Abstract
Angiotensin-converting enzyme (ACE) is an evolutionarily conserved peptidyl dipeptidase. Mammalian ACE converts angiotensin I to the active vasoconstrictor angiotensin II, thus playing a critical role for homeostasis of the renin-angiotensin system. In Drosophila, the ACE homolog Ance is expressed in specific regions of developing organs, but its regulatory mechanism has not been identified. Here we provide evidence that Ance expression is regulated by a combination of Mad and Pannier (Pnr) in imaginal discs. We demonstrate that Ance expression in eye and wing discs depends on Dpp signaling. The Mad binding site of Ance regulatory region is essential for Ance expression. Ance expression in imaginal discs is also regulated by the GATA family transcription factor Pnr. Pnr directly regulates Ance expression by binding to a GATA site of Ance enhancer. In addition, Pnr and Mad physically and genetically interact. Ance null mutants are morphologically normal but show genetic interaction with dpp mutants. Furthermore, we show that human SMAD2 and GATA4 physically interact and ACE expression in HEK293 cells is regulated by SMAD2 and GATA4. Taken together, this study reveals a cooperative mechanism of Ance regulation by Mad and Pnr. Our data also suggest a conserved transcriptional regulation of human ACE.
Similar content being viewed by others
Introduction
Angiotensin converting enzyme (ACE) is a zinc-dependent dipeptidyl carboxypeptidase that converts Angiotensin I to Angiotensin II. In mammals, ACE plays important roles in the regulation of the renin-angiotensin system (RAS) on blood pressure homeostasis by generating Angiotensin II, an active vasoconstrictor that increases blood pressure. ACE has also been implicated in other biological functions including immune response, cytokine expression, and cell proliferation1,2,3,4. ACE knockout mice show not only hypotension but also renal developmental abnormalities, indicating a complex role of ACE5,6. However, the transcriptional regulation and function of ACE in developing tissues in vivo is not well understood.
Ance is a Drosophila homolog of the mammalian Angiotensin-converting enzyme (ACE). Due to the extensive glycosylation of human ACE, Drosophila Ance has been extensively used for three dimensional structure analysis as a model for studying ACE inhibitor binding7,8,9. Despite the evolutionary conservation of Ance, Drosophila does not have angiotensin homologs, raising questions on the biological function of Ance. Interestingly, Ance expression is developmentally regulated by Decapentaplegic (Dpp) signaling during Drosophila embryogenesis10,11. Dpp is the Drosophila homolog of the vertebrate bone morphogenetic proteins (BMPs) and the transforming growth factor beta (TGFβ) that acts as a signaling factor for diverse morphogenetic events. The Dpp signal is mediated by receptor serine/threonine kinases, such as Thickveins (Tkv), Saxophone (Sax), and Punt. The activated receptors induce phosphorylation of the Mad transcription factor leading to activation of downstream target genes12,13,14.
Secreted Dpp forms an extracellular concentration gradient to induce graded responses in cells receiving the signal15,16,17. In embryogenesis, a gradient of Dpp from the dorsal midline leads to differential transcriptional activation in the dorsal ectoderm according to the signal threshold. The highest level of Dpp activity induces Ance expression in the presumptive amnioserosa through Mad binding to the Ance enhancer18,19. On the contrary, lower levels of Dpp signaling lead to the expression of the GATA family transcription factor pannier (pnr) 19,20. Dpp signaling also plays a key role in the patterning and growth of imaginal discs to generate adult organs. For example, Dpp is critical for the progression of the morphogenetic furrow in the eye imaginal disc21,22 and for wing disc growth signaling from the anterior-posterior boundary15,23. Dpp signaling is also required for wing venation during pupal development24,25,26. In eye discs, Pnr is known to be involved in establishment of the dorsoventral (DV) boundary by regulating the dorsal-specific gene expression27, although the relationship with Dpp signaling is unknown. As in embryogenesis, Dpp signaling affects pnr expression in wing discs. The dorsal region of the wing disc is a primordium for the notum, the dorsal part of the thorax. Dpp signaling in this region regulates Wg expression by inducing pnr 28,29. Interestingly, previous studies have shown that Ance is specifically expressed in the dorsal region of the eye disc and the notum region of the wing disc30. We have noticed that the sites of Ance expression in these discs may correspond to the territories of pnr expression in the eye disc. These observations raise questions about whether the Ance expression in these discs is related to the Dpp signaling and the Pnr function.
Despite the region-specific patterns of Ance expression in imaginal discs, little is known about how Ance expression is regulated and what role Ance plays in development. It has been reported that male accessory glands secrete Ance proteins31,32 and that males homozygous for Ance hypomorphic alleles are infertile due to defective spermiogenesis33. Moreover, some of the Ance mutant alleles are lethal10, indicating the requirement of Ance for development. Unexpectedly, new deletion Ance alleles generated in this work are fully fertile and viable, indicating that Ance is not critically required under normal condition. However, it is possible that Ance may become important when an interacting pathway is compromised. Thus, we have addressed two main issues in this study: the regulation of Ance expression and the possible genetic interactions between Ance and its regulators. We also asked if the mechanism for Ance gene regulation is shared in human cells.
Here we demonstrate that Ance expression in the eye and wing discs is regulated by Dpp signaling through Mad binding to the Ance regulatory region. Ance expression in these discs also depends on Pnr binding to one of the GATA sites in the Ance regulatory region. Furthermore, Mad and Pnr show physical interaction. Ance null mutants are fertile and morphologically normal. However, the Ance mutations show genetic interaction with dpp mutants. We also show that human ACE gene expression is regulated by SMAD2 and GATA4 transcription factors. Taken together, Ance is regulated by a combination of Dpp and Pnr and is involved in modulating some of Mad functions, providing insights into the regulation and function of mammalian ACE.
Results
Expression patterns of Ance-lacZ and Ance protein in eye and wing discs
The imaginal disc is a sac-like structure consisting of two epithelial layers, a disc proper (DP) and a peripodial epithelia (PE). While the disc proper is a columnar epithelium, peripodial cells form a thin squamous epithelium. Ance mRNA expression was shown to be enriched in the dorsal region of eye and wing discs30. We have found that this pattern of Ance mRNA is similar to that of pnr expression. Since pnr is selectively expressed in the peripodial cells of eye discs34,35, we investigated whether Ance is also preferentially expressed in the peripodial epithelium.
We utilized an Ance-lacZ reporter that reflects the dorsal expression of Ance mRNA in embryos11. First, we found that Ance-lacZ was dorsally expressed in eye and antenna discs (Fig. 1a,b), which resembled the pattern of Ance mRNA expression shown in a previous report30. Apical confocal sectioning of eye discs indicated that Ance-lacZ expression was preferentially localized to the peripodial epithelium (Fig. 1a’,b’) like that of pnr 34,35. Immunostaining with membrane marker anti-Discs-large (Dlg) antibody showed that Ance-lacZ expressing cells are large peripodial cells (Fig. 1a”’, b”’). We also checked the pattern of Ance protein expression using an anti-Ance polyclonal antibody36. Ance protein was found in most peripodial cells of the eye disc in contrast to the dorsally localized Ance-lacZ reporter expression (Fig. 1a”, b”).
Expression pattern of Ance-lacZ and Ance protein in eye and wing discs. (a–c) Immunostaing of eye discs for Ance-lacZ (green), Ance (red) and Dlg (blue channel in black/white). (a-a”’) Low magnification views of an eye-antenna disc sectioned at the peripodial epithelium (PE) level. (a) Merge. (a’) Ance-lacZ. (a”) Ance protein. (a”’) Dlg. (b-b”’) High magnification views of a dorsal half of the eye disc shown in (a-a”’). (b) Merge. (b’) Ance-lacZ expressed in the dorsal PE. (b”) Ance protein. (b”’) Dlg staining of PE cells. (c-c”’) Dorsal region of the eye disc sectioned at the disc proper (DP) level. (c) Merge. (c’) Ance-lacZ is weakly expressed in the anterior dorsal region of DP. (c”) Ance protein in the lumen between PE and DP in the disc margin (arrow). (c”’) Dlg staining shows differentiating retinal cells posterior to the morphogenetic furrow (MF). It also shows a cross-section view of the lumenal space between the PE and DP layers in the disc margin. (d–f) Immunostaining of wing discs. (d-d”’) Low magnification views of a wing disc sectioned at the PE level. (d) Merge. (d’) Ance-lacZ strongly expressed in the dorsal region. (d”) Ance protein found in most PE cells. (d”’) Dlg staining of PE cells. (e-e”’) High magnification views of dorsal half of the wing disc shown in (d-d”’). (e’) Ance-lacZ expression in dorsal PE cells. (e”) Ance protein. (e”’) Dlg staining of dorsal PE cells. (f-f”’) Dorsal region of the wing disc sectioned at the DP level. (f’) Ance-lacZ expression is weakly detected in the dorsal region of DP. (f”) Ance protein in the lumen (arrow). (f”’) Dlg staining shows DP cells and a cross-section view of the lumen between the PE and DP layers in the disc margin. (g-g”’) High magnification views of the wing pouch region at the PE level shown in (d-d”’). (g’) Ance-lacZ expression (arrow). (g”) Ance protein. (g”’) Dlg staining of dorsal PE cells. (h-h”’) High magnification views of the wing pouch region at the DP level shown in (d-d”’). (h’) No Ance-lacZ expression in the pouch. (h”) Ance protein localized in the disc lumen (arrow). (h”’) Dlg staining of DP cells. Scale bar, 50 μm (a-h”’).
