Abstract
Exosomes are vesicles released by many eukaryotic cells; their cargo includes proteins, mRNA and microRNA (miR) that can be transferred to recipient cells and regulate cellular processes in an autocrine or paracrine manner. While cells of the myoblast lineage secrete exosomes, it is not known whether skeletal muscle fibers (myofibers) release exosomes. In this study, we found that cultured myofibers release nanovesicles that have bilamellar membranes and an average size of 60–130 nm, contain typical exosomal proteins and miRNAs and are taken up by C2C12 cells. miR-133a was found to be the most abundant myomiR in these vesicles while miR-720 was most enriched in exosomes compared to parent myofibers. Treatment of NIH 3T3 cells with myofiber-derived exosomes downregulated the miR-133a targets proteins Smarcd1 and Runx2, confirming that these exosomes have biologically relevant effects on recipient cells. Denervation resulted in a marked increase in miR-206 and reduced expression of miRs 1, 133a, and 133b in myofiber-derived exosomes. These findings demonstrate that skeletal muscle fibers release exosomes which can exert biologically significant effects on recipient cells, and that pathological muscle conditions such as denervation induce alterations in exosomal miR profile which could influence responses to disease states through autocrine or paracrine mechanisms.
Similar content being viewed by others
Introduction
Skeletal muscle is now recognized as a major secretory organ that releases soluble mediators capable of acting locally or on distant tissues1 such as pancreas2, adipose tissue3 and bone4. Recently, it has been recognized that cells can also communicate through the release of exosomes, which are membranous vesicles of about 50–150 nm in diameter that are derived from multivesicular bodies (MVB) during endosome maturation5. Exosomes are released into the extracellular space by fusion of the MVB with the plasma membrane5. In addition to lipids, exosomes contain proteins, some of which are characteristic of exosomes (tetraspanins, HSP70, Alix and TSG101), while others reflect the proteins synthesized in the cell of origin5. Exosomes also contain mRNA and microRNA (miR) cargo which can be transferred to recipient cells and either be translated (mRNAs) or interfere with translation (miRs)6. The diverse cargos delivered by exosomes to recipient cells thus have the potential to affect many different biological processes5.
Exosomes derived from C2C12 myoblasts or myotubes have been isolated and their protein and miR composition has been analyzed7,8,9,10,11. Myotube-derived exosomes influenced myoblast proliferation and differentiation, and exosomal proteins were incorporated into recipient myoblasts7. Profiling of miRs within exosomes released by C2C12 myoblasts and myotubes showed that miRs were selectively incorporated8. Moreover, miRs within exosomes from myotubes silenced Sirtuin1 in myoblasts8. C2C12 derived exosomes were also found to enhance survival and neurite growth of the motor neuron cell line NSC-34, suggesting that muscle could influence motor neuron survival and axon growth12. In a mouse model of high-fat diet-induced insulin resistance, exosome-like vesicles isolated from skeletal muscle were taken up by cultured MIN6B1 cells and modulated their gene expression13. Moreover, C2C12 derived exosomes were taken up by many organs when injected in mice tail veins8. Exosomes secreted by differentiating human skeletal myoblasts were found to induce myogenesis of human adipose-derived stem cells and to enhance regeneration when injected in injured muscle14. Collectively these data suggest that exosomes originating from cells of the myogenic lineage may function as autocrine, paracrine or endocrine mediators.
Due to the difficulty in harvesting exosomes from tissues, most of the work to date to characterize muscle exosomes has been conducted using the C2C12 cell line. Studies of exosome-like particles isolated from muscle have not addressed the fact that skeletal muscle contains many cell types in addition to myofibers, such as nerves, blood vessels, connective tissue, and fat. This point raises concerns regarding any of the exosomal findings due to the evident uncertainty regarding the cells of origin. While it is likely that skeletal muscle fibers (myofibers), a syncytium formed by the fusion of tens to hundreds of myocytes, do indeed release exosomes, this possibility has not been directly addressed. In addition, the question of how pathological states alter the cargo of exosomes derived from myofibers has not been investigated. This study determined whether dispersed mouse myofibers release exosomes, how paralysis induced by nerve transection altered the miR cargo of exosomes released by such fibers and whether such exosomes influence biology of recipient cells. Our findings establish that myofibers release exosomes and modify their cargo during pathological conditions, and that such exosomes regulate expression of Smarcd1 (BAF60a), a structural component of the BAF chromatin remodeling complex which has critical roles in myogenesis15,16, and Runx2, which is a master regulator of osteogenic lineage progression17. The findings have important implications for understanding the mechanisms by which myofibers signal to nearby cells within skeletal muscle, as well other more distant tissues.
Results
Cultured Muscle fibers release exosome-like nanovesicles
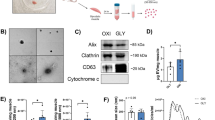
To determine whether myofibers isolated from the mouse hindlimb muscles, namely soleus, plantaris, gastrocnemius and EDL, release nanovesicles, dispersed myofibers were cultured for 48 hours in exosome-depleted medium. The conditioned media was collected and subjected to sequential centrifugation. The pellet obtained by centrifugation at 100,000 × g was analyzed by electron microscopy (Fig. 1) which revealed particles that varied in size from approximately 60 to 130 nm in diameter and had a bilamellar membrane (A and B). By immunogold staining, the vesicles were labeled by antibodies against CD63 (C–F), CD81 (G–J) and CD9 (K).
