Abstract
The elongases of very long chain fatty acid (ELOVL or ELO) are essential in the biosynthesis of fatty acids longer than C14. Here, two ELO full-length cDNAs (TmELO1, TmELO2) from the yellow mealworm (Tenebrio molitor L.) were isolated and the functions were characterized. The open reading frame (ORF) lengths of TmELO1 and TmELO2 were 1005 bp and 972 bp, respectively and the corresponding peptide sequences each contained several conserved motifs including the histidine-box motif HXXHH. Phylogenetic analysis demonstrated high similarity with the ELO of Tribolium castaneum and Drosophila melanogaster. Both TmELO genes were expressed at various levels in eggs, 1st and 2nd instar larvae, mature larvae, pupae, male and female adults. Injection of dsTmELO1 but not dsTmELO2 RNA into mature larvae significantly increased mortality although RNAi did not produce any obvious changes in the fatty acid composition in the survivors. Heterologous expression of TmELO genes in yeast revealed that TmELO1 and TmELO2 function to synthesize long chain and very long chain fatty acids.
Similar content being viewed by others
Introduction
Fatty acids (FAs) are molecules with a variety of biological functions including acting as energy sources and serving as components of cellular lipids and other molecules including eicosanoid hormones such as prostaglandins and leukotriences1. FAs are precursors of sphingolipids, glycerolipids2, hydrocarbons3, 4, fatty alcohols and wax esters5, which can participate in many cell biological processes such as reproduction, growth, migration, differentiation, and apoptosis and are components of pheromones in various arthropod species6,7,8.
Structurally, FAs are composed of long hydrocarbon chains that end in a carboxyl group and are classified based on the chain length and the number of double bonds. Fatty acids are roughly classified by length into the following groups: (1) Short Chain Fatty Acids (SCFA) which have five or fewer carbons, (2) Medium Chain Fatty Acids (MCFAs) which contain 6–12 carbons, (3) Long-Chain Fatty Acids (LCFAs), which contain more than 12 carbon atoms, and (4) Very Long-Chain Fatty Acids (VLCFAs), which contain 22 or more carbon atoms. Each class is associated with unique functions. For example, in mammals LCFAs act as ligands for peroxisome proliferator-activated receptors (PPARs) and regulate energy metabolism9 while VLCFAs have important anti-inflammatory roles10.
Fatty acid synthesis consists of a four-step cycle that includes condensation, reduction, dehydration and reduction steps and occurs primarily in the endoplasmic reticulum (ER)11. Each cycle extends an initial acetyl-CoA by two carbons and can be repeated up to seven times to form palmitic acid (C16:0)2. Further growth requires elongases of long or very long chain fatty acids (ELOVL or ELO) which can elongate C ≥ 14 fatty acids through a fatty acid condensation reaction. Elongases of very long chain fatty acids have been isolated from various organisms, including yeast, mammals, plants and other species12,13,14.
While ELOVLs have been relatively well characterized in vertebrates, little is known about these enzymes in insects, even though LCFAs and VLFAs are widespread among insect taxa15,16,17,18,19,20. Although some ELOVLs were isolated and characterized in a handful of arthropod species including Drosophila melanogaster and Aedes albopictus 21, the ELOs and ELOVLs in insects are poorly understood and require further investigation4, 22, 23. The yellow mealworm beetle, Tenebrio molitor, is a species that has abundant fatty acids including VLCFAs, yet the T. molitor elongases associated with VLCFA synthesis have not been identified. In this study, we identified and characterized two elongases (TmELO1 and TmELO2) of Very Long-Chain Fatty Acids of T. molitor. These two ELOs were chosen out of the 20 putative ELOs identified from a transcriptome analysis due to the availability of full length sequences. Functions of TmELO1 and TmELO2 were analyzed using both an in vitro expression system in yeast and in vivo RNA interference in mature T. molitor larvae.
