Abstract
Oxytocin is a pituitary neuropeptide that affects social behaviour. Single nucleotide polymorphisms (SNPs) in the oxytocin receptor gene (OXTR) have been shown to explain some variability in social abilities in control populations. Whether these variants similarly contribute to the severity of social deficits experienced by children with neurodevelopmental disorders is unclear. Social abilities were assessed in a group of children with autism spectrum disorder (ASD, n = 341) or attention deficit hyperactivity disorder (ADHD, n = 276) using two established social measures. Scores were compared by OXTR genotype (rs53576, rs237887, rs13316193, rs2254298). Unexpectedly, the two most frequently studied OXTR SNPs in the general population (rs53576 and rs2254298) were associated with an increased severity of social deficits in ASD (p < 0.0001 and p = 0.0005), yet fewer social deficits in ADHD (p = 0.007 and p < 0.0001). We conclude that these genetic modifier alleles are not inherently risk-conferring with respect to their impact on social abilities; molecular investigations are greatly needed.
Similar content being viewed by others
Introduction
The past decade has yielded major advances in our understanding of the genetics of human social behaviour. Twin and family studies have shown that traits such as empathy and social cognition have moderate to high heritability1, 2. Genome wide association studies (GWAS) and candidate gene studies now seek to identify specific underlying genes and polymorphisms that influence social abilities1. Oxytocin and vasopressin are pituitary neuropeptides that exert robust effects on social processes in animal models3, 4. In humans, it is hypothesized that these peptides may influence social behaviour by enhancing the saliency of social cues, or by attenuating fear/stress5. Plasma oxytocin levels (whether endogenous, or exogenously administered) have been shown to correlate with facial emotion recognition abilities6,7,8. In control populations, common polymorphisms in oxytocin or vasopressin receptor genes (OXTR and AVPR1a) have been associated with differences in empathy9,10,11, prosocial temperament12, social sensitivity13,14,15,16,17, and stress reactivity in social contexts18, 19. The area on chromosome 3 carrying the OXTR gene (3p24–26) has emerged on GWAS with respect to risk for autism spectrum disorder (ASD) at a genome wide significance level20. Meta-analyses indicate that certain neuropeptide variants are more common in individuals with ASD than in controls21, and may influence social abilities in the general population22.
ASD and attention deficit hyperactivity disorder (ADHD) are common childhood onset neurodevelopmental disorders that are highly heterogeneous, present along a spectrum of severity, and often comorbid23, 24. Both disorders are associated with social deficits, inattention, and hyperactivity to varying degrees23,24,25, leading some to propose that they may be different manifestations of an overarching disorder23. In addition to rare genetic mutations and environmental factors, the additive effects of common genetic variants are hypothesized to contribute to the development of these disorders26, and may explain some of the observed heterogeneity in behaviour, abilities, and psychopathology. Preliminary investigations suggest that single nucleotide polymorphisms (SNPs) in OXTR may act as modifier alleles with respect to the severity of social deficits experienced by individuals with ASD (For a review of the literature, see Supplementary Table S1). Two studies have also examined OXTR SNPs in small groups of children with ADHD27, 28. It remains unclear whether OXTR SNPs act as: 1) general risk factors for social impairment across populations (i.e. effects span diagnostic boundaries), or 2) specific modifier alleles in certain behavioural or biological context (i.e. disorder specific effects). Two recent studies have shown that OXTR variants were associated with a similar magnitude and direction of social differences in children with ASD and their unaffected relatives6, 29. This question, however, has not yet been examined across different diagnostic groups, where the environmental, behavioural, and genetic architecture differ. Given the increasing number of studies employing trans-diagnostic approaches in psychiatry30, it is important to examine whether behavioural risk variants function similarly across disorders.
In this study, we examined the association between four SNPs in OXTR and social abilities in a large sample of children and youth with ASD or ADHD and present the first cross disorder comparison of genotype effects. To extend on preliminary research to date, we used two validated continuous social metrics, and restricted initial hypotheses to Caucasian participants only, given evidence of possible ancestral differences in the effects of OXTR on social abilities31, 32. First, in our group of children with ASD, we confirmed several associations between social deficits and OXTR variants (OXTR rs53576 G-allele, rs237887 A-allele, and rs2254298 A-allele) that had been previously shown in smaller samples. New associations were detected in the group of children with ADHD. Second, we compared the magnitude and direction of the impact of common variation in OXTR on social abilities between diagnostic groups (ASD vs. ADHD). Based on the family studies described above, and the high degree of symptom overlap and comorbidity between ASD and ADHD, we hypothesized that genotype effects would not differ by diagnosis. However, unexpectedly, highly significant diagnostic differences were detected, with effectively a reciprocal direction of association between genotype and social abilities between the two diagnostic groups. Our findings indicate that common polymorphisms in neuropeptide receptor genes likely represent a specific and not a general risk modifier, with effects that vary by diagnosis in the context of different biological, behavioural, or environmental conditions. These results have implications for all analyses involving common alleles and complex traits.