EM characterization of nanovesicles released by dispersed muscle fibers. Dispersed mouse hindlimb muscle fibers were incubated for 48 hours. Nanovesicles were isolated by ultracentrifugation of conditioned medium then stained with uranyl acetate (A,B) or labeled with 10-nm immunogold particles using antibodies against the exosomal membrane markers CD63 (C–F), CD81 (G–J) and CD9 (K), and stained with uranyl acetate. Nanovesicles were imaged by electron microscopy. Scale bar: 100 nm.
To further analyze the size distribution of these nanovesicles, the particles were subjected to nanoparticle tracking analysis. Approximately 1.9 × 109 vesicles were obtained from the media conditioned by incubation of about 300 myofibers for 48 hours (Fig. 2A and B). Nanovesicles had a mean size of 121 + /− 3.7 nm.
Size distribution and uptake by cells of nanovesicles released by dispersed muscle fibers. Dispersed mouse muscle fibers were incubated for 48 hours, after which nanovesicles were isolated by differential centrifugation. (A and B) The size distribution and concentration of particles was analyzed by nanoparticle tracking. (C and E) Nanovesicles were labeled with PKH67 dye and overlaid on cultures of proliferating C2C12 cells. The control cells were treated with an equal volume of dye-treated PBS. After 5 hours, the cells were fixed, stained with phalloidin to visualize cell bodies, counterstained with DAPI to label nuclei and imaged by confocal microscopy. Panel (C) shows control cells incubated in media containing PKH67-treated PBS; panels (D and E) show cells incubated with PKH67-labeled nanoparticles. In these images, nanoparticles are green, phalloidin is red and nuclei are blue (DAPI).
Collectively, the data indicate that dispersed mouse hindlimb muscle fibers release nanoparticles with the size, biochemical markers and biological properties characteristic of exosomes.
Uptake of labeled nanovesicles by C2C12 myoblasts
A fundamental property of exosomes is that they are taken up by recipient cells thereby delivering their cargo and influencing biological processes5,6. To determine whether the nanovesicles isolated from media conditioned by culture with dispersed myofibers were taken up by recipient cells, we treated C2C12 cells with vesicles released by myofibers that had been labeled with the fluorescent dye PKH67. C2C12 myoblasts showed uptake of PKH67-labeled vesicles (Fig. 2D and E) while cells that received the dye-only control had negligible fluorescence (Fig. 2C).
Influence of denervation on miR profiles in myofibers
Denervation atrophy has been found to result in significant changes in miR expression profiles in muscle tissue including a significant increase in miR-206 levels18. To determine miR expression profiles in myofibers and understand how such profiles were altered by denervation, and how miR levels in exosomes released by myofibers differ from those of the parent fiber, miR expression profiles of denervated or sham-denervated EDL myofibers were determined at 7 days post-denervation or sham-denervation using the nCounter mouse miR assay. This technique profiles expression levels of 578 mouse microRNAs. Using a mean of 100 counts in the sham-denervated group as a lower limit for inclusion, data for 58 miRs were evaluated in subsequent analysis (Tables 1 and S1). Expression levels ranged from a low of 100 (miR-148a) to a high of ~21,000 counts (miR-1). The most highly expressed miRs present in myofibers were miR-1, miR-133a, miR-22, miR-378 and miR-720 (Table 1 and S1). When comparing the profiles of miRs from denervated and sham-denervated groups, significant differences were observed for 20 miRs (Table 1 and S1), including decreased expression of miR-1 and 133a and a 25-fold increase in the expression of miR-206 (Table 1).
Relative abundance of selected miRs in exosomes from myofibers
Based on the above results, qPCR was used to investigate the expression of myomiRs and other selected miR targets in nanovesicles and parent myofibers from control muscle (Fig. 3A and B). This analysis showed that miRs could be detected in fiber-derived nanovesicles and that miR-133a and miR-1 were highly expressed in exosomes relative to miR-22 (Fig. 3B), a miR which was present at similar levels in control and denervated myofibers based on counts observed by nCounter analysis (Table S1). Compared to parent myofibers, miR-720 was enriched in exosomes while the opposite was true for miR-1 (Fig. 3A and B).
Expression of myomiRs and other selected miR targets in fiber-derived exosomes. Fibers derived from mixed calf muscle were cultured for 48 hours in exosome- free medium. RNA was isolated from fibers (A) and corresponding nanovesicles (B) and miR expression analyzed by qPCR. The expression is relative to that of miR-22 which was the lowest expressed among the targets tested. The data are from three independent fiber/exosomes preparations.