Results
Sequence Analysis of ELOs from T. molitor
The open reading frames (ORFs) of full-length TmELO1 (GenBank accession no. MF279188) and TmELO2 (GenBank accession no. MF279189) were identified by DNAStar and the physical and chemical properties of the deduced proteins were calculated using the ProtParam tool of ExPASy24 (Table 1). The instability index suggested that TmELO2 was a relatively stable peptide of 323 aa, and TmELO1 was of similar length at 334 aa but more unstable than TmELO2. The grand average of hydropathicity (GRAVY) shows that both TmELO1 and TmELO2 are hydrophilic. Further analysis indicates the presence of 5 putative transmembrane regions in TmELO1 (27–46, 66–88, 171–193, 205–224, 234–251 amino acids) and 7 for TmELO2 (25–47, 68–90, 116–135, 142–161, 171–193, 205–227, 237–254 amino acids). Prediction of subcellular locations using Euk-mPLoc 2.025,26,27 suggests that both TmELO1 and TmELO2 were localized to the endoplasmic reticulum, the primary site of LCFA and VLCFA synthesis. Secondary structures of both TmELO1 andTmELO2 were predicted to be relatively similar to one another. Protein secondary structure prediction of TmELO1 showed the percentage of α-helixes, β-turns, random coils, and extended strands among the total amino acids to be 34.7%, 6.9%, 29.3% and 29.0%, respectively. The percentage in TmElO2 was 38.4% α-helixes, 8.1% β-turns, 23.2% random coils, and 30.3% extended strands.
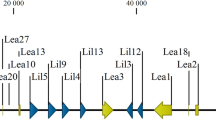
Multi-sequencing alignment of the TmELOs with ELOs from other species showed that the proteins had some similar motifs such as KXXEXXDT, HXXMYXYY, TXXQXXQ and HXXHH, a histidine-box motif that is conserved in all elongases28 (Fig. 1). The TmELO1 and TmELO2 had the highest identity at 92.51% with Tribolium castaneum LOC660197 and 88.62% with T. castaneum LOC660257 respectively, two uncharacterized predicted elognases. Compared with D. melanogaster elongases, TmELO had identity at 60.17% with CG31523, an uncharacterized, predicted elongase in D. melanogaster (Genbank accession no. NP_649474.1). The TmELO sequences shared 17.18–43.79% identity with Homo sapiens with HsELOVL7 being the most similar. The TmELO sequences had low identity to S. cerevisiae with the highest identity being only 18.67%.
Multi-sequencing alignment of TmELO1 and TmELO2 with H. sapiens ELOVL1 and ELOVL7 (Genbank accession no. KJ894579.1 and AB181393.1), S. cerevisiae ELO2 (Genbank accession no. NM_001178748.1), D. melanogaster elo68α (CG32072), eloF (CG16905), CG31523 (Genbank accession no. AJ871925.1, AM292552.1 and NP_649474.1), T, castaneum LOC660197 (Genbank accession no. XM_966451.3). The similar motifs which were marked by black lines are distinctive signs to the ELONGASE protein families. The conserved histidine motif HXXHH in the centre of the protein is boxed.
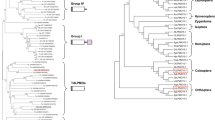
A phylogenetic tree was constructed comparing the amino acid sequences of T. molitor elongases 1 and 2 with elongases from other organisms (Fig. 2). The phylogenetic analysis shows that TmELO1 clustered with T. castaneum LOC660197 and D. melanogaster CG31523 while TmELO2 clustered with T. castaneum LOC660257 and D. melanogaster CG2781, another predicted member of the ELO family.
Phylogenetic tree of ELO included H. sapiens ELOVL1-7(marked by Purple squares), S. cerevisiae ELO1-3, all 20 D. melanogaster ELOs, 18 T. castaneum ELOs and TmELO1-2. Blue triangles indicate ELO which have been studied in insects. The phylogenetic tree was constructed using Neighbor-Joining method with 1000 bootstrap replicates. Each species was followed by its Genbank accession number.
Relative transcript level at developmental stages
Expression profiles of TmELO1 and TmELO2 at various developmental stages were generated using qRT-PCR (Fig. 3). Expression of both elongases was detected throughout all developmental stages examined. Expression of TmELO1 peaked in 1st instar larvae and pupae while TmELO2 peaked in embryos and was somewhat reduced throughout the remaining developmental stages. During embryonic development, expression of TmELO2 was significantly higher than TmELO1. Expression of TmELO1 was significantly higher than TmELO2 throughout all other stages examined.