Results
Study sample
The sample consisted of 617 participants with either ASD or ADHD (n = 404 Caucasian). The ASD group had lower mean intelligence quotients (IQs), and higher anxiety levels (see Table 1). Genotype frequencies and linkage analyses are presented in Supplementary Tables S2 and S3, compared to/based on reference genome data from the 1000 Genomes Project33. OXTR allele frequencies did not deviate from Hardy Weinberg equilibrium (Supplementary Table S4), except for rs237887 in ADHD.
Measures
Severity of social deficits were quantified by examining the number of incorrect items selected by the participants on the Reading the Mind in the Eyes Test (RMET)34, and through their parent/caregiver’s ratings of concerns on the Social Communication Questionnaire (SCQ) (see methods)35. Therefore, throughout the manuscript, higher scores on either metric suggest greater social deficits. Correlations between measures in ASD and ADHD are shown in Supplementary Tables S5 and S6. Twelve percent of the participants with ADHD had SCQ scores that fell above the cut-off for concern for ASD (Total SCQ > 15). For comparison, 23% of the participants with ASD fell above the cut-off for clinical concern regarding ADHD on the Child Behavior Checklist (T-score > 70).
Aim 1
Confirmation of previous OXTR genotype effects on social abilities
Using logistic regression, we initially examined OXTR SNPs with prior evidence of genotype effects on social abilities within each diagnostic group (in ASD: rs53576, rs2254298, and rs237887; in ADHD: rs53576 and rs13316193), restricting the sample to those of Caucasian ancestry only, and adjusting for other possible confounding factors (see Methods). When considering multiple testing, five comparisons (3 in ASD and 2 in ADHD) across two social metrics yielded a corrected p-value of 0.005 as the threshold for significance for an overall alpha of 0.05.
In the Caucasian ASD cohort, we hypothesized that the OXTR rs53576 G-allele carriers, rs2254298 A-allele carriers, and the rs237887 AA carriers would have greater social deficits, based on previous literature. In line with our hypotheses, rs53576 G-allele carriers (GG/GA) had higher SCQ scores [Odds Ratio (OR) of an increase in SCQ score for the high-risk genotype group compared to low-risk genotype group: 1.4 (95% CI: 1.2–1.6), Z-value = 4.2, p < 0.0001 uncorrected]. Differences were non-significant on the RMET for this SNP (Z = 1.2, p = 0.2) (Table 2, left panel). Similarly, for OXTR rs2254298, the hypothesized high-risk group (AA/AG) also had higher SCQ scores in the ASD cohort (OR 1.2, 95% CI: 1.1–1.4, Z = 3.5, p = 0.0005), suggesting greater social deficits (Table 2). There were no differences on the RMET (Z = −1.2, p = 0.2). As hypothesized, OXTR rs237887 genotype was associated with differences in social abilities, with the AA-allele group having greater social deficits than the GG group on the both the SCQ (OR: 1.2, 95% CI: 1.0–1.4) and the RMET (OR: 1.2, 95% CI: 1.0–1.5), although neither finding would survive correction for multiple comparisons (SCQ: p = 0.05; RMET: p = 0.06; Table 2).
In the Caucasian ADHD cohort, we hypothesized that OXTR rs13316193 CC genotype group would have greater social deficits, as would the rs53576 G-allele carriers. In our sample, OXTR rs13316193 genotype was not associated with differences in social abilities on either metric (SCQ: Z = 1.2, p = 0.2; RMET: Z = 0.5, p = 0.6, Table 2). OXTR rs53576 genotype was associated with differences on both the SCQ (Z = −2.7, p = 0.007) and RMET (Z = −2.3; p = 0.02); the direction of deficits was congruent between metrics [OR for SCQ: 0.7 (95% CI: 0.6–0.9); OR for RMET: 0.7 (95% CI: 0.5–1.0)], yet opposite to the direction hypothesized, and detected in the ASD group.
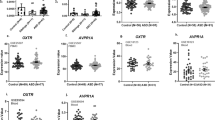
The results from our primary hypotheses are displayed in Fig. 1 (white panels); values are indicated in Table 2 for genotype risk group comparisons, and mean scores across all genotypes are shown in Supplementary Table S7. Covariates from these categorical logistic regression models in the Caucasian cohort are displayed in Supplementary Tables S8 and S9. In summary, several previous associations between OXTR SNPs and social abilities were confirmed in the group of children with ASD, but not ADHD.