In more basal sections, Dlg was detected in an array of differentiating photoreceptor cells in the disc proper (Fig. 1c”’) where Ance-lacZ was weakly detected in the anterior dorsal region (Fig. 1c’). Tangential sectioning at the disc proper level also shows a cross-section view along the margin of the disc. In this marginal area, Ance was highly localized in the lumenal space between the peripodial epithelium and the disc proper (Fig. 1c, c”, c”’). Although Ance protein was not clearly detected in the differentiating retina posterior to the morphogenetic furrow (Fig. 1c”), closer examination revealed weak levels of Ance in photoreceptor cells (arrowheads in Fig. S1e). Since Ance-lacZ is preferentially expressed in the dorsal peripodial cells, it can be inferred that the Ance protein detected widely in other areas of the eye disc probably originates from the secretion from cells expressing Ance.
Next, we examined the Ance expression in the wing disc. Ance-lacZ expression was abundant in the peripodial cells of the notum area (Fig. 1d–e”’), which is similar to the pnr expression pattern27,34,35. Weak expression of Ance-lacZ was also detected in the notum disc proper cells (Fig. 1f’). In contrast to the high levels of Ance-lacZ expression in the notum region, Ance-lacZ was weakly detected in the peripodial cells that cover the wing pouch region (Fig. 1g’). Ance protein in the wing disc was detected more broadly than the region of Ance-lacZ expression (Fig. 1d,e,g). Ance was abundant in the lumenal space (Fig. 1f”, h”), thus in contact with most disc proper cells including the notum and the wing pouch (Fig. S2). Taken together, the expression patterns of Ance-lacZ and Ance protein suggest that Ance mRNA is highly expressed in the dorsal peripodial region, while Ance protein is secreted to neighboring peripodial cells, lumenal space, and most disc proper cells.
Ance expression is regulated by Dpp signaling during larval development
Ance expression is known to be regulated by Dpp signaling during embryogenesis10,11. Because Dpp signaling can activate different transcriptional target genes in different tissues and stages18,19,37, it is important to check whether Dpp signaling is required for the specific pattern of Ance expression in larval imaginal discs. We therefore examined the effects of loss of Mad, the transcription factor that mediates Dpp signaling, on Ance-lacZ expression. Ance-lacZ reporter expression was nearly undetectable in Mad mutant clones generated in the peripodial cells of wing discs (Fig. 2a,b and S3a). In addition, loss of Tkv, the receptor serine/threonine kinase for the Dpp ligand, resulted in reduced Ance-lacZ reporter expression in peripodial cells of eye disc (Fig. S4a,b).
Dpp signaling is required for Ance expression in eye and wing discs. (a,b) Wing disc containing Mad 1-2 mutant clones stained for GFP clone marker and Ance-lacZ. Large Mad 1-2 mutant clones (GFP-negative) are shown in the wing pouch region (the box area in Fig. 1d’). Ance-lacZ expression is nearly absent in Mad 1-2 clones (arrow). (a) Merge of (a’-a”) at the PE level. (a’) GFP. (a”) Ance-lacZ. (b-b”) Z-section view of the dashed line in (a). Loss of Ance-lacZ expression in Mad 1-2 clone is indicated by the arrow. (c,d) Eye disc containing Mad 1-2 mutant clones stained for GFP clone marker and Ance protein. (c) Merge of (c’-c”) at the EP level. (c’) GFP. (c”) Ance protein. Mad 1-2 clone located away from the Ance-lacZ expressing area (marked by arrow) shows normal level of Ance protein. A Mad 1-2 mutant clone in the dorsal PE region (the box area in Fig. 1a’) shows a low level of Ance staining (dotted circle). (d-d”) High magnification views of the box area in (c). A low level of Ance protein can be detected (Arrow). (e-e”) Wing disc with GFP overexpression by ptc-Gal4. (e) Merge of an apical section images (e’-e”). (e’) GFP in the ptc-expressing region. (e”) No ectopic Ance expression in the ptc area of the wing pouch (circle). (f–f”) Wing disc with tkv QD overexpression by ptc-Gal4. (f) Merge of apical section images (f’-f”). (f’) GFP in the ptc-expressing region. (f”) Ectopic Ance expression in the proximal sites (arrows) in the ptc area of the wing pouch. Scale bars, 50μm (a–f”).
We also tested the effects of loss of Mad on Ance protein expression. When Mad 1-2 mutant clones were generated in the regions distant from the dorsal domain of Ance-lacZ expression in eye disc, they showed normal level of Ance protein (arrow in Fig. 2c). This result suggests that Ance protein in these clones is provided non-autonomously from the Mad + Ance-expressing cells. Mad 1-2 clones located within the Ance-expressing region showed Ance proteins within the clones but at a reduced level (Fig. 2c,d).
We also checked whether ectopic Dpp signaling can activate Ance expression using Gal4-UAS system38. When GFP was overexpressed along the anterior-posterior boundary of wing disc using ptc-Gal4 driver (labelled as ptc > GFP), there was no ectopic Ance expression in the ptc-expressing area (Figs 2e and S5a). However, overexpression of tkv QD, a constitutively active form of tkv, led to ectopic expression of Ance in the ptc region of wing disc (Figs 2f and S5b). Interestingly, the ectopic Ance expression was mainly localized to two proximal spots in the ptc region of the wing pouch (Fig. S5b), suggesting that Dpp signaling can activate Ance expression in specific regions. Together, these results suggest that Dpp signaling is necessary and sufficient for Ance expression in specific territories of imaginal discs.
Ance is dispensable for development
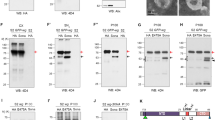
Since Ance is expressed in specific domains of discs, we tested whether Ance plays a role in the development of these organs. To examine the function of Ance, we generated two Ance-deletion mutants, Ance d110 and Ance d128, by imprecise excision of a P-element inserted in 5′ UTR of the Ance gene. In both mutants, the first exon and adjacent regions were deleted (1247 and 976 bp deletion in Ance d110 and Ance d128, respectively), which was confirmed by genomic PCR and sequencing (Fig. 3a,b). Ance mRNA and proteins were not detected in these deletion mutants, which were checked by RT-PCR (Fig. 3c), western blot (Fig. 3d), and immunostaining of imaginal discs (Fig. 3e), indicating that these two mutations are null alleles. These Ance-deletion mutants were viable and fertile with no obvious morphological defects in adult eyes and wings (Fig. 3f,g). This was unexpected because it was previously reported that Ance mutants show lethality or infertility10,33. Interestingly, Ance null mutants generated in this work fully complemented the Ance mutants (Ance 34Eb–1 and Ance 34Eb–2) described in earlier reports (data not shown). In contrast, the lethality of Ance 34Eb–1 and Ance 34Eb–2 failed to complement a Smg5 mutation (Smg5 e04233), which is located approximately 1 kb upstream from the 5′ region of the Ance gene. These results indicate that the lethality of Ance 34Eb–1 and Ance 34Eb–2 is likely due to a mutation(s) in Smg5 rather than the Ance gene (data not shown). Taken together, these data suggest that Ance is dispensable for development.
Ance-deletion mutants are viable. (a) Schematic structure of the Ance gene. The G3223 line has a P-element inserted in the 5′ UTR of Ance. Ance d110 and Ance d128 mutants were generated by imprecise excision of G3223. Two mutants lack the first exon of Ance gene. Red arrows and green box indicate genomic PCR primers used in (b) and RT-PCR in (c), respectively. (b) Confirmation of deletions in Ance mutants. Genomic PCR results of w 1118, Ance d110, and Ance d128 show smaller PCR products in two Ance mutants compared to w 1118 control. (c) RT-PCR results of w 1118, Ance d110, and Ance d128. No PCR product is detected in two Ance mutants. (d) Ance protein is not detected in Ance d110 and Ance d128 mutants. (e) Absence of Ance staining in Ance mutants. Eye discs of w 1118, Ance d110, and Ance d128 stained for Ance and Dlg. Scale bar, 100 μm. (f) Adult eye morphology of w 1118, Ance d110, and Ance d128. w 1118 control shows the normal eye. Ance d110 and Ance d128 show normal eye morphology. (g) Adult wing morphology of w 1118, Ance d110, and Ance d128. w 1118 control shows the normal wing. Ance d110 and Ance d128 show normal wing morphology.