Relative transcript level of TmELO1 and TmELO2 at different development stages of T. molitor. The transcript levels of TmELO1 and TmELO2 at different developmental stages were conducted by qRT-PCR using RpS3 as a reference housekeeping gene. Y axis values are mean ± SE of relative expression levels; Lowercase letters (a–d) represent significant differences (P < 0.05) according to Tukey’s multiple range test.
Fatty acid compositions and effects of RNAi
Total fatty acid compositions of mature larvae are listed in Table 2. The major fatty acids of T. molitor were C14:0, C16:0, C18:0, C18:1 and C18:2. Saturated fatty acids (SFAs), monounsaturated fatty acids (MUFAs) and polyunsaturaed fatty acids (PUFAs) were 25.55%, 32.26% and 41.25%, respectively. The fatty acids (C > 16) were 81.2%.
Double-stranded RNA (dsRNA) fragments were generated for gene silencing using RNA interference. The dsRNA fragment lengths of TmELO1 were 361 bp (dsELO1-1) and 298 bp (dsELO1-2), and TmELO2 were 401 bp (dsELO2-1) and 342 bp (dsELO2-2), respectively. The relative transcript levels of TmELO1 and TmELO2 in dsRNAs-injected mature larvae decreased significantly at 1d, 2d, 3d and 4d after injection (Fig. 4). One day after injection, the transcript level of TmELO1 was significantly reduced by 76.5–81.3% and TmELO2 was significantly reduced by 75.8–86.7%. Both TmELO genes were largely suppressed after 1d, 2d, 3d and 4d, which showed that the dsRNA had a long-lasting effects on the TmELO1 and TmELO2 expression. Fatty acid compositions in the survivors after RNAi injections were not significantly different than controls (data not shown), however, dsRNA injection was associated with increased larval mortality (Fig. 4). Compared to the control, mortality after TmELO1 RNAi treatment significantly increased to 50.8%, while, the mortality following TmELO2 RNAi treatment was slightly, but not significantly higher than controls.
The dsRNA-mediated suppression of (A) TmELO1 and (B) TmELO2 transcrpts in mature larvae. 5000 ng dsRNA was injected in every larva. Students t test was used in data analysis. Y axis values are mean ± SE of relative transcript level. Uppercase letters (A–C) represent significant differences (P < 0.01) according to Tukey’s multiple range test. (C). Mature larvae of T. molitor were injected with dsRNA of EGFP or TmELO1 or TmELO2. There were three replicates and each replicate included 40 mature larvae. Y axis values are mean ± SE of the death rate. Uppercase letters (A,B) represent significant differences (P < 0.01) according to Tukey’s multiple range test.
In-vitro functional characterization of TmELOs
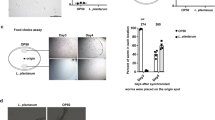
The ORFs of TmELO1 and TmELO2 cDNA were isolated from T. molitor, cloned into a pYES2 yeast expression vector, and the recombinant plasmids were introduced into yeast strain INVSc1. Transcripts of two TmELOs were detected by RT-PCR in the TmELO-transformed yeast (Fig. 5). The fatty acid analysis of transformed yeast showed TmELO1 expression increased relative amounts of C14:0, C16:0, C16:1 and C 20:0, and C24:0 fatty acids and reduced C18:0 fatty acids when compared with controls (Table 3). Expression of TmELO2 in yeast increased relative amounts of C14:0, C14:1, C16:0, C16:1 fatty acids and reduced C18:1 fatty acids. In general, our observations suggest that in yeast, TmELO1 could produce C20:0 and elongate SFA to C24; TmELO2 primarily increases the percentage of C16:0 and C16:1. (Table 3).