Comparison of the association between OXTR genotype and social deficits between diagnostic groups in participants with Caucasian ancestry. Predicted scores at age 11. SCQ: Social Communication Questionnaire- score out of 28 social communication/interaction items only, RMET: Reading the Mind in the Eyes Test, number of incorrect items out of 28, ASD: Autism spectrum disorder, ADHD: Attention deficit hyperactivity disorder. Higher scores across both metrics suggest greater social deficits. All analyses were adjusted for age and sex differences, and those involving the RMET were adjusted for IQ as well. The high-risk/ low-risk groupings were based on the patterns observed in previous studies. P-values are uncorrected. Error bars represent 95% confidence intervals (CI). White panels indicate primary hypotheses from the literature (aim 1). Exact sample sizes detailed in Tables 1 and 2.
Aim 2
Examination across diagnostic groups
Next, we combined data across both diagnostic groups, and tested for genotype*diagnosis interactions in the Caucasian participants. After correcting for multiple comparisons (n = 8), a p-value of 0.006 was used for significance regarding the interaction term. In this context, we also tested for genotypic differences in social abilities across all SNPs in the remaining diagnostic groups (i.e. those not previously tested as part of our primary aims, including OXTR rs2254298 and rs237887 in ADHD, as well rs13316193 in ASD) (Fig. 1, grey panels, Table 2).
For rs53576, the diagnostic interaction terms were significant on both the SCQ and the RMET (SCQ: Wald χ2 = 25.0, p < 0.0001; RMET: Wald χ2 = 11.4, p = 0.0007), indicating that the ‘risk variant’ in ASD showed a significantly different effect in ADHD (as shown above, the OR of social deficits with the hypothesized risk variant was >1 on both metrics in ASD, but <1 on both metrics in ADHD, see Table 2). For OXTR rs2254298, the genotype*diagnosis interaction term was significant for the SCQ only (Wald χ2 = 31.8, p < 0.0001). In ADHD, the AA/GA-allele was associated with fewer social deficits on the SCQ [Z = −4.7; p < 0.0001, OR 0.7 (0.6–0.8)], opposite to the direction initially detected in the ASD group (OR = 1.2 in ASD). The diagnostic interaction term for OXTR rs2254298 was not significant on the RMET (Wald χ2 = 0.3, p = 0.6), where the risk allele had no significant effect in ADHD or ASD (Table 2). Likewise, there were no significant differences in social abilities on either metric for rs13316193 in ASD nor in ADHD (Table 2), resulting in non-significant SNP*diagnosis interactions. Although no significant allele effects were found for rs237887 after correcting for multiple comparisons (Table 2), the trend towards effects in opposite directions across the two diagnoses resulted in similarly borderline interaction effects across both outcomes (SCQ: Wald χ2 = 5.9, p = 0.02; RMET: Wald χ2 = 3.0, p = 0.08). In summary, across multiple OXTR variants, there was evidence that the hypothesized risk allele was associated with more significant social deficits in ASD, but fewer social deficits in ADHD.
Secondary aims
As secondary aims, we also examined (1) whether social abilities varied by arginine vasopressin receptor 1a (AVPR1a) RS3 microsatellite length, and (2) the impact of ancestry on OXTR findings. Greater length of the AVPR1a RS3 microsatellite was previously associated with greater receptor expression levels and superior social abilities in typically developing populations36,37,38, although no effect was detected in a large sample of children with ASD29. Previous studies have also found evidence for a differential direction of effects of OXTR polymorphisms in Caucasian vs. non-Caucasian individuals, particularly for rs225429831, 32.
(1) AVPR1a RS3
AVPR1a is the gene coding for the corresponding vasopressin receptor. Upstream of the exon is a microsatellite promoter region, which includes an RS3 segment of CT, TT, and GT repeats. The RS3 segment varies in length; in our sample the range was 310–346 base pairs (bp), with a median of 328. We categorized each individual’s promoter length as ‘long’ or ‘short, with the cut-point placed at 328 bp, as this was both the median in our sample, and similar to cut-points used in previous studies (e.g. 326 and 32836, 32639, or median value40,). We considered those carrying two ‘short’ alleles to be the high-risk group, and compared them to those with two ‘long’ alleles as the low-risk group. In the group of Caucasian participants with ASD, there were no differences in social abilities based on microsatellite length on the SCQ [short/short vs. long/long, Z = −0.9, p = 0.3, OR = 0.9 (95% CI: 0.8–1.1)], or the RMET [Z = 1–0.6, p = 0.1, OR = 0.8 (95% CI: 0.7–1.0)]. Similarly, in the ADHD group, there were no differences in social abilities by microsatellite length on the SCQ [Z = 0.7, p = 0.4, OR = 1.1 (0.9–1.2)] or on the RMET [Z = −0.6, p = 0.6, OR: 0.9 (0.7–1.2)].