Ance mutations show genetic interaction with Dpp signaling
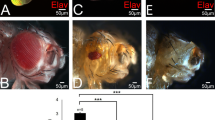
Although Ance deletion mutants and Ance RNAi flies show no discernible phenotype in the normal condition, it is possible that Ance mutations might show synthetic phenotypes with other mutations in the functionally related genes. Since Ance expression is regulated by Dpp signaling, we reasoned that Ance might show genetic interaction with Dpp signaling components. To detect genetic interaction, we examined the effects of altering the Ance level on the phenotypes of dpp overexpression or dpp mutations. First, we used flies that overexpress dpp in the eye. As shown in Fig. 4a, overexpression of dpp under GMR-Gal4 driver (GMR > dpp) resulted in roughening of the eye surface. Under this background, reducing the Ance gene dosage by one copy of Ance mutation partially suppressed the rough eye phenotype, showing improved arrays of ommatidia (Fig. 4c). In third instar eye disc, dpp overexpression by GMR-Gal4 resulted in irregular wrinkling of developing retina, as shown by immunostaining for the pan-neuronal marker Elav and the cell membrane marker Dlg (Fig. 4b-b”). One copy of Ance mutation significantly suppressed the abnormalities caused by dpp overexpression in eye disc (Fig. 4d-d”).
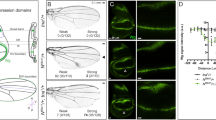
Genetic interaction between Dpp signaling and Ance. (a) Effects of GMR > dpp in adult eye. Ectopic Dpp results in roughening of the eye surface. (b) Effects of GMR > dpp in developing eye disc. Ectopic Dpp results in irregular wrinkling of developing retina. (b) Merge. (b’) Elav. (b”) Dlg. Scale bar, 50 μm. (c) Heterozygous Ance d128 mutation partially suppresses GMR > dpp phenotype in adult eye. (d) Heterozygous Ance d128 mutation partially suppresses GMR > dpp phenotype in eye disc, resulting in relatively regular arrays of photoreceptor clusters. (e–g) Genetic interaction between dpp and Ance mutations in the wing. (e) Ance d128/Ance d128 homozygote. (f) dpp s1 dpp d-ho/dpp s1 dpp d-ho homozygote. (g) dpp s1 dpp d-ho Ance d128/dpp s1 dpp d-ho Ance d128 mutants. dpp s1 dpp d-ho Ance d128/dpp s1 dpp d-ho Ance d128 mutants show frequent partial loss of the second vein (L2 vein) as indicated by circled area. (h–j) Genetic interaction between tkv and Ance mutations. (h) tkv a12/+ heterozygote. (i) tkv a12/Ance d110 transheterozygote. (j) tkv a12/Ance d128 transheterozygote. tkv a12/Ance d110 and tkv a12/Ance d128 transheterozygotes result in abnormal thickvein phenotype at the distal tip of the L4 vein indicated by arrow and inset. (k) Quantification of L2 wing vein defects in dpp and Ance mutations. Percent indicates the frequency of partial loss of L2 vein. 1 = 0% (n = 0/105), 2 = 7.3% (n = 7/95), 3 = 49.5% (n = 53/103), 4 = 70.1% (n = 68/97). (l) Quantification of L4 vein defects in tkv and Ance mutations. Percent indicates the frequency of the L4 vein defect. 1 = 4.1% (n = 3/71), 2 = 54.7% (n = 87/159), 3 = 49.2% (n = 67/132), 4 = 76.3% (n = 103/135), 5 = 71.8% (n = 102/142).
We also checked whether Ance mutations genetically interact with dpp hypomorphic mutation. Ance d128 mutation does not affect any vein (Fig. 4e). dpp s1 dpp d-ho homozygotes show loss of distal part of 4th longitudinal wing vein (L4) (Fig. 4f), besides the partial loss of the L2 vein at a low frequency (7.3%) as seen in Fig. 4g. Interestingly, Ance d128 mutation strongly enhanced the frequency of L2 vein loss phenotypes of dpp s1 dpp d-ho to 70.1% of flies examined (Fig. 4g,k). This result indicates that there is a synergistic interaction between Ance and dpp mutations. In addition, we tested whether mutations in Ance and tkv a12 can show genetic interaction in the wing. tkv a12/+ heterozygote showed normal veins (Fig. 4h). However, tkv a12/Ance d110 trans-heterozygote resulted in a weak thickvein phenotype at the distal tip of the L4 vein (Fig. 4i, 54.7%). tkv a12/Ance d128 trans-heterozygote showed nearly identical genetic interaction as tkv a12/Ance d110 (Fig. 4j, 49.2%). In addition, tkv a12 heterozygotes under Ance d110 (or Ance d128) homozygote condition showed increased frequencies of the thickvein phenotype (Fig. 4i, 76.3% and 71.8%, respectively). Taken together, these data show that dpp mutant phenotypes can be modified by altering the level of Ance function, although loss of Ance alone is insufficient to cause visible developmental defects.
Ance expression is directly controlled by pannier
As described earlier (Fig. 1), the Ance-lacZ reporter shows preferential dorsal expression similar to the pattern of pannier (pnr) expression27,34,35. To check whether pnr is involved in regulating Ance expression during larval development, we generated clones of pnr null mutant cells in eye imaginal discs. Because Ance-lacZ is expressed near the dorsal margin of the eye disc, pnr VX6 clones outside the Ance-lacZ expressing region did not show any lacZ expression. Conversely, pnr VX6 mutant clones within the Ance-expressing dorsal area of the eye disc showed little or no expression of Ance-lacZ (Figs 5a-a” and S6). In contrast, variable levels of Ance protein were detected in pnr VX6 mutant clones (Fig. S7), consistent with non-cell autonomous distribution of Ance protein. We also tested the effects of Pnr overexpression using various Gal4 drivers but could not check the effects due to early lethality. Nevertheless, our analysis of mutant clones clearly indicates that pnr is required for the dorsal-specific Ance expression.
Pnr is required for Ance expression and binds to Ance regulatory region. (a) pnr VX6 mutant clones result in reduction of Ance-lacZ expression in the dorsal eye disc. (a) Merge. (a’) GFP. (a”) Ance-lacZ. The areas marked by dotted lines show clear decrease in the level of Ance-lacZ. PE: peripodial membrane. Scale bars, 50 μm. (b) The scheme of a 533 bp regulatory region of Ance-lacZ reporter. This regulatory region of Ance-lacZ has three potential GATA sites, as shown in the GATA region sequences in (d). (c) Pnr binds to Ance regulatory region. Electrophoretic mobility shift assay (EMSA) shows that Ance 533 bp enhancer is bound by increasing amounts of GST-Pnr-DBD. Only 3′ part of Ance enhancer is bound by GST-Pnr-DBD. (d) Sequences of GATA regions in Ance enhancer. In Ance 3′ regulatory part, three potential GATA sites exist. Three oligonucleotides (oligo 1 to 3) and a mutated oligo were used in (e,f). ‘2 Mt’ indicates an oligonucleotides mutated in the GATA 2 region. (e) Pnr binds to a second GATA site in the Ance regulatory region. EMSA with biotin-labeled potential GATA sites and GST-Pnr-DBD shows considerable band upshift by the second GATA site. This band shift is inhibited by unlabeled competitor DNA. (f) Mutation of the second GATA site in the Ance regulatory region (biotin-labeled Mt DNA) results in no binding with Pnr-DBD (lane 1). Addition of the oligo mutated in the second GATA (unlabeled Mt DNA) in the lane 1 reaction has no effect (lane 2). Pnr-DBD could bind to biotin-labeled wild-type (Wt) DNA in the presence of unlabeled Mt DNA (lane 3). Unlabeled Wt oligo reduced the binding between Pnr-DB and biotin-labeled Wt DNA (lane 4).
As Ance-lacZ expression was lost in pnr VX6 mutant clones, we hypothesized that Pnr might be directly required for Ance expression. To check this possibility, we tested whether Pnr can bind to the Ance enhancer. The Ance-lacZ reporter contains a 533 bp Ance regulatory region which has three potential GATA binding sites in the 3′ half. First, we carried out electrophoretic mobility shift assay (EMSA) using purified GST-Pnr-DBD (DNA binding domain) and the 533 bp Ance regulatory region. EMSA using GST-Pnr-DBD and the 533 bp Ance enhancer showed an upward band shift when GST-Pnr was added, indicating that Pnr binds to the Ance region (Fig. 5c). The 3′ region of the Ance enhancer containing three GATA sequences showed similar band shifting, but the 5′ region did not, suggesting that one or more of the GATA sites might be involved in binding with GST-Pnr-DBD (Fig. 5c). Additional EMSA assays with three DNA fragments containing different GATA sites showed that the second GATA site was strongly bound to GST-Pnr-DBD (Fig. 5e). These data suggest that Pnr directly regulates Ance expression at transcriptional level by binding to the second GATA site of the Ance enhancer. To confirm that the second GATA site in Ance enhancer is important, we generated a mutant form of GATA2 fragment, which has AAAA instead of GATA in the second GATA site. In EMSA assays, Pnr binding to the GATA2 DNA was strongly reduced by the addition of unlabeled wild-type GATA2 DNA but not by the mutated GATA2 DNA (Fig. 5f). This further supports the specific binding of Pnr to the GATA2-containing DNA.
In addition, we carried out luciferase assay in S2 cells using the wild-type and the mutated Ance GATA enhancer. Single transfection of either Mad or pnr weakly increased luciferase activity by 8.8% and 20.6%, respectively. Double transfection of Mad and pnr significantly enhanced luciferase activity to 40.3%, which was higher than the combined effects of Mad and pnr (Fig. 6a). Next, we performed similar luciferase assays with Ance GATAmt, the Ance regulatory region with a mutation in the GATA2 site. In this assay, Mad alone could weakly activate luciferase activity (11.4%), but Pnr alone did not significantly enhance the luciferase activity due to the GATA2 mutation. Interestingly, when both Mad and pnr were overexpressed, the effect of Mad was consistently inhibited (3.8% lower than the control level) (Fig. 6b). This suggests that overexpressed Pnr may interact with Mad but fail to bind to the mutated Ance GATAmt regulatory region, perhaps dominantly inhibiting the Mad effects.