Discussion
Tenebrio molitor larvae contain a large proportion of C ≥ 16 fatty acids including LCFAs and VLCFAs and here we characterize two elongases, TmELO1 and TmELO2 that are involved in their synthesis. Both TmELO1 and TmELO2 are expressed throughout the lifetime of T. molitor and do not exhibit sex-specific expression. During embryonic stages, TmELO2 appears to be the predominant form while TmELO1 is expressed at higher levels throughout the remaining developmental stages. Our results are consistent with findings in mammals that demonstrate developmental regulation of different elongases, and that expression is regulated not only by developmental signals but also by diet29. The effect of diet on T. molitor ELOVLs during development is intriguing but is outside the scope of the current study.
In the present study, expression of T. molitor elongases in yeast cells demonstrates differences in the fatty acids produced by TmELO1 and TmELO2 (Table 3). Expression of TmELO2 led to significant increases of fatty acids up to 16 carbons in length, suggesting that TmELO2 function is limited to long-chain fatty acid synthesis. Meanwhile, TmELO1 expression led to similar increases in long-chain fatty acids but also produced a significant increase in C20 fatty acids and slight, but not significant increases in C22, and C24 fatty acids, suggesting that TmELO1 may possess an additional role in the synthesis of very long-chain fatty acids. Surprisingly, RNAi-mediated knockdown of TmELO1 and TmELO2 in T. molitor did not significantly alter the fatty acid composition (data not shown). The lack of significant changes suggests that residual TmELO1 and TmELO2 transcript remained following RNAi treatment. It is also likely that T. molitor elongases have redundant roles and can maintain fatty acid levels in the absence of TmELO1 or TmELO2 activity. In support of the idea of redundant roles of elongases, ELOs in other species also have broad and somewhat overlapping functions. For example, in humans, HsELOVL1 can elongate C20–C26 SFAs30, while HsELOVL3 can elongate FAs from C16 to C222.
Our observation that the two TmELOs generate FAs of different lengths is not unprecedented. In mammals, ELOVL1-7 play different roles in the elongation cycle of fatty acids2, 31, 32. ELOVL1 produces C20 to C26 SFAs and MUFAs30. ELOVL2 can elongate C20 to C22 PUFAs33. ELOVL3 can elongate FAs from C16 to C22, with the highest activity toward C18-CoAs2. ELOVL4 is specialized to elongate ultra long-chain fatty acids (ULCFAs) which are longer than C2634, 35. ELOVL5 elongates FAs from C18 to C2036. ELOVL6 can elongate C12:0 FAs to C16:037. Finally, ELOVL7 has highest activity toward C18:3n-3 FA and C18:3n-6 FA38, 39.
Three ELOs of S. cerevisiae also showed variable functions. ScELO1 can elongate C14 FAs to C16 FAs12 while ScELO2 is involved in the synthesis of SFAs and MUFAs to C24 and ScELO3 is essential for the synthesis of C24:0 to C26:0 and can also elongate a wide range of SFAs and MUFAs13. Similar results were also observed in Drosophila. Chertemps et al. (2005) reported an elongase gene named elo68α in Drosophila males which can elongate myristoleic and palmitoleic acids (C14:1n-9 and C16:1n-9) in-vtiro expression in yeast22. Chertemps et al. (2007) found that eloF cDNA of D. melanogaster expressed in yeast could elongate medium-chain saturated and unsaturated fatty acids up to C304.
In T. molitor, silencing of TmELO1 via RNAi resulted in an increased mortality rate indicating that TmELO1 is essential for mealworm survival. Similar requirements for elongases in organism survival have been reported in other species. For example, in Drosophila, RNAi to CG6660, which encodes a predicted elongase, induced a similar lethal phenotype40. Elongases have also been shown to be required for neonatal survival in mice41, survival of the protozoan parasite, Toxoplasma gondii 42, and growth and survival of cancer cells43. Together, these studies emphasize that the requirement for elongases in growth, development, and survival is likely conserved among metazoans.
Materials and Methods
Insects
T. molitor were obtained from Shandong Agricultural University and have been reared in our laboratory at Zhejang A&F University for two years. The artificial climate chambers were maintained at 26 ± 1 °C, 65 ± 5% relative humidity and a 8:16 (L:D) photoperiod. The mealworm were reared on wheat bran mixed with cabbage.
