0.14; http://hannonlab.cshl.edu/fastx_toolkit/). Eight million paired-end reads were used for assembling the genome using SPAdes ver. 3.1.024. Genome scaffolding with assembled contigs was performed with SSPACE25. To obtain candidate spheroid body genome sequences from the final scaffold pool, we performed BLASTn search using the EtSB genome3 as a query. Eleven large scaffolds with high sequence similarity to the EtSB genome were retrieved, and gaps between those scaffolds were closed by PCR with information from pairs of paired-end reads.
Genome annotation
Predictions of the ORFs on the completed circular genome sequence were performed by using GeneMarkS26. The gene models were carefully inspected and refined manually. Transfer RNA genes were detected by tRNAscan-SE27, and rRNA genes were predicted based on nucleotide similarity. Putative pseudogenes on the spheroid body genome were identified by tBLASTn against non-coding regions of the genome using cyanobacterial proteins in the NCBI RefSeq database as queries. Coding regions interrupted by stop codons and/or disrupted by frame shifts as well as severely truncated ORFs were tagged as putative pseudogenes. IS elements in the genomes of the RgSB and EtSB were initially identified by ISsaga, an IS identification system for prokaryotic genomes28. The output from ISsaga was manually checked and refined. A list for the detected IS-related sequences in our analysis is shown in Table S4.
The circular genome map was generated by using DNAplotter29. Structural comparisons between the EtSB and RgSB genomes were performed with Mauve30. The initial KO ID assignment was performed by using the KEGG Automatic Annotation Server31 and then manually refined. The annotated genome data is available in DNA databases in Japan/GenBank/European Molecular Biology Laboratory under BioProject accession number PRJDB4388.
Gene evolutionary rate
We estimated dN values of protein-coding genes in the RgSB genome. Nucleotide sequences for 1,563 orthologous genes were extracted from genomes of the RgSB, Cyanothece spp. PCC 8801, PCC 8802, and ATCC 51142 and then were aligned based on codons using an in-house ruby script and MAFFT32 with the L-INS-i option. The dN values were estimated with the CODEML implemented in PAML33 (settings: runmode = 0, seqtype = 1, CodonFreq = 2, model = 1) based on the organismal phylogenetic relationship among the RgSB and its free-living relatives (i.e. Cyanothece spp; see Figure S3A). The dN values of the RgSB genes, which are values for a branch from the node representing the most recent common ancestor of Cyanothece spp. PCC 8801, PCC 8802, and the RgSB (branch X in Figure S3A), were standardized by the corresponding dN values for branch of Cyanothece spp., that is, the values from the node shared by the RgSB to a node of the most recent common ancestor of two Cyanothece spp. (branch Y in Figure S3A; Table S5). The dN values that were impossible to standardize (i.e., dN values for the RgSB or Cyanothece spp. gene = 0) were omitted from downstream analyses. Outliers among the dN values for the RgSB genes were detected using the Grubbs’ test. Differences in the normalized dN values between the gene sets of the two biosynthetic pathways for Chl-a and vitamin B12, and total genes in the genome was tested by the Wilcoxon rank-sum test. In addition, we estimated non-synonymous substitution rates of protein-coding genes shared between the EtSB and RgSB genomes. Nucleotide sequences for 1,450 orthologous genes were extracted from the two spheroid body genomes, Cyanothece spp. PCC 8801, PCC 8802, and ATCC 51142, and then analysed as described above (the tree topology used for the estimation is presented in Figure S3B). The dN values of the RgSB and EtSB sequences for branches from the node representing the latest common ancestor (branches X and Y in Figure S3B) were compared (Table S6). To test the bias of the difference of dN values between RgSB and EtSB genes, a Wilcoxon signed-rank test was performed.
References
Nowack, E. C. M. Paulinella chromatophora – rethinking the transition from endosymbiont to organelle. Acta Soc. Bot. Pol. 83, 387–397 (2014).
Adler, S., Trapp, E. M., Dede, C., Maier, U.-G. & Zauner, S. In Endosymbiosis (Ed. Löffelhardt, W.) 167–179 (Springer, 2014).
Nakayama, T. et al. Complete genome of a nonphotosynthetic cyanobacterium in a diatom reveals recent adaptations to an intracellular lifestyle. Proc. Natl. Acad. Sci. 111, 11407–11412 (2014).
Nakayama, T. & Inagaki, Y. Unique genome evolution in an intracellular N2-fixing symbiont of a rhopalodiacean diatom. Acta Soc. Bot. Pol. 83, 409–413 (2014).
Round, F. E., Crawford, R. M. & Mann, D. G. The Diatoms: Biology & Morphology of the Genera. (Cambridge University Press, 1990).
Nakayama, T., Ikegami, Y., Ishida, K., Inagaki, Y. & Inouye, I. Spheroid bodies in rhopalodiacean diatoms were derived from a single endosymbiotic cyanobacterium. J. Plant Res. 124, 93–97 (2011).
Drum, R. W. & Pankratz, S. Fine structure of an unusual cytoplasmic inclusion in the diatom genus. Rhopalodia. Protoplasma 60, 141–149 (1965).