Ance expression is controlled by Mad and Pnr in vivo. (a) Luciferase assay using Ance WT-luc in S2 cells. Double transfection of Mad and pnr induces stronger enhancement in luciferase activity compared to single transfection of Mad or pnr. (b) Luciferase assay using Ance GATAmt-luc in S2 cells. Double transfection of Mad and pnr fails to induce luciferase activity compared to single transfection of Mad or pnr. Luciferase activity was normalized to control Renilla luciferase activity. All data represent the mean ± s.e.m. (error bars), and P-values were calculated by using Student’s t-test. n.s, not significant. ***P < 0.001. ****P < 0.0001. (c-c”) Expression pattern of Ance-lacZ in eye disc. (c) Merge of (c’) and (c”). (c’) Ance-lacZ. (c”) Dlg staining. (d-d”) Expression pattern of Ance-lacZ in wing disc. (d) Merge of (d’) and (d”). (d’) Ance-lacZ. (d”) Dlg staining. (e-e”) Expression pattern of MDel-lacZ in eye disc. Loss of Mad binding sites in Ance enhancer nearly abolishes lacZ expression in eye disc (e’). (e) Merge of (e’) and (e”). (e’) MDel-lacZ. (e”) Dlg staining. (f) Expression pattern of MDel-lacZ in wing disc. Loss of Mad binding sites in Ance enhancer nearly abolishes lacZ expression. (f) Merge. (f’) MDel-lacZ. (f”) Dlg. (g-g”) Expression pattern of Ance GATAmt-lacZ in eye disc. Mutation of the second GATA site in Ance enhancer results in strong reduction of lacZ expression. (g) Merge. (g’) Ance GATAmt-lacZ. (g”) Dlg. (h) Expression pattern of Ance GATAmt-lacZ in wing disc. Mutation of the second GATA site in Ance enhancer results in strong reduction of lacZ expression. (h) Merge. (h’) Ance GATAmt-lacZ. (h”) Dlg. Scale bars, 100 μm (c–h”).
Our data from the luciferase assays in S2 cells showed that Mad and Pnr can bind to the Ance regulatory region to activate transcription. However, the level of activation was relatively weak, and it was difficult to determine whether the combined effects of Mad and Pnr were additive or synergistic. Thus, we tested the effects of Mad and Pnr in vivo using transgenic reporter lines. Firstly, to determine the effects of Dpp signaling on the Ance expression, we utilized an MDel-lacZ reporter, which is a modified version of Ance-lacZ mutated in the Mad-binding sites19. As shown in Fig. 6e and f, MDel-lacZ expression was significantly reduced in both eye and wing imaginal discs compared to Ance-lacZ (Fig. 6c,d). In addition, we checked the possibility that Ance regulated by Dpp signaling might have a positive or negative feedback during development. As indicated in Fig. S8, there was no difference in the level of phospho-Mad (p-Mad) between wild-type and Ance-deletion mutant, indicating that there is no obvious feedback regulation between Dpp signaling and Ance.
Secondly, to test whether the second GATA site in the Ance enhancer is important in vivo, we generated an Ance GATAmt-lacZ line, which consists of AAAA instead of GATA in the second GATA site and checked its expression pattern in eye and wing discs. As indicated in Fig. 6g and h, lacZ expression was significantly reduced compared to Ance-lacZ expression (Fig. 6c,d), indicating that the second GATA site bound by Pnr is required for Ance expression. Since loss of either Mad or Pnr binding site severely reduces Ance expression in imaginal discs, these data suggest that Mad and Pnr act together in vivo rather than acting independently.
Physical and genetic interaction between Pnr and Dpp signaling
Our data described above show that Mad and Pnr might function together for Ance expression in imaginal discs. While pnr expression is restricted to the dorsal domain27,34,35, dpp mRNA expression in third instar eye disc is restricted to the morphogenetic furrow and part of the dorsal/ventral margins21,39. However, Dpp is secreted to broader regions of the eye disc. Therefore, although mRNA expression patterns of pnr and dpp are not identical in the eye disc, cells that receive the Dpp signal might overlap in the dorsal margin region of pnr expression. Thus, we examined the pattern of p-Mad expression in third instar eye disc. p-Mad expression was enriched along the MF, but it was also elevated at the dorsal margin of eye disc where pnr is expressed (Fig. 7a-a”, arrow). Therefore, Pnr and Mad may function together to regulate Ance expression. To address this possibility, we tested whether Pnr and Dpp signaling genetically interact (Fig. 7b). When tkv a12 and pnr VX6 flies were crossed, trans-heterozygote progeny (tkv/+; pnr/+) showed extra vein formation from the posterior crossvein (Fig. 7b,c). Although the extra vein phenotype was mild, it was consistently observed in 60.4% transheterozygotes, while such phenotype was rarely detected in tkv/+ or pnr/+ heterozygotes (7.9% and 0%, respectively) (Fig. 7d).
Physical and genetic interaction between Pnr and Dpp signaling. (a) Immunostaining of pnr > GFP eye disc for GFP and phospho-Mad (p-Mad). GFP is selectively expressed in the dorsal region. p-Mad expression is ubiquitously distributed but is relatively stronger in the dorsal region. (a) Merge. (a’) pnr > GFP. (a”) pMad. Scale bars, 100 μm. (b-d) Genetic interaction between tkv a12 and pnr VX6. tkv a12/+ heterozygote shows normal posterior crossvein (c). tkv a12/pnr VX6 transheterozygote results in extra vein formation (indicated by arrow) from the posterior crossvein (c). (d) Quantification of the extra vein phenotype. Percentage of the extra vein phenotype in tkv a12/+, pnr VX6/+, and tkv a12/pnr VX6 flies. tkv a12/+ = 7.9% (n = 6/76), pnr VX6/+ = 0% (n = 0/75), tkv a12/pnr VX6 = 60.4% (n = 58/96). (e) Mad and Pnr form a complex. Pnr is co-immunoprecipitated by anti-Mad antibody in larval extracts. (f) Mad and Pnr directly interact. GST-pulldown assay shows direct interaction between Pnr and Mad. N-terminal part of Pnr physically interacts with MBP-Mad. Coomassie blue-stained SDS-PAGE gel shows GST, GST-PnrN, GST-PnrC, and GST-PnrFull. (g) A model for the relationships among Ance, Dpp signaling, and Pnr in imaginal discs. Dorsal expression of Ance is regulated by a combination of three ways. Firstly, Mad activated by Dpp signaling binds to a Mad binding site of the Ance regulatory region. Secondly, Pnr directly activates Ance expression by binding to a GATA site of Ance regulatory region. Thirdly, Mad physically interacts with dorsal-specific Pnr, which may provide the dorsal specificity of Ance expression. Mad is required for wing vein and eye differentiation by activating unknown targets (dashed arrows). Genetic interactions with Mad suggest that Ance may affect the Mad function in eye and wing vein development (see Discussion for details). It is unknown whether pnr is regulated by Dpp signaling as in embryogenesis.
The overlapping localization of Pnr and Mad and their genetic interaction suggested that they might be associated together in a protein complex. To test this possibility, we carried out co-immunoprecipitation between Pnr and Mad using larval extracts. As shown in Fig. 7e, Pnr was co-immunoprecipitated by anti-Mad antibody, suggesting that Pnr and Mad physically interact. We also checked whether their interaction is direct. GST-pulldown assay using bacterially purified GST-Pnr and MBP-Mad showed that N-terminal part of Pnr binds to MBP-Mad (Fig. 7f), indicating that Pnr and Mad directly interact. Taken together, these data support physical and functional interaction between Pnr and Mad, as outlined in the scheme shown in Fig. 7g (See Discussion).
SMAD2 and GATA4 are required for ACE regulation in human cells
Mammalian Angiotensin-converting enzyme (ACE) plays key roles in regulating renin-angiotensin system. Since our data showed that Drosophila Mad and Pnr are required for Ance expression, we hypothesized that mammalian SMAD and GATA factors, homologs of Drosophila Mad and Pnr, respectively, might be involved in ACE expression. To test this possibility, we examined the effects of RNAi of SMAD1/2 and GATA2/4 in HEK293 cells (human embryonic kidney 293 cells). SMAD1 and GATA2 siRNAs did not affect ACE expression (Fig. S9a), but SMAD2 and GATA4 siRNAs were effective in reducing ACE expression (Fig. 8a,b). The levels of knockdown for each siRNA for SMAD1/2 and GATA2/4 were approximately 70–90% (Fig. S9b,c). Furthermore, double siRNA treatments for both SMAD2 and GATA4 were more effective in reducing ACE expression compared to single siRNA for SMAD2 or GATA4 (Fig. 8c). We also tested whether overexpressing SMAD2 and GATA4 is sufficient to induce the expression of ACE. Overexpression of either SMAD2 or GATA4 showed slight increase in the ACE mRNA level, whereas overexpression of both SMAD2 and GATA4 resulted in further increase (Fig. 8d), which was similar to the effects of overexpressing Mad and Pnr in Drosophila S2 cells (Fig. 6a).