Cloning of TmELO cDNAs
Total RNA was extracted from different stages of T. molitor using RNAiso Plus (TAKARA, Dalian, China), per the manufacturer’s protocol and resulting RNA was stored at −80 °C. The concentration and quality of total RNA was measured with a UV/VIS spectrophotometer (BioDrop μLite, Cambridge, UK) and agarose gel electrophoresis was used to verify integrity of the RNA. The 500ng total RNA was used in cDNA synthesis reactions using the PrimeScriptTM 1st strand cDNA Synthesis kit (TAKARA, Dalian, China) and resulting cDNA was stored at −20 °C. De novo transcriptome sequencing was conducted at Tianke High-Tech Development Co., Ltd. (Zhengjiang, China) and the transcriptome analysis of T. molitor was performed in our laboratory (data not shown). The specific primers used to amplify two putative ELO genes are listed in Table 4. The PCR conditions for all these amplicons were 35 cycles of 94 °C for 15 s, 55 °C for 30 s and 72 °C for 1 min. A 3′A-overhang was generated by Taq DNA polymerase (TAKARA), and amplified products were gel purified with an E.Z.N.A gel extraction kit (Omega bio-tek, Norcross, GA) and linked with a plasmid pMD19-T vector (TAKARA) to form a recombinant plasmid, which was then transformed into DH5α (E. coli) for 10 hrs cultivation in LB solid medium with 50 μg/ml ampicillin. The plasmids were sequenced with sequence-specific primer M13 and maintained as glycerol stocks at −80 °C.
Sequence characterization of TmELOs
Full-length cDNA, ELO nucleotide sequences were translated into amino acid sequences and chemical and physical characteristics of TmELO genes were determined by the Expert Protein Analysis System program (http://web.expasy.org/protparam/)24. Trans-membrane structures were calculated using the TMHMM Server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/). Subcellular locations were predicted by Euk-mPLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/euk-multi-2/)25,26,27. SOMPA44 was used for the prediction of protein secondary structure. Amino acid sequences of other species ELOs were obtained from NCBI (http://www.ncbi.nlm.nih.gov/) and a phylogenetic tree was constructed and analyzed with respect to its homolog using the Neighbor Joining method. A bootstrap consensus tree of 1000 replicates was used to evaluate branch strength for analysis using MEGA 6.06.
Preparation of dsRNA and injection
The dsRNA fragments of two ELO genes which were used to silence the corresponding mRNA transcripts were synthesized using the in vitro Transcription T7 Kit (TAKARA)45 and the specific primers used are listed in Table 4. Plasmid pMD19-T vectors linked with amplified TmELO1 and TmELO2 were the templates. The quality and length of the dsRNA fragments were measured by BioDrop μLite (BioDrop, Cambridge, England) and electrophoresis. The fragments were purified and diluted into a final concentration of 1 μg/μl. All dsRNAs were stored at −20 °C. Each of the two ELO dsRNA were injected separately into mature larvae of T. molitor and EGFP dsRNA was used as the control. The larvae were collected 1d, 2d, 3d and 4d after injection, flash-frozen in liquid nitrogen and stored at −80 °C. Animals were observed daily and the death rate was recorded through the pupal and adult stages till two weeks after injection. Every sample contained 20 larvae (n = 3) which were injected with 5 μg dsRNA fragments.
Quantitative real-time PCR
Total RNA extraction from T. molitor and cDNA synthesis was conducted using the same procedures as Cloning of TmELO cDNAs. The cDNA products were diluted to 5 ng/μl. Primers (Table 4) for PCR and dsRNA synthesis were designed according to TmELO1 and TmELO2 sequences. The qRT-PCR reaction consisted of 20 μl including 10 μl SYBR® Premix Ex Taq TM (TAKARA), 1 μl each of 10 μM forward and reverse primer, 1 μl of diluted cDNA and dd H2O. Ribosomal protein S3 (TmRpS3) (Genbank No.KJ868729.1) was selected as a housekeeping gene for normalization. Two-step qRT-PCR was conducted on Bio-Rad CFX96 (Bio-Rad, Hercules, California, USA) under the following conditions: 95 °C for 3 min, followed by 40 amplification cycles of 15 s at 95 °C and 30 s at 55 °C. Melting curves were used to verify the specificity of amplifications. Three biological replicates were carried out per treatment. Relative quantification analysis was then calculated using the 2−ΔΔCt formula46.