Alteration in miR expression in exosomes from denervated fibers
Exosomes released by the denervated EDL myofibers also showed a marked increase in miR-206 levels (about 15-fold) and a reduction in the levels of miR-1 (Fig. 4B), as in myofibers (Fig. 4A). In contrast to myofibers, exosomes showed significantly reduced levels of miR-133a and miR-133b, a change which was not detected in the corresponding fibers (Fig. 4A and B).
qPCR evaluation of the effect of denervation on miRs expression. Dispersed muscle fibers were isolated from mouse EDL muscle 7 days after sciatic nerve transection or sham transection and maintained in culture in exosome-free medium for 48 hours. RNA was extracted from dispersed fibers (A) and corresponding nanovesicles preparations (B) and miRs expression levels were determined by qPCR. Data are mean values ± SD for 5 animals per group except for the exosomes produced by denervated fibers (n = 4). *p < 0.05; **p < 0.01; ***p < 0.001.
mRNA targets of miRs altered in exosomes from denervated fibers
To understand potential biological effects of the changes in miR profiles observed, experimentally confirmed mRNA targets of miRs that were altered in exosomes produced by denervated myofibers were identified by Ingenuity Pathways Analysis. This analysis revealed 214 mRNA targets which included growth factors and their receptors (IGF-1, IGF-1R, CTGF, EGFR), molecules involved in calcium-calmodulin signaling (Calm1, NFATC4) and transcriptional regulation (MEF2A, HDAC4, RUNX2) (Tables 2 and S2). Ingenuity Pathways Analysis was then used to identify canonical pathways represented by these mRNAs. The most significantly represented pathways were Myc, IGF-1 and calcium signaling (Tables 2 and S2). Also significant were STAT3, a downstream mediator of IL-6 signaling, and PTEN, which exerts negative feedback on IGF-1, insulin and other pathways downstream of receptor tyrosine kinases. An analysis of upstream regulators of these pathways showed that the most significant correlations were with myomirs 1 and 133, but that TGF-β and estradiol signaling were also highly significantly related.
Myofiber-derived exosomes downregulate Smarcd1 and Runx2
To determine whether exosomes released by myofibers exert biological effects on recipient cells, we tested whether treatment of NIH 3T3 cells with exosomes released by myofibers from healthy muscle altered protein levels of either of two known targets of miR-133a, Smarcd1, and Runx2. Smarcd116 participates in programs that specify cardiac and skeletal muscle development15,16. Runx2 is a master regulator of osteogenic lineage progression17. These targets were chosen for analysis because, as indicated above, miR-133a is the most abundant miR found in fiber-derived exosomes in our analysis and NIH 3T3 cells are ideal recipient cells to study the biological effects of miR-133a because they express very low levels of this miR16,19. Treatment of NIH 3T3 cells with myofiber-derived exosomes for 48 hours significantly and reproducibly reduced Smarcd1 and Runx2 protein levels (Fig. 5A and B). The findings confirm that cargos transferred to recipient cells by exosomes derived from myofibers exert biological effects.
Fiber-derived exosomes modulate Smarcd1 and Runx2 expression. NIH3T3 grown in exosome- depleted medium were treated with fiber-derived exosomes (4 μg/ml) or PBS for 48 hours. The expression of Smarcd1 and Runx2 was analyzed by Western blot. Representative blots are shown in panel (A). Panel (B) shows the quantitation of Smarcd1 and Runx2 expression relative to Gapdh for three separate experiments, each with 3 wells per condition for a total of 9 samples for each condition. **p < 0.005, ****p < 0.0001. Full-length blots are presented in Supplementary Figure S1.
Discussion
We investigated whether isolated skeletal muscle fibers release exosome-like nanovesicles when cultured in vitro. This approach extends prior work examining exosomes released by myoblasts or myotubes derived from C2C12 cells7,8,9,10,11,12, differentiating human skeletal myoblasts14 or present in preparations of skeletal muscle13. Full-length muscle fibers remain viable for at least 1 week in culture which affords an opportunity to examine their properties outside of the confines of an individual muscle, thus reducing confounding effects of other cellular elements that compose skeletal muscle that is blood vessels, connective tissue, nerves and fat. Dispersed fibers have been used to study a wide variety of properties of skeletal muscle, including membrane permeability20, calcium handling21 and sarcomere function22.
The findings presented above demonstrate that cultured myofibers release nanovesicles that possess many of the characteristic features of exosomes described in the literature5,23: sizes of 60 to 130 nm, a bilamellar membrane and positive immunogold staining for the tetraspanins CD63, CD81 and CD95,24. The myofiber-derived nanovesicles were taken up by C2C12 myoblasts and contained microRNAs. The exosomes released by cultured myofibers could arise from the myofiber itself and/or from the satellite cells located between the sarcolemma and basement membrane surrounding the fiber. Given the much greater mass of the muscle fiber and the more limited number of satellite cells it is likely that the majority of exosomes evaluated in our studies were derived from the skeletal muscle fiber.
Taken together, our data indicate that intact muscle fibers release exosomes and provide a means to study the properties of exosomes released by such fibers from healthy and diseased muscle without contamination from exosomes released from other cell types present in the muscle.
The exosomes contained microRNAs that were also present in the parent myofibers, with miR-133a being by far the most abundant. Consistent with prior profiling studies in different cell types25,26, there appeared to be some selectivity in packaging of miRs in fiber-derived exosomes. For example, the relative levels of miR-720 were enriched in exosomes as compared to those present in the fibers, while miR-1 was, to a degree, excluded.
The exosomes released by cultured muscle fibers not only could be taken up by target C2C12 cells but also could downregulate the expression of Smarcd1 and Runx2 proteins in NIH3T3, providing evidence that such exosomes can signal through their cargo to nearby cells in a paracrine manner. In the case of Smarcd1 and Runx2 proteins, their downregulation by fiber-derived exosomes was likely to be mediated by miR-133a because both have been reported as validated targets of this miR16,17, it is the most abundant miR found in these exosomes, and evidence exists that only the most highly-expressed miRs regulate target mRNAs27.