SMAD2 and GATA4 are required for ACE expression in HEK293 cells. (a–c) Effects of silencing SMAD2 or GATA4 on ACE expression. Y-axis indicates relative expression compared with control siRNA (siSCR). (a) SMAD2 knockdown results in reduction of ACE level (49.4%). (b) GATA4 knockdown reduces ACE level (64.9%). (c) Single knockdown of SMAD2 or GATA4 decreases ACE expression (54.1% or 68.9%), respectively. Double siRNAs for SMAD2 and GATA4 result in further inhibition of ACE level (26.5%). (d) ACE expression upon overexpressing SMAD2 and/or GATA4. Double transfection of SMAD2 and GATA4-expressing vectors induces stronger enhancement in ACE expression compared to single transfection of SMAD2 or GATA4. (e) Enrichment of SMAD2 and GATA4 on ACE promoter. Left: Schematic diagram of human ACE promoter. Transcription start site (TSS) and primer sets used for qPCR are indicated. Right: Relative enrichment of SMAD2 and GATA4 to IgG control. (f) Reciprocal co-immunoprecipitation between SMAD2 and GATA4 in HEK293 cells. Overexpressed GATA4 (GATA4-V5) is co-immunoprecipitated with endogenous SMAD2 and vice versa. GATA4 is detected by anti-V5 antibody. (g) ACE overexpression suppresses the increased level of phospho-ERK (p-ERK) in SMAD2/GATA4-knockdown HEK293 cells. siSMAD2/siGATA4 in HEK293 cells increases p-ERK compared to siSCR control. ACE overexpression decreases p-ERK in siSMAD2/siGATA4-treated cells. (h) Angiotensin II (Ang II) treatment reduces the level of p-ERK in SMAD2/GATA4-knockdown HEK293 cells. siSMAD2/siGATA4 in HEK293 cells increases p-ERK compared to siSCR control. Ang II treatment decreases p-ERK in a dose-dependent manner. (i) Ang II rescues the reduced cell viability in SMAD2/GATA4-knockdown HEK293 cells in a dose-dependent manner. All data represent the mean ± s.e.m. (error bars) and P-values were calculated by using Student’s t-test. n.s, not significant, *P < 0.05. **P < 0.01. ***P < 0.001. ****P < 0.0001.
To check whether SMAD2 and GATA4 are directly involved in regulation of ACE expression, we performed chromatin immunoprecipitation (ChIP) assays (Fig. 8e). Our data showed that GATA4 and SMAD2 bind to the first exon of ACE gene, indicating their transcriptional roles in the regulation of ACE expression. Taken together, both SMAD2 and GATA4 are required for ACE expression in HEK293 cells as in Drosophila.
The regulation of ACE expression by SMAD2 and GATA4 suggests that SMAD2 and GATA4 might be associated in a protein complex to regulate their target gene expression as in Drosophila. To test this possibility, we carried out co-immunoprecipitation experiments between SMAD2 and GATA4 using GATA4-overexpressing HEK293 cell lysates. Overexpressed GATA4 was co-immunoprecipitated by endogenous SMAD2 immunoprecipitation, and endogenous SMAD2 was also co-immunoprecipitated by overexpressed GATA4 (Fig. 8f). These results suggest that SMAD2 and GATA4 form a protein complex in human as seen in the Mad-Pnr co-immunoprecipitation in fly.
To look for clues to the function of SMAD2 and GATA4, we examined their effects on the extracellular signal-regulated kinase (ERK) signaling that regulates a variety of cell physiology. First, we checked the level of phosphorylation of ERK (p-ERK) and found that the treatment of siSMAD2/siGATA4 increased p-ERK compared to the siScramble (siSCR) control (Fig. 8g). Interestingly, the increased p-ERK level in SMAD2/GATA4-knockdown HEK293 cells was suppressed when their target gene ACE was overexpressed. We also tested whether an addition of Angiotensin II (Ang II) can have similar effects. Indeed, the increased p-ERK level in SMAD2/GATA4-knockdown cells was antagonized by Ang II in a dose-dependent manner, indicating that ACE overexpression and Ang II treatment could rescue the effects of silencing SMAD2 and GATA4. Next, we checked viability in HEK293 cells. As shown in Fig. 8i, SMAD2 and GATA4 siRNAs decreased cell viability compared to control siRNA. An addition of Ang II did not affect the viability in siSCR-treated HEK293 cells, but it partially rescued reduced cell viability of SMAD2/GATA4-knockdown cells. Taken together, we conclude that SMAD2 and GATA4 directly regulates ACE expression as in Drosophila and loss-of-function phonotypes of SMAD2 and GATA4 can be restored in part by ACE overexpression or Ang II treatment.
Discussion
Expression of the Drosophila ACE homolog Ance is developmentally regulated in larval imaginal discs. Here we have provided evidence that Mad and Pnr transcription factors play key roles in Ance regulation.
Previously, it has been shown that the Ance expression during embryogenesis is activated by Dpp signaling from the ectodermal dorsal midline10,11. Our data show that Dpp signaling is essential for Ance expression in imaginal discs, consistent with the role of Dpp for Ance expression in embryogenesis. Transcriptional activation in eukaryotes is often regulated by synergistic effects of other enhancer binding proteins40. It has been shown that Mad is a partner of the Fos transcription factor for a context-dependent transcriptional activation in Drosophila 41. Mammalian Smad proteins can also promote different TGFβ target gene expression in cooperation with cell type-specific transcription factors42,43. In this regard, it is worth noting that both Mad and the GATA factor Pnr are required for Ance expression during larval development. In embryogenesis, Ance and pnr are independent targets of Dpp signaling with different thresholds for Dpp activity. In contrast, Ance expression in imaginal discs is regulated by a combination of Dpp and Pnr.
According to the clonal analysis, loss of either Mad or Pnr almost completely abolishes the Ance-lacZ expression in the eye and wing discs. This indicates that Mad and Pnr regulate Ance expression in a cooperative manner, consistent with their physical interaction. We also found a weak but consistent genetic interaction in wing venation between tkv and pnr mutations (Fig. 7b–d). Based on these results, we propose a model in which Mad activated by Dpp signaling can interact with Pnr for tissue-specific Ance expression (Fig. 7g). Secreted Dpp activates Mad in broad regions of imaginal discs. In contrast, Ance expression is enriched in the dorsal area of the eye and wing discs, indicating that additional dorsal factors are required for the restricted Ance expression. In our model, one of such dorsal factors might be Pnr, thus allowing context-dependent expression of the Ance gene in the dorsal-specific pattern.
The Ance deletion mutants generated in this study develop normally to fertile adults. This result was somewhat unexpected because Ance mutants were reported to be lethal or sterile10,31,33. Our molecular and genetic analysis of Ance null mutants shows that Ance is dispensable for development and fertility. It is possible that other Ance homologs might be able to compensate the loss of Ance. Ance and its homolog, Angiotensin-converting enzyme-related (Acer), can catalyze the same peptide, although they have different affinities for the substrates44. Therefore, there is a possibility that the Acer function could compensate for loss of Ance during development. However, double mutants for Ance and Acer were also found to be developmentally normal and fertile (data not shown). Analysis of the Ance function is more complicated due to the presence of four other Ance-related genes in the fly genome; Ance-2, Ance-3, Ance-4, and Ance-5. Due to their potentially redundant functions, loss of one or more Ance homologs may not affect animal development, unless the functions of all the Ance homologs are disrupted. Alternatively, other peptidases may also be used in the development of imaginal discs to circumvent the absence of Ance peptidase activity. Imaginal discs contain a variety of peptidases that hydrolyze a wide range of peptides and proteins45. The hydrolysis of peptides and proteins by some of these peptidases might be important for the production of dipeptides and free amino acids for protein synthesis required for morphogenesis. Active forms of signaling peptides might also be generated by peptidases for development of imaginal discs44,45. Thus, the absence of Ance can be compensated by other peptidases that can hydrolyze unidentified substrates of Ance to produce necessary peptides for development.