Heterologous expression of TmELO ORFs in yeast
The open reading frames (ORFs) of TmELO1 and TmELO2 were amplified by eTmELO-F and eTmELO-R primers. Primer F and Primer R separately contain a Xho I site and a BamH I site (Table 4). After PCR, the products were digested with Xho I and BamH I, ligated into the yeast expression vector pYES2 (Invitrogen), and used to transform E. coli DH5α. The E. coli were cultured and recombinant pYES2-TmELO1, and pYES2-TmELO2 plasmids were extracted from DH5α using a Endo-free Plasmid Mini Kit I (OMEGA) according to manufacturer instructions. The pYES2, pYES2-TmELO1, and pYES2-TmELO2 plasmids were introduced into INVSc1 (Saccharomyces cerevisiae) using the lithium acetate method47. A single colony was selected from INVSc1 strains transformed with pYES2 (as control), pYES2-TmELO1 or pYES2-TmELO2 plasmids, introduced into 15 ml of SC-U medium containing 2% glucose, and maintained overnight, with shaking, at 30 °C. The overnight cultured yeast was resuspended in 50 ml of SC-U medium containing 2% galactose and grown to an optical density (OD600) reached 0.4. Cells were harvested after 24 h at 30 °C with shaking, washed twice by dd H2O and stored at −80 °C until the FA were analyzed.
FAs analysis
T. molitor larvae were homogenized and trans-methylated in 1% H2SO4 in methanol (v:v) at 80 °C for 2 h to prepare mealworm fatty acid methyl ester (FAME)(n = 4). Total lipid was extracted from yeast cells by using acidified glass beads to break cell wall for 10 min, and using chloroform:methanol (2:1, v-v) and 1% H2SO4 in methanol (v:v) at 80 °C for 2 h to prepare yeast FAME (n = 4). Mealworms and yeast FAME were added to 2 mL 0.9% NaCl and extracted twice with 2 ml of hexane. Hexane was subsequently removed by nitrogen gas and the total FAME was resuspended in 300 ml of hexane. After mealworms or yeast were homogenized, 100 μg C17:0 was added as an internal control. FAME were then extracted and analyzed by GC. The samples were analyzed on an Agilent 6890 N Gas Chromatograph (GC) equipped with a DB-23 column (60 m × 0.25 mm) with 0.25 μm film thickness. The following temperature program was employed: 160 °C for 1 min, then 10 °C/min to 240 °C, with He as carrier gas. F.A.M.E. Mix, C4-C24 (Supelco®) was used as the external standard.
References
Calder, P. C. Functional Roles of Fatty Acids and Their Effects on Human Health. J. Parenter. Enteral Nutr. 39, 18S–32S (2015).
Sassa, T. & Kihara, A. Metabolism of very long-chain fatty acids: genes and pathophysiology. Biomol. Ther. 22, 83–92 (2014).
Juárez, M. P. Fatty Acyl-CoA Elongation in Blatella germanica integumental microsomes. Arch. Insect Biochem. 56, 170–178 (2004).
Chertemps, T. et al. A female-biased expressed elongase involved in long-chain hydrocarbon biosynthesis and courtship behavior in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 104, 4273–4278 (2007).
Teerawanichpan, P., Robertson, A. J. & Qiu, X. A fatty acyl-CoA reductase highly expressed in the head of honey bee (Apis mellifera) involves biosynthesis of a wide range of aliphatic fatty alcohols. Insect Biochem. Mole. 40, 641–649 (2010).
Cuvillier, O. Sphingosine in apoptosis signaling. Biochim. Biophys. Acta. 1585, 153–162 (2002).
Hannun, Y. A. & Luberto, C. Ceramide in the eukaryotic stress response. Trends Cell Biol. 10, 73–80 (2000).
Hannun, Y. A. & Obeid, L. M. The Ceramide-centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind. J. Biol. Chem. 277, 25847–25850 (2002).
Nakamura, M. T., Yudell, B. E. & Loor, J. J. Regulation of energy metabolism by long-chain fatty acids. Prog. Lipid Res. 53, 124–144 (2014).