Prechtl, J., Kneip, C., Lockhart, P., Wenderoth, K. & Maier, U.-G. Intracellular spheroid bodies of Rhopalodia gibba have nitrogen-fixing apparatus of cyanobacterial origin. Mol. Biol. Evol. 21, 1477–1481 (2004).
Hagino, K., Onuma, R., Kawachi, M. & Horiguchi, T. Discovery of an endosymbiotic nitrogen-fixing cyanobacterium UCYN-A in Braarudosphaera bigelowii (Prymnesiophyceae). PLoS One 8, e81749 (2013).
Kies, L. in Algae and symbioses (ed. Reisser W) 353–377 (Biopress, 1992).
Geitler, L. Z. E. der Epithemiaceen Epithemia, Rhopalodia und Denticula (Diatomophyceae) und ihre vermutlich symbiontischen Sphäroidkörper. Plant Syst. Evol. 128, 259–275 (1977).
Kneip, C., Voss, C., Lockhart, P. J. & Maier, U.-G. The cyanobacterial endosymbiont of the unicellular algae Rhopalodia gibba shows reductive genome evolution. BMC Evol. Biol. 8, 30 (2008).
Bandyopadhyay, A. et al. Novel metabolic attributes of the genus Cyanothece, comprising a group of unicellular nitrogen-fixing cyanobacteria. mBio 2, e00214–11 (2011).
Welsh, E. A. et al. The genome of Cyanothece 51142, a unicellular diazotrophic cyanobacterium important in the marine nitrogen cycle. Proc. Natl. Acad. Sci. USA 105, 15094–150949 (2008).
Moran, N. A. & Plague, G. R. Genomic changes following host restriction in bacteria. Curr. Opin. Genet. Dev. 14, 627–33 (2004).
Islam, M. R. et al. slr1923 of Synechocystis sp. PCC6803 is essential for conversion of 3,8-divinyl(proto)chlorophyll(ide) to 3-monovinyl(proto)chlorophyll(ide). Plant Physiol. 148, 1068–1081 (2008).
Korkhov, V. M., Mireku, S. A. & Locher, K. P. Structure of AMP-PNP-bound vitamin B12 transporter BtuCD–F. Nature 490, 367–372 (2012).
Archibald, J. M. Evolution: Gene transfer in complex cells. Nature 524, 423–424 (2015).
Lhee, D. et al. Diversity of the photosynthetic Paulinella species, with the description of Paulinella micropora sp. nov. and the chromatophore genome sequence for strain KR01. Protist 168, 155–170 (2017).
Nowack, E. C., Melkonian, M. & Glockner, G. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr. Biol. 18, 410–418 (2008).
Reyes-Prieto, A. et al. Differential gene retention in plastids of common recent origin. Mol. Biol. Evol. 27, 1530–1537 (2010).
Bombar, D., Heller, P., Sanchez-Baracaldo, P., Carter, B. J. & Zehr, J. P. Comparative genomics reveals surprising divergence of two closely related strains of uncultivated UCYN-A cyanobacteria. ISME J. https://doi.org/10.1038/ismej.2014.167 (2014).
Tripp, H. J. et al. Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium. Nature 464, 90–94 (2010).
Nurk, S. et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 20, 714–737 (2013).
Boetzer, M., Henkel, C. V., Jansen, H. J., Butler, D. & Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–9 (2011).
Besemer, J., Lomsadze, A. & Borodovsky, M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29, 2607–2618 (2001).
Schattner, P., Brooks, A. N. & Lowe, T. M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33, W686–W689 (2005).
Varani, A. M. et al. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 12, R30 (2011).
Carver, T., Thomson, N., Bleasby, A., Berriman, M. & Parkhill, J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25, 119–120 (2009).
Darling, A. C. E., Mau, B., Blattner, F. R. & Perna, N. T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403 (2004).
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C. & Kanehisa, M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185 (2007).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–80 (2013).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–91 (2007).
Acknowledgements
This work was supported by grants from the Ministry of Education, Culture, Sports, Science & Technology of Japan (Grant-in-Aid for Scientific Research on Innovative Areas: 3308) and those from the Japanese Society for the Promotion of Science (23117006, 16H04826, 16H01703 and K17K151640).
Author information
Authors and Affiliations
Contributions
T.N. and Y.I. designed the study; T.N. and Y.I. performed the study; and T.N. and Y.I. analysed data and wrote the paper.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nakayama, T., Inagaki, Y. Genomic divergence within non-photosynthetic cyanobacterial endosymbionts in rhopalodiacean diatoms. Sci Rep 7, 13075 (2017). https://doi.org/10.1038/s41598-017-13578-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-13578-8
This article is cited by
-
The marine nitrogen cycle: new developments and global change
Nature Reviews Microbiology (2022)
-
Overlooked and widespread pennate diatom-diazotroph symbioses in the sea
Nature Communications (2022)
-
Global distribution patterns of marine nitrogen-fixers by imaging and molecular methods
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.