Although Ance null mutants do not exhibit obvious developmental defects, it is important to note that loss of Ance can modulate the phenotypes of impaired Dpp signaling. It has been shown that overexpression of dpp by GMR-Gal4 causes severe roughness and bulging of the adult eye46. We showed that reduced Ance partially suppressed the effects of Dpp overexpression (Fig. 4a–d). These results suggest that the effects of dpp gain-of-function in the eye are mediated at least in part by Ance, consistent with the induction of Ance gene expression by Dpp signaling. Because Ance overexpression alone is insufficient to affect the eye, Ance seems to modulate the function of an unknown target of Dpp signaling to modify the Dpp-induced eye phenotype (Fig. 7g). Because Ance protein is detected in the eye disc lumen as well as disc proper cells, it may produce peptide factors that modify the effects of Dpp shown in Fig. 4c,d. Furthermore, our data show genetic interaction between hypomorphic dpp (or tkv) mutations and Ance mutation in wing veins. Ance-lacZ is enriched in the dorsal part of wing disc, but high levels of Ance protein are broadly detected in the lumen of the wing pouch area (Fig. 1d’,d”, Fig. S2k,l). Ance protein abundant in the wing pouch, either in the lumen or disc proper cells by uptake, may produce active peptides to affect vein differentiation, thus providing a possible explanation for the genetic interactions in vein phenotypes. These genetic interactions in dpp Ance double mutants (dpp Ance) are synergistic, since dpp or Ance single mutants show little deficiency in the second vein (Fig. 4e–g,k). Under dpp (or tkv) hypomorphic conditions, the Ance expression level is expected to be reduced partially. The introduction of an Ance null allele under this condition further reduces the Ance level. However, because Ance null mutants develop normally, the vein phenotype in dpp Ance double mutants cannot simply be explained by a reduction of the Ance level. Instead, Ance may modulate a Mad function in vein patterning. When Dpp signaling is fully functional, the Mad function in vein development may not be affected by loss of Ance alone. In tkv heterozygotes or dpp hypomorphic conditions, however, the reduction of Ance may critically impair such pathways as seen in the double mutant conditions. Thus, despite the lack of visible phenotypes in Ance null mutants, Ance seems to play modulatory roles when Dpp signaling is partially compromised. These genetic results raise a possibility that Ance expression is not only regulated by Mad but also interacts with specific Mad signaling outputs that affect diverse tissues, as proposed in Fig. 7g. The identification of related substrates for Ance dipeptidase activity will help to understand the mechanism underlying these genetic interactions.
The human somatic ACE, a zinc-dependent dipeptidyl carboxypeptidase, plays an important role in the regulation of the renin-angiotensin system. ACE has long been targeted for effective treatment of hypertension and related cardiovascular diseases through ACE inhibitors47,48,49. TGFβ is also associated with activity of ACE in renal fibrosis and disease progression. Thus, anti-TGFβ antibody can improve renal fibrosis in mouse model50. Kidney diseases can be treated more effectively when both ACE and TGFβ are blocked by inhibitor and antibody, respectively51, suggesting an association between ACE and TGFβ signaling. In addition, GATA4 activates atrioventricular canal-specific enhancer that has synergistic effects with Smad during heart development52, although the mechanism underlying the synergy is unknown. Our data show that human SMAD2 and GATA4 are not only required for ACE expression but can also induce it in HEK293 cells. Our ChIP data and co-immunoprecipitation data suggest that SMAD2 and GATA4 physically interact each other and bind to the first exon of ACE gene to play a transcriptional role in ACE expression.
The Ras/Raf/ERK pathway plays oncogenic roles in many tumors53,54. However, ERK activation can also lead to anti-proliferative events depending on the conditions of signal activity55,56. According to our data, downregulation of SMAD2 and GATA4 in HEK293 cells reduces cell viability, possibly due to increased cell death. Increased p-ERK levels in SMAD2/GATA4-knockdown cells suggest that cell viability may be reduced by ERK-dependent apoptosis. These data are consistent with recent studies in which ERK signaling induces apoptosis in HEK 293 cells57. Interestingly, Angiotensin II treatment on SMAD2/GATA4-knockdown cells can rescue reduced cell viability and restore the phosphorylation level of ERK. In addition, ACE overexpression decreased p-ERK in SMAD2/GATA4-knockdown cells. Together, our work in Drosophila and human cells provides insight into the possible regulation of ACE expression by these two transcription factors in mammals.
Materials and Methods
Fly genetics
All Drosophila strains were grown and maintained at 25 °C. UAS-dpp, UAS-tkv QD, UAS-GFP, GMR-Gal4, and ptc-Gal4 were used for GAL4-UAS system38. Ance-lacZ and Mdel-lacZ lines were provided by Mike Levine (Princeton University, USA) and Christine Rushlow (New York University, USA), respectively. dpp s1 dpp d-ho (Bloomington, stock #398), tkv a12 FRT40A/CyO (gift from Christian Dahmann, Technische Universität, Dresden, Germany), Mad 1-2 FRT40A/CyO, hsflp; ubi-GFP FRT40A, FRT82B pnr VX6/TM6B, Tb, hsflp; FRT82B ubi-GFP, hsflp; FRT82B arm-lacZ lines were used for checking genetic interaction and generating mutant clones. For generation of Ance deletion lines (Ance d110 and Ance d128), P(EP)G3223 was imprecisely excised by crossing with Bc/CyO, Δ2-3 flies. For generation of Ance GATAmt-lacZ line, Ance promoter with mutation in GATA site was cloned into pH-Pelican vector using one-step SLIC method58 and the cloned vector was used for generating transgenic lines.
Immunostaining
Eye and wing imaginal discs were fixed with 4% paraformaldehyde in PBS for 15 min on ice. After washing twice with PBS, fixed discs were blocked in 5% normal goat serum/PBT (PBS with 0.3% Triton-X100) for 30 min. Samples were incubated at 4 °C overnight with primary antibodies at the following concentrations: rabbit anti-Ance (from R. Elwyn Isaac, University of Leeds, UK) at 1:2000, rabbit anti-Dlg (from Kyungok Cho, Korea Advanced Institute of Science and Technology, Korea) at 1:500, rat anti-Elav (Developmental Studies Hybridoma Bank, DSHB) at 1:100, mouse anti-lacZ (DSHB) at 1:100, sheep anti-GFP (Bio-Rad, 4745-1051) at 1:100, rabbit phospho-Smad (from Carl-Henrik Heldin, Uppsala University, Sweden) at 1:1000. After washing three times with PBT, secondary antibodies conjugated with Cy3 (1:600), Cy5 (1:400), or FITC (1:100) (Alexa Fluor, Molecular Probes) were used. Confocal fluorescent images were acquired using a Carl Zeiss LSM710 confocal microscope.
Electrophoretic Mobility Shift Assays (EMSA)
EMSA was carried out with GST-Pnr-DBD, PCR products of Ance 533 bp enhancer, 5′ Ance enhancer, 3′ Ance enhancer, and biotin-labeled oligonucleotides with potential GATA binding sites. DNA binding domain of pnr cDNA was PCR-amplified and cloned into pGEX-4T1 vector using one-step SLIC method58. After transformation of pGEX-4T1-Pnr-DBD into BL21 competent cells, GST-Pnr-DBD protein was purified by using GST bead. 533 bp full-length enhancer, 5′ enhancer, and 3′ enhancer of Ance-lacZ enhancer were amplified by PCR reaction. 1 ng of Ance enhancer PCR products was incubated with 0.5 or 1 μg of GST-Pnr-DBD proteins in binding buffer (24 mM HEPES pH 7.9, 8 mM Tris pH 8, 2 mM EDTA, 12% glycerol, and 50 ng/μl Salmon sperm DNA) for 30 min at room temperature. After incubation, DNA-protein complexes were resolved on a 6% native PAGE gel for 1 h at 70 V in 0.5X TBE. DNA-protein complexes on native gel were visualized by RedSafe.
For EMSA with biotin-labeled oligonucleotides and GST-Pnr-DBD, biotin-labeled probes with potential GATA sites were synthesized from Bioneer (Korea). EMSA was carried out according to the method described above. After electrophoresis, DNA-protein complexes were transferred to nylon membrane. Further steps were proceeded using LightShift Chemiluminescent EMSA kit (Thermo Scientific) according to the manufacturer’s instruction.
Luciferase assay
533 bp of wild-type or mutant Ance regulatory region from Ance-lacZ enhancer was cloned into pGL4.20-luc2 vector, and pAc5.1-FLAG-Mad and pAc5.1-Pnr-V5/His were generated by using pAc5.1-V5/HisA vector, Mad cDNA and pnr cDNA. Luciferase assay was performed in S2 cells using 0.1 μg pGL4.20-Ance-luc2 (or pGL4.20-AnceGATAmt-luc2), 0.1 μg pGL4.82-hRluc, 0.4 μg pAc5.1-FLAG-Mad, 0.4 μg pAc5.1-Pnr-V5, or 0.4 μg or 0.8 μg pAc5.1-lacZ. A total of 1 μg DNA was used for each transfection. Dual Luciferase Reporter Assay System (Promega) was used according to manufacturer’s instructions. Firefly luciferase activity was normalized to control Renilla luciferase activity.
Immunoprecipitation
Drosophila larval lysates were prepared in CHAPS buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% CHAPS, and protease inhibitor cocktail), and the lysates were cleared by centrifugation. The cleared lysate was incubated with 1 μg of goat anti-Mad antibody (Santa Cruz) for 1 hour at 4 °C. The mixture of lysate and antibody was incubated with 30 μl of Protein G Dynabead (Thermo Scientific) for overnight at 4 °C. After several rounds of washing in CHAPS buffer, proteins were eluted in SDS sample buffer by boiling at 94 °C for 5 min. Mouse anti-Pnr antibody and secondary anti-mouse antibody conjugated with HRP (Jackson ImmunoResearch) were used for western blotting.