Smarcd1 is involved in skeletal muscle myogenesis15, possibly by binding MyoD28, which raises the possibility that myofiber-derived exosomes may modulate Smarcd1 protein levels of nearby satellite cells and/or other myogenic precursors and influence their biology. In a healthy muscle, myogenic precursors are largely comprised of quiescent satellite cells which reside between the sarcolemma and basement membrane, placing satellite cells very close to muscle fibers. Thus, satellite cells are bathed in extracellular fluid enriched in substances secreted by the fiber including exosomes. These observations raise the possibility that the increased miR-206 present in exosomes from denervated fibers may play a role in the activation and/or proliferation of satellite cells known to occur after nerve transection29,30,31. In addition, muscle levels of myomirs have been shown to be altered by older age, gender and testosterone levels32. Advanced age reduces satellite cell proliferation33 leading to the question of whether age-related changes in exosomal myomir cargo contributes to this satellite cell abnormality. Future studies are needed to address these provocative questions.
The possibility that miRs in exosomes released by either healthy or paralyzed muscle might have distant effects should also be considered. In support of this proposal, and as noted above, changes in miR profiles of exosomes derived from skeletal muscle have been suggested to regulate pancreatic β-cell gene expression13. Because myofiber-derived exosomes can modulate levels of Runx2, an osteoblast differentiation factor, the possibility that such exosomes could also influence the fate of mesenchymal stromal cells, and thus bone biology, must be considered.
The most dramatic denervation-related change in miR expression levels was the marked increase in both myofibers and exosomes of miR-206. miR-206 is a skeletal muscle specific miR that is greatly upregulated during myoblast differentiation and promotes satellite cell differentiation by regulating the expression of HDAC434, as well as that of Pax3, Pax 7, utrophin, DNA polymerase-alpha and connexin 4335,36,37. Several lines of evidence indicate that miR-206 is involved in muscle regeneration in both mouse and rat models38,39,40 although there is also evidence implicating miR-206 in growth-inhibitory and/or muscle atrophy programs18. Regardless of whether miR-206 promotes or reduces atrophy, the marked increase in miR-206 in exosomes released by denervated muscle fibers has obvious implications for inter-cellular signaling during denervation atrophy. Since potential targets of miR-206 include molecules such as BDNF, NGFR, IGF-1 and IGFBP-5, it has been suggested that miR-206 could also be involved in the regulation of muscle mass and in synapse formation during re-innervation38,41. Thus, upregulation of miR-206 in fiber-derived exosomes after denervation could be a teleologic strategy to aid in local muscle repair by providing readymade miR-206 in a paracrine manner to neighboring cells. The large increase in miR-206 observed in exosomes produced by denervated myofibers could have significant effects on other types of recipient cells such as motor nerve terminals at the motor end plate and neighboring capillaries, arteries or veins and, extending the biological action of this myomiR, on non-muscle cells. For example, miR-206 could also affect bone because it inhibits osteoblast differentiation via downregulation of connexin 4342.
Mechanisms by which denervation drives alterations of miR expression profiles are poorly understood. MyoD, a critical myogenic differentiation factor from the basic helix-loop-helix family of transcription factors, drives expression of several myomiRs43 and is persistently elevated in denervated muscle44. Thus, increased MyoD-induced transcription may explain some changes in miR expression in denervated muscle, such as upregulation of miR-206, but seems less likely to contribute to reduced expression of miR-1 or miR-133. Critical upstream signals responsible for denervation atrophy involve activation of non-classical NF-kB signaling through p65 and cRel45 which appears to be downstream of the de novo sarcolemmal expression of connexin 43 and 45 hemichannels20. Further study will be required to understand how these signaling pathways directly or indirectly modulate transcription and stability of miRNA transcripts.
The clinical significance of our findings remains speculative. There are several reports in which blood miR levels have been proposed as a biomarker for a variety of diseases including those affecting skeletal muscle46,47. The myomirs have been identified in the circulation where their levels have been shown to change with exercise48,49,50,51 or muscular dystrophies46,47. The origin of circulating myomirs is not well understood. In one study, extracellular vesicles isolated from blood by density gradient separation were positive for the markers CD81, TSG101 and alpha-sarcoglycan and contained miR-206, suggesting that they arose from cells of the myogenic lineage52. In a separate study, a bout of exercise was found to increase the number of small extracellular vesicles in blood53. The possibility that myofibers signal to distant targets via release of exosomes is an interesting topic for future studies.
In summary, our findings provide direct support for the conclusion that myofibers release exosomes that contain microRNA as cargo and that exosomal miR profiles are altered by denervation. The data above supports the conclusion that myofiber-derived exosomes modulate protein levels of key factors in myogenic or osteogenic differentiation of mesenchymal progenitor cells, and likely other cell types, pointing to the exciting possibility of a role of such exosomes in paracrine regulation by myofibers of nearby cells. Our findings also raise the possibility that myofiber-derived exosomes are an important source of circulating myomirs and extracellular vesicles, providing support for the use of circulating myomiRs levels as biomarkers for muscle health and disease.