Bannenberg, G. & Serhan, C. N. Specialized pro-resolving lipid mediators in the inflammatory response: an update. Biochim. Biophys. Acta. 1801, 1260–1273 (2010).
Nugteren, D. H. The enzymic chain elongation of fatty acids by rat-liver microsomes. Biochim. Biophys. Acta. 106, 280–90 (1965).
Toke, D. A. & Martin, C. E. Isolation and characterization of gene affecting fatty acid elongation in Saccharomyces cerevisiae. J. Boil. Chem. 271, 18413–18422 (1996).
Oh, C. S., Toke, D. A., Mandala, S. & Martin, C. E. ELO2 and ELO3, Homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Boil. Chem. 272, 17376–17384 (1997).
Leonard, A. E., Pereira, S. L., Sprecher, H. & Huang, Y. S. Elongation of long-chain fatty acids. Prog. Lipid Res. 43, 36–54 (2004).
Stanley-Samuelson, D. W. & Dadd, R. H. Long-chain polyunsaturated fatty acids: Patterns of occurrence in insects. Insect Biochem. 13, 549–558 (1983).
Howard, R. W. & Stanley-Samuelson, D. W. Fatty acid composition of fat body and malpighian tubules of the tenebrionid beetle, Zophobas atratus: Significance in eicosanoid-mediated physiology. Comp. Biochem. Phys. Part B: Biochem. Mole. Biol. 115, 429–437 (1996).
Spike, B. P., Wright, R. J., Danielson, S. D. & Stanley-Samuelson, D. W. The fatty acid compositions of phospholipids and triacylglycerols from two chinch bug species Blissus leucopterus leucopterus and B. iowensis (Insecta: Hemiptera; Lygaeidae) are similar to the characteristic dipteran pattern. Comp. Biochem. Phys. Part B: Biochem. Mole. Biol. 99, 799–802 (1991).
Buckner, J. S. & Hagen, M. M. Triacylglycerol and phospholipid fatty acids of the silverleaf whitefly: Composition and biosynthesis. Arch. Insect Biochem. 53, 66–79 (2003).
Stanley-Samuelson, D. W., Loher, W. & Blomquist, G. J. Biosynthesis of polyunsaturated fatty acids by the australian field cricket, Teleogryllus commodus. Insect Biochem. 16, 387–393 (1986).
Shipley, M. M., Dillwith, J. W., Bowman, A. S., Essenberg, R. C. & Sauer, J. R. Changes in lipids of the salivary glands of the lone star tick, Amblyomma americanum, during feeding. J. Parasitol. 79, 834–842 (1993).
Urbanski, J. M., Benoit, J. B., Michaud, M. R., Denlinger, D. L. & Armbruster, P. The molecular physiology of increased egg desiccation resistance during diapause in the invasive mosquito, Aedes albopictus. Proc. Biol. Sci. 277, 2683–2692 (2010).
Chertemps, T., Duportets, L., Labeur, C. & Wicker-Thomas, C. A new elongase selectively expressed in Drosophila male reproductive system. Biochem. Bioph. Res. Co. 333, 1066–1072 (2005).
Szafer-Glusman, E. et al. A Role for very-long-chain fatty acids in furrow ingression during cytokinesis in Drosophila Spermatocytes. Curr. Biol. 18, 1426–1431 (2008).
Gasteiger, E. et al. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook (eds Walker, J. M.) 571–607 (Humana Press, 2005).
Chou, K. C. & Shen, H. B. Euk-mPLoc: A fusion classifier for large-scale eukaryotic protein subcellular location prediction by incorporating multiple sites. J. Proteome Res. 6, 1728–1734 (2007).
Chou, K. C. & Shen, H. B. A new method for predicting the subcellular localization of eukaryotic proteins with both single and multiple sites: Euk-mPLoc 2.0. PLOS One 5, e9931 (2010).
Chou, K. C. & Shen, H. B. Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 3, 153–162 (2008).
Moon, Y. A., Shah, N. A., Mohapatra, S., Warrington, J. A. & Horton, J. D. Identification of a mammalian long chain fatty acyl elongase regulated by sterol regulatory element-binding proteins. J. Biol. Chem. 276, 45358–45366 (2001).