HEK293 cell lysates were prepared in lysis buffer (40 mM Tris-Cl pH7.5, 120 mM NaCl, 1 mM EDTA, 0.3% CHAPS, protease inhibitors). The cleared lysate was incubated with 2 μg of rabbit anti-SMAD2 antibody (Cell Signaling, 5339 P) or mouse anti-GATA4 antibody (Santa Cruz, sc-25310X) for overnight at 4 °C. The mixture of lysate and antibody was incubated with 30 μl of Protein A or G SureBead (Bio-Rad) for 2 hours. Mouse anti-V5 (Invitrogen, R960-25) and Rabbit anti-SMAD2 were used for western blot.
GST pull-down assay
Full-length, N-terminal, and C-terminal parts of pnr cDNA were cloned into pGEX-4T1 vector using one-step SLIC method. Full-length Mad cDNA was cloned into pMal-c2 vector. For GST-pulldown, IPTG-inducible R2 cells (BL21 derivative) were transformed with plasmids for GST-PnrFull, GST-PnrN-term, GST-PnrC-term, and MBP-Mad. GST- or MBP-proteins were purified using a standard method. Equal amount of glutathione Sepharose 4B beads (Bioprogen) with GST- and MBP-fusion proteins were incubated in pull down buffer (20 mM TrisHCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 10% glycerol, 1% Triton X-100, 1 mM DTT, protease inhibitor cocktail, and 1 mM PMSF). Mouse anti-MBP antibody (NEB, E8032) and secondary anti-rabbit antibody (Jackson ImmunoResearch) were used for western blotting.
Cell culture and transfection
Drosophila S2 cells were cultured in M3 media (Sigma) with 10% insect medium supplement (Sigma). Transfection was carried out with Effectene reagent (Qiagen, Hilden, Germany) according to manufacturer’s instructions.
HEK293 cells were cultured in Dulbecco’s modified Eagles medium (DMEM) with 10% fetal bovine serum (FBS). For siRNA-mediated gene silencing, siRNA was transfected into HEK293 cells by reverse transfection using Lipofectamin 2000 to a final concentration of 10 μM. siRNAs were generated from Bioneer (Korea). siRNA sequences are shown in Table S1. Total RNA of siRNA-transfected cells was extracted 48 hours after transfection by Qiazol (Qiagen, UAS) according to the manufacturer’s instruction. For plasmid transfection, pCEP4 SMAD2 (Addgene, #16484) and tetO-GATA4 (Addgene, #46030) were used. cDNA was synthesized using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Realtime PCR Master Mix (Toyobo, Japan) was used to perform qRT-PCR assays. qRT-PCR primer sequences are shown in Table S2.
ACE vector (Sino Biological, HG11598-UT) was used for overexpression experiment and western blot analysis, and Angiotensin II (Sigma-Aldrich, A9525) was treated for western blot analysis and cell viability assay. For western blot analysis, rabbit anti-ERK (Cell Signaling, 4595 T) and rabbit anti-phospho ERK (Cell Signaling, 3101 S) were used.
ChIP assays
ChIP assays were performed by using a standard protocol (Millipore) with mouse anti-GATA4 (Santa Cruz, sc-25310 X), control mouse IgG, rabbit anti-SMAD2 (Cell Signaling, 5339 S), or control rabbit IgG. Eluted DNAs were analyzed by qPCR. The primer sequences are provided in Supplementary Table S3.
Cell viability assay
Approximately 1000 cells were plated onto 96 well plates in 10% FBS media. The culture media were exchanged using 0.5% FBS media, and Ang II was treated at indicated concentration 24 hours after plating. Relative cell viability was measured 24 hours after Ang II treatment using CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to the manufacturer’s instructions.
Statistical analysis
Statistical significance was determined by unpaired one-tailed Student’s t-test. P-values of <0.05 were considered as statistically significant. All data represent the mean ± s.e.m. (error bars).
References
Powell, J. et al. Inhibitors of angiotensin-converting enzyme prevent myointimal proliferation after vascular injury. Science 245, 186–188, https://doi.org/10.1126/science.2526370 (1989).
Navarro, J. F., Milena, F. J., Mora, C., Leon, C. & Garcia, J. Renal pro-inflammatory cytokine gene expression in diabetic nephropathy: effect of angiotensin-converting enzyme inhibition and pentoxifylline administration. Am J Nephrol 26, 562–570, https://doi.org/10.1159/000098004 (2006).
Heeneman, S., Sluimer, J. C. & Daemen, M. J. A. P. Angiotensin-converting enzyme and vascular remodeling. Circulation Research 101, 441–454, https://doi.org/10.1161/circresaha.107.148338 (2007).
Platten, M. et al. Blocking angiotensin-converting enzyme induces potent regulatory T cells and modulates TH1- and TH17-mediated autoimmunity. P Natl Acad Sci USA 106, 14948–14953, https://doi.org/10.1073/pnas.0903958106 (2009).
Krege, J. H. et al. Male-female differences in fertility and blood pressure in ACE-deficient mice. Nature 375, 146–148, https://doi.org/10.1038/375146a0 (1995).
Esther, C. R. et al. Mice lacking angiotensin-converting enzyme have low blood pressure, renal pathology, and reduced male fertility. Laboratory Investigation 74, 953–965 (1996).
Kim, H. M., Shin, D. R., Yoo, O. J., Lee, H. & Lee, J.-O. Crystal structure of Drosophila angiotensin I-converting enzyme bound to captopril and lisinopril1. FEBS Letters 538, 65–70, https://doi.org/10.1016/s0014-5793(03)00128-5 (2003).
Akif, M. et al. High-resolution crystal structures of Drosophila melanogaster angiotensin-converting enzyme in complex with novel inhibitors and antihypertensive drugs. J Mol Biol 400, 502–517, https://doi.org/10.1016/j.jmb.2010.05.024 (2010).
Akif, M. et al. Structural characterization of angiotensin I-converting enzyme in complex with a selenium analogue of captopril. The FEBS Journal 278, 3644–3650, https://doi.org/10.1111/j.1742-4658.2011.08276.x (2011).
Tatei, K., Cai, H., Ip, Y. T. & Levine, M. Race: a Drosophila homologue of the angiotensin converting enzyme. Mech Dev 51, 157–168, https://doi.org/10.1016/0925-4773(95)00349-5 (1995).
Rusch, J. & Levine, M. Regulation of a dpp target gene in the Drosophila embryo. Development 124, 303–311 (1997).
Gurdon, J. B. & Bourillot, P. Y. Morphogen gradient interpretation. Nature 413, 797–803, https://doi.org/10.1038/35101500 (2001).
Ashe, H. L. & Briscoe, J. The interpretation of morphogen gradients. Development 133, 385–394, https://doi.org/10.1242/dev.02238 (2006).
O’Connor, M. B., Umulis, D., Othmer, H. G. & Blair, S. S. Shaping BMP morphogen gradients in the Drosophila embryo and pupal wing. Development 133, 183–193, https://doi.org/10.1242/dev.02214 (2006).
Teleman, A. A. & Cohen, S. M. Dpp gradient formation in the Drosophila wing imaginal disc. Cell 103, 971–980, https://doi.org/10.1016/S0092-8674(00)00199-9 (2000).
Ashe, H. L. BMP signalling: synergy and feedback create a step gradient. Curr Biol 15, R375–377, https://doi.org/10.1016/j.cub.2005.05.003 (2005).
Affolter, M. & Basler, K. The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nat Rev Genet 8, 663–674, https://doi.org/10.1038/nrg2166 (2007).
Wharton, S. J., Basu, S. P. & Ashe, H. L. Smad affinity can direct distinct readouts of the embryonic extracellular Dpp gradient in Drosophila. Current Biology 14, 1550–1558, https://doi.org/10.1016/j.cub.2004.08.053 (2004).
Xu, M., Kirov, N. & Rushlow, C. Peak levels of BMP in the Drosophila embryo control target genes by a feed-forward mechanism. Development 132, 1637–1647, https://doi.org/10.1242/dev.01722 (2005).
Liang, H. L., Xu, M., Chuang, Y. C. & Rushlow, C. Response to the BMP gradient requires highly combinatorial inputs from multiple patterning systems in the Drosophila embryo. Development 139, 1956–1964, https://doi.org/10.1242/dev.079772 (2012).
Heberlein, U., Wolff, T. & Rubin, G. M. The TGf-beta-Homolog Dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila Retina. Cell 75, 913–926, https://doi.org/10.1016/0092-8674(93)90535-X (1993).
Chanut, F. & Heberlein, U. Role of decapentaplegic in initiation and progression of the morphogenetic furrow in the developing Drosophila retina. Development 124, 559–567 (1997).
Burke, R. & Basler, K. Dpp receptors are autonomously required for cell proliferation in the entire developing Drosophila wing. Development 122, 2261–2269 (1996).
Affolter, M., Nellen, D., Nussbaumer, U. & Basler, K. Multiple requirements for the receptor serine/threonine kinase thick veins reveal novel functions of TGF beta homologs during Drosophila embryogenesis. Development 120, 3105–3117 (1994).
Sturtevant, M. A. & Bier, E. Analysis of the genetic hierarchy guiding wing vein development in Drosophila. Development 121, 785–801 (1995).
Celis de, J. F. Expression and function of decapentaplegic and thick veins during the differentiation of the veins in the Drosophila wing. Development 124, 1007–1018 (1997).
Maurel-Zaffran, C. & Treisman, J. E. pannier acts upstream of wingless to direct dorsal eye disc development in Drosophila. Development 127, 1007–1016 (2000).