Materials and Methods
Animals
Male C57B/6 J mice weighing 25 g were obtained from Jackson Labs (Bar Harbor, ME) and housed in rooms with 12:12 hour light: dark cycles and controlled temperature and humidity. Animals were provided food and water ad libitum. Denervation was performed as previously described with some modifications20. The animals were anesthetized by inhalation of isofluorane, hair was removed with a clipper and the skin was cleaned with betadine solution and ethanol. A small incision was made just posterior to the head of the left femur and the sciatic nerve was exposed by careful dissection. A 2 mm piece of the nerve was removed and the wounds were closed with suture and surgical glue. Animals were administered carprofen for 3 days postoperatively. Some animals received a sham nerve transection in which the nerve was exposed but not manipulated. At 7 days after nerve or sham transection, animals were anesthetized by inhalation of isofluorane and hindlimb muscles were removed by careful dissection, minimizing any pulling on the muscle. All studies with animals were approved by the IACUC at the James J. Peters VA Medical Center and were carried out in accordance with the NIH Guide for Care and Use of Laboratory Animals.
Isolation and culture of skeletal muscle fibers
Muscle fibers were isolated and maintained, as previously described54. Briefly, muscle tissue (gastrocnemius, plantaris, EDL and soleus, or EDL alone, as indicated in the figure legends) was removed from mouse hindlimbs as above described and immediately digested with 400 U/ml of collagenase Type I (Worthington) in DMEM-F12 medium (1:1) (ThermoFisher) at 37 °C for 45–60 minutes, depending on the collagenase lot. The digested tissue was transferred to DMEM/F12 (1:1) supplemented with Penicillin/Streptomycin/Amphotericin B (Antibiotics-Antimycotic,ThermoFisher) and 15% exosome-depleted horse serum prepared by overnight centrifugation at 100,00 × g55. Digested muscle tissue was triturated with a wide bore pipette to release single fibers. The released fibers were collected with a fire-polished glass pipette and transferred to a pre-warmed dish containing the above medium. The process was repeated until the required number of fibers was collected. Fibers were passaged 2–3 times in fresh pre-warmed medium to remove debris and cultured for 48 hours. Typically, 150–600 fibers were cultured with a survival of about 90%.
Isolation of nanovesicles
Nanovesicles were isolated by differential centrifugation55. The fiber conditioned medium was collected and centrifuged at 1800 rpm for 30 minutes, followed by centrifugation at 10,000 rpm for 45 minutes. The resulting supernatant was centrifuged at 100,000 × g for 70 minutes (Beckman Coulter Optima, type SW-41 rotor). The pellet containing nanovesicles was resuspended in PBS and ultracentrifuged, as described above.
Extraction of RNA
Total RNA was isolated from the cultured fibers and corresponding nanovesicle preparations using the miRNeasy microRNA isolation kit (Qiagen, Germantown, MD). Briefly, fibers pellets or nanovesicles prepared as above were extracted with Qiazol reagent (Qiagen) and the RNA was further purified according to the protocol provided by the manufacturer (Qiagen). In the case of nanovesicles, cel-miR-39 (8 pg) (Qiagen) was added to the Qiazol extract for internal normalization prior to miR purification.
microRNA profiling
The profile of microRNA levels present in skeletal muscle fibers was examined using nCounter mouse miRNA arrays (NanoStrings Technologies, Seattle, WA) which survey the expression levels of 578 mouse microRNAs. miRNA profiling was performed through the POP service of the manufacturer (NanoStrings Technologies). Raw data were normalized using the average intensity of the 100 most highly expressed miRNAs. Average counts of negative controls +2 standard deviations were subtracted from the individual counts. miRs for which counts were greater than or equal to 100 after these corrections were included in subsequent analysis and fold-change in expression levels between sham and denervated samples was calculated.
Quantitative RT-PCR
Total RNA was reverse transcribed using a pool of the specific primers for the miRs of interest provided with the TaqMan miR assays. The primers were pooled and diluted to a final 1:100 dilution. Reverse transcription was performed with the TaqMan MicroRNA Reverse transcription kit (Life Technologies-ThermoFisher) according to the protocol provided by the manufacturer (Life Technology protocol #4465407). Due to the low amount of starting material (22 ng of RNA for fibers and 3 μl/20 μl of RNA for the nanovesicles) a pre-amplification step was performed with the pre-AMP Master mix according to the manufacturer’s protocol (Life Technologies protocol #4465407). The pre-AMP product was diluted 1:8 and 0.8 μl of preamplification product was amplified using specific miR TaqMan assays and the TaqMan Universal PCR Master mix (Life Technologies-ThermoFisher) in a 10 μl reaction. Expression levels were normalized against U6sn for muscle fibers, and cel-miR-39 for nanovesicles. Relative expression was calculated with the 2−∆∆Ct method56 using the sham-denervated samples as a control.
Size distribution of nanovesicles released by cultured muscle fibers
Nanovesicle tracking analysis was performed with a NanoSight LM10-HS instrument (Malvern Instruments, Westborough, MA). An aliquot of the nanovesicle preparation (derived from 700 fibers) was diluted 1:25 in PBS and loaded onto the NanoSight instrument after calibration with 100 μm polystyrene latex microbeads (Malvern Instruments). Five, 60 seconds videos were recorded at a camera level set at 14. The videos were analyzed with the NS500 software to determine the particles size distribution and concentration using the following parameters: auto-blur, track length and size and a threshold detection of 10.