Wang, Y. et al. Tissue-specific, nutritional, and developmental regulation of rat fatty acid elongases. J. Lipid Res. 46, 706–715 (2005).
Ohno, Y. et al. ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proc. Natl. Acad. Sci. USA 107, 18439–18444 (2010).
Jakobsson, A., Westerberg, R. & Jacobsson, A. Fatty acid elongases in mammals: Their regulation and roles in metabolism. Prog. Lipid. Res. 45, 237–249 (2006).
Guilou, H., Zadravec, D., Martin, P. G. & Jacobsson, A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid. Res. 49, 186–199 (2010).
Zadravec, D. et al. ELOVL2 controls the level of n-6 28:5 and 30:5 fatty acids in testis, a prerequisite for male fertility and sperm maturation in mice. Chem. Phys. Lipids 52, 245–255 (2011).
Li, W. et al. Depletion of ceramides with very long chain fatty acids causes defective skin permeability barrier function, and neonatal lethality in ELOVL4 deficient mice. Int. J. Biol. Sci. 3, 120–128 (2007).
Vasireddy, V. et al. Elovl4 5-bp-deletion knock-in mice develop progressive photoreceptor degeneration. Invest. Ophthalmol Vis. Sci. 47, 4558–4568 (2006).
Moon, Y. A., Hammer, R. E. & Horton, J. D. Deletion of ELOVL5 leads to fatty liver through activation of SREBP-1c in mice. J. Lipid Res. 50, 412–423 (2009).
Matsuzaka, T. et al. Crucial role of a long-chain fatty acid elongase, ELOVL6, inobesity-induced insulin resistance. Nat. Med. 13, 1193–1202 (2007).
Naganuma, T., Sato, Y., Sassa, T., Ohno, Y. & Kihara, A. Biochemical characterization of the very long-chain fatty acid elongase ELOVL7. FEBS Lett. 585, 3337–3341 (2011).
Purdy, J. G., Shenk, T. & Rabinowitz, J. D. Fatty acid elongase 7 catalyzes lipidome remodeling essential for human cytomegalovirus replication. Cell Rep. 10, 1375–1385 (2015).
Parvy, J. P. et al. Drosophila melanogaster Acetyl-CoA-Carboxylase sustains a fatty acid-dependent remote signal to waterproof the respiratory system. PLOS Genet. 8, e1002925 (2012).
Cameron, D. J. et al. Essential role of Elovl4 in very long chain fatty acid synthesis, skin permeability barrier function, and neonatal survival. Int. J. Biol. Sci. 3, 111–119 (2007).
Mazumdar, J., H Wilson, E., Masek, K., A Hunter, C. & Striepen, B. Apicoplast fatty acid synthesis is essential for organelle biogenesis and parasite survival in Toxoplasma gondii. Proc. Natl. Acad. Sci. USA 103, 13192–13197 (2006).
Röhrig, F. & Schulze, A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 16, 732–749 (2016).
Geourjon, C. & Deléage, G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci. 11, 681–684 (1995).
Davanloo, P., Rosenberg, A. H., Dunn, J. J. & Studier, F. W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 81, 2035–2039 (1984).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Gietz, R. D., Schiestl, R. H., Willems, A. R. & Woods, R. A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355–360 (1995).
Acknowledgements
The authors would like to thank for the financial support from DEAN-ZAFU cooperative project (2045200233) and Developmental Fund of Zhejiang A&F University (2012FR087).
Author information
Authors and Affiliations
Contributions
T.X.Z., M.Z.W. and D.Y.Z. conceived and designed the experiments; T.X.Z. and H.S.L. performed the experiments; T.X.Z., N.H., S.Y. W. J.H.P. and D.Y.Z. analyzed the data; T.X.Z. J.H.P. and D.Y.Z. wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zheng, T., Li, H., Han, N. et al. Functional Characterization of Two Elongases of Very Long-Chain Fatty Acid from Tenebrio molitor L. (Coleoptera: Tenebrionidae). Sci Rep 7, 10990 (2017). https://doi.org/10.1038/s41598-017-11134-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-11134-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.