Tomoyasu, Y., Ueno, N. & Nakamura, M. The Decapentaplegic morphogen gradient regulates the notal wingless expression through induction of pannier and u-shaped in Drosophila. Mechanisms of Development 96, 37–49, https://doi.org/10.1016/S0925-4773(00)00374-9 (2000).
Sato, M. & Saigo, K. Involvement of pannier and u-shaped in regulation of decapentaplegic-dependent wingless expression in developing Drosophila notum. Mech Dev 93, 127–138, https://doi.org/10.1016/S0925-4773(00)00282-3 (2000).
Siviter, R. J. et al. Ance, a Drosophila angiotensin-converting enzyme homologue, is expressed in imaginal cells during metamorphosis and is regulated by the steroid, 20-hydroxyecdysone. Biochem J 367, 187–193, https://doi.org/10.1042/BJ20020567 (2002).
Rylett, C. M., Walker, M. J., Howell, G. J., Shirras, A. D. & Isaac, R. E. Male accessory glands of Drosophila melanogaster make a secreted angiotensin I-converting enzyme (ANCE), suggesting a role for the peptide-processing enzyme in seminal fluid. J Exp Biol 210, 3601–3606, https://doi.org/10.1242/jeb.009035 (2007).
Corrigan, L. et al. BMP-regulated exosomes from Drosophila male reproductive glands reprogram female behavior. J Cell Biol 206, 671–688, https://doi.org/10.1083/jcb.201401072 (2014).
Hurst, D., Rylett, C. M., Isaac, R. E. & Shirras, A. D. The Drosophila angiotensin-converting enzyme homologue Ance is required for spermiogenesis. Dev Biol 254, 238–247, https://doi.org/10.1016/S0012-1606(02)00082-9 (2003).
Pereira, P. S., Pinho, S., Johnson, K., Couso, J. P. & Casares, F. A 3′ cis-regulatory region controls wingless expression in the Drosophila eye and leg primordia. Developmental Dynamics 235, 225–234, https://doi.org/10.1002/dvdy.20606 (2006).
Oros, S. M., Tare, M., Kango-Singh, M. & Singh, A. Dorsal eye selector pannier (pnr) suppresses the eye fate to define dorsal margin of the Drosophila eye. Dev Biol 346, 258–271, https://doi.org/10.1016/j.ydbio.2010.07.030 (2010).
Houard, X. et al. The Drosophila melanogaster-related angiotensin-I-converting enzymes Acer and Ance–distinct enzymic characteristics and alternative expression during pupal development. Eur J Biochem 257, 599–606, https://doi.org/10.1046/j.1432-1327.1998.2570599.x (1998).
Hamaratoglu, F., Affolter, M. & Pyrowolakis, G. Dpp/BMP signaling in flies: from molecules to biology. Semin Cell Dev Biol 32, 128–136, https://doi.org/10.1016/j.semcdb.2014.04.036 (2014).
Brand, A. H. & Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993).
Blackman, R. K., Sanicola, M., Raftery, L. A., Gillevet, T. & Gelbart, W. M. An extensive 3′ cis-regulatory region directs the imaginal disk expression of decapentaplegic, a member of the TGF-beta family in Drosophila. Development 111, 657–666 (1991).
Long, H. K., Prescott, S. L. & Wysocka, J. Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell 167, 1170–1187, https://doi.org/10.1016/j.cell.2016.09.018 (2016).
Szuts, D. & Bienz, M. LexA chimeras reveal the function of Drosophila Fos as a context-dependent transcriptional activator. P Natl Acad Sci USA 97, 5351–5356, https://doi.org/10.1073/pnas.97.10.5351 (2000).
Mullen, A. C. et al. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell 147, 565–576, https://doi.org/10.1016/j.cell.2011.08.050 (2011).
Bier, E. & De Robertis, E. M. Embryo Development. BMP gradients: A paradigm for morphogen-mediated developmental patterning. Science 348, aaa5838, https://doi.org/10.1126/science.aaa5838 (2015).
Siviter, R. J. et al. Peptidyl dipeptidases (Ance and Acer) of Drosophila melanogaster: major differences in the substrate specificity of two homologs of human angiotensin I-converting enzyme. Peptides 23, 2025–2034, https://doi.org/10.1016/S0196-9781(02)00190-0 (2002).
Wilson, C. L., Shirras, A. D. & Isaac, R. E. Extracellular peptidases of imaginal discs of Drosophila melanogaster. Peptides 23, 2007–2014, https://doi.org/10.1016/S0196-9781(02)00188-2 (2002).
Kang, J., Yeom, E., Lim, J. & Choi, K. W. Bar represses dPax2 and decapentaplegic to regulate cell fate and morphogenetic cell death in Drosophila eye. PLoS One 9, e88171, https://doi.org/10.1371/journal.pone.0088171 (2014).
Ishidoya, S., Morrissey, J., McCracken, R., Reyes, A. & Klahr, S. Angiotensin II receptor antagonist ameliorates renal tubulointerstitial fibrosis caused by unilateral ureteral obstruction. Kidney Int 47, 1285–1294, https://doi.org/10.1038/ki.1995.183 (1995).
Ruiz-Ortega, M. et al. ACE inhibition reduces proteinuria, glomerular lesions and extracellular matrix production in a normotensive rat model of immune complex nephritis. Kidney Int 48, 1778–1791, https://doi.org/10.1038/ki.1995.476 (1995).
Nakamura, A. et al. Combination therapy with an angiotensin-converting-enzyme inhibitor and an angiotensin II receptor antagonist ameliorates microinflammation and oxidative stress in patients with diabetic nephropathy. J Diabetes Investig 4, 195–201, https://doi.org/10.1111/jdi.12004 (2013).
Liang, X. et al. Anti-TGF-beta antibody, 1D11, ameliorates glomerular fibrosis in mouse models after the onset of proteinuria. PLoS One 11, e0155534, https://doi.org/10.1371/journal.pone.0155534 (2016).
Benigni, A. et al. Add-on anti-TGF-beta antibody to ACE inhibitor arrests progressive diabetic nephropathy in the rat. J Am Soc Nephrol 14, 1816–1824, https://doi.org/10.1097/01.ASN.0000074238.61967.B7 (2003).
Stefanovic, S. et al. GATA-dependent regulatory switches establish atrioventricular canal specificity during heart development. Nature Communications 5, 3680, https://doi.org/10.1038/ncomms4680 (2014).
Blume-Jensen, P. & Hunter, T. Oncogenic kinase signalling. Nature 411, 355–365 (2001).
Leicht, D. T. et al. Raf Kinases: Function, regulation and role in hman cancer. Biochimica et biophysica acta 1773, 1196–1212, https://doi.org/10.1016/j.bbamcr.2007.05.001 (2007).
Murphy, L. O. & Blenis, J. MAPK signal specificity: the right place at the right time. Trends in Biochemical Sciences 31, 268–275, https://doi.org/10.1016/j.tibs.2006.03.009 (2006).
Cagnol, S. & Chambard, J.-C. ERK and cell death: Mechanisms of ERK-induced cell death – apoptosis, autophagy and senescence. FEBS Journal 277, 2–21, https://doi.org/10.1111/j.1742-4658.2009.07366.x (2010).
Cagnol, S., Van Obberghen-Schilling, E. & Chambard, J.-C. Prolonged activation of ERK1,2 induces FADD-independent caspase 8 activation and cell death. Apoptosis 11, 337–346, https://doi.org/10.1007/s10495-006-4065-y (2006).
Jeong, J. Y. et al. One-step sequence- and ligation-independent cloning as a rapid and versatile cloning method for functional genomics studies. Appl Environ Microbiol 78, 5440–5443, https://doi.org/10.1128/AEM.00844-12 (2012).
Acknowledgements
We are grateful to Elwyn Isaac, Michael Levine, Christine Rushlow, Christian Dahmann, Carl-Henrik Heldin, Kyungok Cho, the Bloomington Drosophila Stock Center, the Drosophila Genomics Resource Center, and the Developmental Studies Hybridoma Bank for reagents and fly stocks. We also thank Kyungok Cho and Jean Jung for commenting on the manuscript. This research was supported by NRF-2014K1A1A2042982 and NRF-2017R1A2B3007516 to K.-W.C., NRF-2017038671 and NRF-2016M3A9B4915818 to M.-Y.K. through the National Research Foundation of Korea, funded by the Korean, Ministry of Education, Science & Technology, and N10170001 and N11170166 to M.-Y.K. through KAIST Institute for the BioCentury, Cancer Metastasis Control Center.
Author information
Authors and Affiliations
Contributions
A.-R.K. and K.-W.C. designed the study. A.-R.K., M.-Y.K. and E.-B.C. designed and A.-R.K. and E.-B.C. performed HEK293 experiments. A.-R.K. did all the rest. All authors discussed and interpreted the results. A.-R.K. and K.-W.C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, AR., Choi, EB., Kim, MY. et al. Angiotensin-converting enzyme Ance is cooperatively regulated by Mad and Pannier in Drosophila imaginal discs. Sci Rep 7, 13174 (2017). https://doi.org/10.1038/s41598-017-13487-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-13487-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.