Electron Microscopy
Negative staining of exosomes with uranyl acetate
Nanovesicles were isolated from the conditioned cultured medium of mouse calf muscle fibers (about 240 fibers), as described above. The 100,000 × g pellet was suspended in 2% paraformaldehyde in PBS and 5-μl aliquots deposited for 20 minutes on formvar- and carbon- coated 200-mesh copper grids (Electron Microscopy Sciences, Hatfield, PA). Samples on grids were then fixed with 1% glutaraldehyde for 5 minutes, and negatively stained with uranyl acetate-oxalate solution for 5 minutes followed by embedding in methyl cellulose as described55. Samples on grids were viewed using a Hitachi H7000 electron microscope (Tokyo, Japan) at 75 kV. The electron microscope was equipped with an AMT Advantage HS digital camera (Danvers, MA, USA) and micrographs were digitally recorded.
Immunogold labeling of exosomes
Five-μl aliquots of exosomes suspended in 2% paraformaldehyde in PBS were deposited for 20 minutes on formvar- and carbon-coated 200-mesh copper grids and immunogold labeled as described55 with the following modifications. Blocking media consisted of PBS containing 0.1% gelatin and 0.1% bovine serum albumin (PBS+) for 10 minutes followed by 5% goat serum in PBS+ for 20 minutes. Samples on grids were incubated with primary antibodies diluted 1:100 in 1% goat serum in PBS+ for 1 hour then with secondary antibodies conjugated to 10-nm immunogold (Electron Microscopy Sciences, Hatfield, PA) diluted 1:25 in PBS+ for 1 hour. Immunogold-labeled samples were post-fixed with 1% glutaraldehyde for 5 minutes, and negatively stained with uranyl acetate-oxalate solution for 5 minutes followed by embedding in methyl cellulose. Primary antibodies against CD63, CD81 and CD9 were purchased from System Biosciences (SBI, Palo Alto, CA). Samples on grids were imaged with the electron microscope as described above.
Uptake of nanovesicles by C2C12 myoblasts
To study uptake, nanovesicles isolated from cultures of ~300–400 fibers were labeled with the fluorescent dye PKH67 (Sigma Aldrich, St Louis, MO). The 100,000 × g pellet was resuspended in 150 μl diluent C (Sigma Aldrich, St Louis, MO) and the suspension diluted with an equal volume of diluent C. The PKH67 dye stock was diluted 60-fold in 250 μl diluent C (Sigma Aldrich, St Louis, MO) and the nanovesicle preparation or an equal volume of PBS was added to the dye. After incubation at room temperature for 5 minutes, an equal volume of 1% BSA was added to quench the reaction. The mixture was diluted with PBS and centrifuged for 70 minutes at 100,000 × g. The pellet was resuspended in PBS and centrifuged as described above.
C2C12 myoblasts were seeded in 4 well slide chambers and allowed to reach confluence. The labeled vesicles or control dye were resuspended in 300 μl of exosome-depleted DMEM/2% horse serum and added to C2C12 myoblasts. After incubation for 5 hours, the cells were fixed in 4% paraformaldehyde in PBS for 15 minutes and washed with PBS. The cells were permeabilized with 0.1% Triton in PBS for 3 minutes, washed with PBS and blocked with 10% normal goat serum (Sigma Aldrich) in 50 mM Tris HCl buffer pH 7.4, 0.15 M NaCl (TBS) for 30 minutes. Alexa-568 labeled phalloidin (Life Technologies-ThermoFisher) was added at 1:1000 dilution in blocking solution for 30 minutes. Nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI). The slides were mounted in Fluorogel mounting medium (Electron Microscopy Sciences, Hatfield, PA) and imaged with a Zeiss confocal microscope. Images were processed with Adobe Photoshop CC.
Treatment of NIH 3T3 cells with fiber-derived exosomes
Exosomes were prepared as above, resuspended in PBS and the protein concentration determined by the Micro BCA assay (Pierce-ThermoFisher). NIH3T3 were seeded in 24 wells plates (6–7 × 104 cells/well) and cultured in DMEM supplemented with 10% heat-denatured calf serum and with Antibiotics-Antimycotic (ThermoFisher) and depleted of exosome by ultracentrifugation as described above. Exosomes were added at a concentration of 4 μg protein/ml medium for 48 hours, while control wells received the same volume of PBS. Cells were washed twice with cold PBS and lysed with 10 mM Na phosphate pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.5% Na deoxycholate, 0.5% SDS supplemented with protease and phosphatase inhibitor cocktails (Sigma Aldrich, St Louis, MO). Lysates were briefly sonicated, centrifuged at 14,000 rpm for 15 minutes at 4 °C and the supernatant saved. Protein concentration were determined with the BCA reagent (Pierce-ThermoFisher). Proteins (20–25 μg) were separated by SDS PAGE, and the gels blotted onto a PVDF membrane. The membrane was blocked with 0.5% non-fat dry milk in Tris buffer saline (TBS) and incubated overnight at 4 °C with the primary antibody diluted in blocking solution. The blot was incubated with the appropriate HRP conjugated secondary antibody (1:5000–1:7500, GE Healthcare Life Sciences, Marlborough, MA) in blocking solution, exposed to ECL Prime Western Blot detection reagent (GE Healthcare Life Sciences) and imaged with Amersham Imager 600 (GE Healthcare Life Sciences). The following antibodies were used: anti- Smarcd1 rabbit polyclonal antibody (1:500 dilution; AB83208, Abcam, Cambridge, MA), anti-Runx2 mouse monoclonal antibody (1:500 dilution; Santa Cruz Biotechnology, Dallas, TX,) and a mouse monoclonal antibody against GAPDH (1:4000 dilution; GeneTex, Irvine, CA) as loading control. Quantification of the bands was performed with Image Quant software (GE Healthcare).
Data Analysis
All values are expressed as means +/− SD. Statistical analysis was performed with Graph Pad Prism v 7.0 software. An unpaired, two-tailed t-test was used to test the significance of differences between means. P < 0.05 was considered significant.
References
Pedersen, B. K. Muscle as a secretory organ. Compr Physiol 3, 1337–1362 (2013).
Handschin, C. et al. Abnormal glucose homeostasis in skeletal muscle-specific PGC-1alpha knockout mice reveals skeletal muscle-pancreatic beta cell crosstalk. J Clin Invest 117, 3463–3474 (2007).
Bostrom, P. et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 481, 463–468 (2012).
Colaianni, G. et al. The myokine irisin increases cortical bone mass. Proc Natl Acad Sci USA 112, 12157–12162 (2015).
Colombo, M., Raposo, G. & Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 30, 255–289 (2014).
Valadi, H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9, 654–659 (2007).
Forterre, A. et al. Myotube-derived exosomal miRNAs downregulate Sirtuin1 in myoblasts during muscle cell differentiation. Cell Cycle 13, 78–89 (2014).
Aswad, H. et al. Exosomes participate in the alteration of muscle homeostasis during lipid-induced insulin resistance in mice. Diabetologia 57, 2155–2164 (2014).
Guescini, M. et al. C2C12 myoblasts release micro-vesicles containing mtDNA and proteins involved in signal transduction. Exp Cell Res 316, 1977–1984 (2010).
Le Bihan, M. C. et al. In-depth analysis of the secretome identifies three major independent secretory pathways in differentiating human myoblasts. J Proteomics 77, 344–356 (2012).
Romancino, D. P. et al. Identification and characterization of the nano-sized vesicles released by muscle cells. FEBS Lett 587, 1379–1384 (2013).
Madison, R. D., McGee, C., Rawson, R. & Robinson, G. A. Extracellular vesicles from a muscle cell line (C2C12) enhance cell survival and neurite outgrowth of a motor neuron cell line (NSC-34). J Extracell Vesicles 3 (2014).
Jalabert, A. et al. Exosome-like vesicles released from lipid-induced insulin-resistant muscles modulate gene expression and proliferation of beta recipient cells in mice. Diabetologia 59, 1049–1058 (2016).
Choi, J. S. et al. Exosomes from differentiating human skeletal muscle cells trigger myogenesis of stem cells and provide biochemical cues for skeletal muscle regeneration. J Control Release 222, 107–115 (2016).
Forcales, S. V. et al. Signal-dependent incorporation of MyoD-BAF60c into Brg1-based SWI/SNF chromatin-remodelling complex. EMBO J 31, 301–316 (2012).
Goljanek-Whysall, K. et al. myomiR-dependent switching of BAF60 variant incorporation into Brg1 chromatin remodeling complexes during embryo myogenesis. Development 141, 3378–3387 (2014).
Zhang, Y. et al. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci USA 108, 9863–9868 (2011).
Soares, R. J. et al. Involvement of microRNAs in the regulation of muscle wasting during catabolic conditions. J Biol Chem 289, 21909–21925 (2014).
Zhang, Y. et al. Control of mesenchymal lineage progression by microRNAs targeting skeletal gene regulators Trps1 and Runx2. J Biol Chem 287, 21926–21935 (2012).
Cea, L. A. et al. De novo expression of connexin hemichannels in denervated fast skeletal muscles leads to atrophy. Proc Natl Acad Sci USA 110, 16229–16234 (2013).
Jin, J. P. et al. Truncation by Glu180 nonsense mutation results in complete loss of slow skeletal muscle troponin T in a lethal nemaline myopathy. J Biol Chem 278, 26159–26165 (2003).
Andrews, J. L. et al. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc Natl Acad Sci USA 107, 19090–19095 (2010).
Raposo, G. & Stoorvogel, W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200, 373–383 (2013).
Mathivanan, S., Ji, H. & Simpson, R. J. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 73, 1907–1920 (2010).
Guduric-Fuchs, J. et al. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics 13, 357 (2012).
Ohshima, K. et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One 5, e13247 (2010).
Mullokandov, G. et al. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods 9, 840–846 (2012).
Forcales, S. V. The BAF60c-MyoD complex poises chromatin for rapid transcription. Bioarchitecture 2, 104–109 (2012).
McGeachie, J. K. The fate of proliferating cells in skeletal muscle after denervation or tenotomy: an autoradiographic study. Neuroscience 15, 499–506 (1985).
Murray, M. A. & Robbins, N. Cell proliferation in denervated muscle: identity and origin of dividing cells. Neuroscience 7, 1823–1833 (1982).
Snow, M. H. A quantitative ultrastructural analysis of satellite cells in denervated fast and slow muscles of the mouse. Anat Rec 207, 593–604 (1983).
Nielsen, S. et al. Muscle specific miRNAs are induced by testosterone and independently upregulated by age. Front Physiol 4, 394 (2013).
Brack, A. S. & Munoz-Canoves, P. The ins and outs of muscle stem cell aging. Skelet Muscle 6, 1 (2016).
Winbanks, C. E. et al. TGF-beta regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J Biol Chem 286, 13805–13814 (2011).
Anderson, C., Catoe, H. & Werner, R. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res 34, 5863–5871 (2006).
Kim, H. K., Lee, Y. S., Sivaprasad, U., Malhotra, A. & Dutta, A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol 174, 677–687 (2006).
Rosenberg, M. I., Georges, S. A., Asawachaicharn, A., Analau, E. & Tapscott, S. J. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol 175, 77–85 (2006).
Yuasa, K. et al. MicroRNA-206 is highly expressed in newly formed muscle fibers: implications regarding potential for muscle regeneration and maturation in muscular dystrophy. Cell Struct Funct 33, 163–169 (2008).
Liu, N. et al. microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J Clin Invest 122, 2054–2065 (2012).
Huang, Q. K. et al. MiR-206 Attenuates Denervation-Induced Skeletal Muscle Atrophy in Rats Through Regulation of Satellite Cell Differentiation via TGF-beta1, Smad3, and HDAC4 Signaling. Med Sci Monit 22, 1161–1170 (2016).
McCarthy, J. J. MicroRNA-206: the skeletal muscle-specific myomiR. Biochim Biophys Acta 1779, 682–691 (2008).
Inose, H. et al. A microRNA regulatory mechanism of osteoblast differentiation. Proc Natl Acad Sci USA 106, 20794–20799 (2009).
Sweetman, D. et al. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev Biol 321, 491–499 (2008).
Ishido, M., Kami, K. & Masuhara, M. In vivo expression patterns of MyoD, p21, and Rb proteins in myonuclei and satellite cells of denervated rat skeletal muscle. Am J Physiol Cell Physiol 287, C484–493 (2004).
Jackman, R. W., Cornwell, E. W., Wu, C. L. & Kandarian, S. C. Nuclear factor-kappaB signalling and transcriptional regulation in skeletal muscle atrophy. Exp Physiol 98, 19–24 (2013).
Israeli, D. et al. Circulating miRNAs are generic and versatile therapeutic monitoring biomarkers in muscular dystrophies. Sci Rep 6, 28097 (2016).
Koutsoulidou, A. et al. Elevated Muscle-Specific miRNAs in Serum of Myotonic Dystrophy Patients Relate to Muscle Disease Progress. PLoS One 10, e0125341 (2015).
Baggish, A. L. et al. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J Physiol 589, 3983–3994 (2011).
Banzet, S. et al. Changes in circulating microRNAs levels with exercise modality. J Appl Physiol (1985) 115, 1237–1244 (2013).
Mooren, F. C., Viereck, J., Kruger, K. & Thum, T. Circulating microRNAs as potential biomarkers of aerobic exercise capacity. Am J Physiol Heart Circ Physiol 306, H557–563 (2014).
Nielsen, S. et al. The miRNA plasma signature in response to acute aerobic exercise and endurance training. PLoS One 9, e87308 (2014).
Guescini, M. et al. Muscle Releases Alpha-Sarcoglycan Positive Extracellular Vesicles Carrying miRNAs in the Bloodstream. PLoS One 10, e0125094 (2015).
Fruhbeis, C., Helmig, S., Tug, S., Simon, P. & Kramer-Albers, E. M. Physical exercise induces rapid release of small extracellular vesicles into the circulation. J Extracell Vesicles 4, 28239 (2015).
Cornelison, D. D. & Wold, B. J. Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev Biol 191, 270–283 (1997).
Thery, C., Amigorena, S., Raposo, G. & Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3, Unit3.22 (2006).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta CT) Method. Methods 25, 402–408 (2001).
Acknowledgements
The research reported here was supported by the Department of Veterans Affair, Veterans Health Administration, Rehabilitation Research and Development Service (B-9212-C and B-2020-C) and the James J. Peters VA Medical Center.
Author information
Authors and Affiliations
Contributions
R.D.G. was involved in the experimental design, execution and data analysis of the experiments described in this paper; L.H. was involved in the denervation experiments; S.H. was involved in the denervation experiments as well as muscle fiber cultures; the E.M. analysis was performed by H.K.-R.; C.P.C. and W.A.B. were involved in the experimental design and data analysis. All authors were involved in the drafting and revision of the manuscript
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
De Gasperi, R., Hamidi, S., Harlow, L.M. et al. Denervation-related alterations and biological activity of miRNAs contained in exosomes released by skeletal muscle fibers. Sci Rep 7, 12888 (2017). https://doi.org/10.1038/s41598-017-13105-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-13105-9
This article is cited by
-
Exercise-derived skeletal myogenic exosomes as mediators of intercellular crosstalk: a major player in health, disease, and exercise
Journal of Physiology and Biochemistry (2023)
-
Influence of microRNAs and exosomes in muscle health and diseases
Journal of Muscle Research and Cell Motility (2020)
-
Secreted miRNAs in the tripartite neuromuscular junction
ExRNA (2019)
-
Exosomes and Extracellular RNA in Muscle and Bone Aging and Crosstalk
Current Osteoporosis Reports (2019)
-
Therapeutic potential of exosomes in rotator cuff tendon healing
Journal of Bone and Mineral Metabolism (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.