(2) Examination of OXTR across ancestry groups
We subsequently combined data across Caucasian and non-Caucasian participants, and tested for ancestry*genotype interactions. The aim was to determine whether the direction of genotype effects was sufficiently similar between the Caucasian and non-Caucasian groups to justify performing the genotype group comparisons on the entire combined sample, irrespective of ancestry. For the ASD cohort, ancestry*genotype interaction terms were non-significant for all OXTR SNPS, on the SCQ and on the RMET (p > 0.05 for all). In the ADHD group, there were no significant ancestry*genotype interactions on the RMET (p > 0.05 for all), but significant effects were observed on the SCQ for rs2254298 (Wald χ2 = 29.1, p < 0.0001), where the opposite direction of an association between the risk allele and social deficits was detected in the non-Caucasian cohort [OR = 1.4 (95% CI: 1.1–1.7), p = 0.002] compared to the Caucasian cohort [OR = 0.7 (95% CI: 0.6–0.8) p < 0.001]. Interaction terms were also significant in ADHD for rs53576 (Wald χ2 = 7.6, p = 0.006) and rs13316193 (Wald χ2 = 5.1, p = 0.02) on the SCQ.
For SNPs where the ancestry*genotype interactions were conservatively non-significant (p > 0.05), we pooled data across ancestry groups, and calculated the combined odds ratios in the larger sample (Table 2, right side). Overall, the direction of effects was very similar in the combined and Caucasian only samples, although the larger combined sample size led to more significant associations.
Exploratory analyses
The finding of differential effects of genotype on social deficits between diagnostic groups was unexpected. To further explore and confirm this pattern, we repeated the analyses using additive genotype risk models (GG vs. GA vs. AA), as opposed to categorical (GG/GA vs. AA). Results were similar with either model (see Supplementary Table S10).
Next, given an emerging hypothesis that oxytocin may affect social behaviour by decreasing anxiety or fear responses, potentially at the level of the amygdala41, 42, and given the high rate of comorbid anxiety disorders in ASD (up to 40%)43, we post-hoc hypothesized that differential anxiety levels between the two clinical groups could potentially explain the reciprocal pattern. Anxiety was quantified through parent report on the Child Behavior Checklist anxiety subscale (CBCL-anxiety) and the Revised Children’s Anxiety and Depression Scale (RCADS). We compared anxiety scores by diagnosis, then by genotype, and subsequently included anxiety as an additional covariate in the original SNP-SCQ models. Anxiety analyses were restricted to participants who also had SCQ scores available and were of Caucasian ancestry (CBCL n = 377, RCADS n = 235), to facilitate comparison to previous findings.
On the CBCL-anxiety, the ASD group had higher mean T-scores (Table 1) and higher total anxiety raw scores compared to the ADHD group [mean raw score = 4.4 (95% CI: 3.5–4.6) vs. 2.9 (2.1–3.8), t = −4.0, p < 0.0001]. On the RCADS, we found higher social anxiety in the ASD group compared to the ADHD group [mean T-score of 58.0 (54.5–61.5) vs. 53.1 (48.6–57.6), t = −2.2, p = 0.03]. There were no differences in total anxiety on the RCADS between diagnostic groups [52.9 (49.8–56.0) vs. 51.9 (47.8–55.9), p = 0.6]. Results were unchanged after adjusting for age and sex.
For genotype group comparisons, we tested for effects of rs53576 and rs2254298 on anxiety measures in ASD and ADHD separately. No genotype effects were detected on the RCADS for either SNP (p > 0.05). For rs53576, a modest genotype effect was detected on the CBCL-anxiety raw score in the ASD group in line with the direction of effects seen with the SCQ [AA = 2.9 (95% CI: 1.8–4.0); GG/GA = 4.6 (4.2–5.1), t = −2.9, p = 0.004]. There were no significant effects of rs53576 on CBCL-anxiety in the ADHD group (difference: −0.2, p = 0.7). The SNP*diagnosis interaction term was non-signification (p = 0.09), suggesting a lack of differential effects of genotype on anxiety between the two diagnostic groups. For rs2254298, CBCL-anxiety scores were modestly affected by genotype in the ADHD group only (AA/GA vs. GG: −1.0 points, p = 0.03). The diagnostic interaction term was again non-significant (p = 0.3).
We subsequently included CBCL-anxiety score as an additional covariate in the SCQ models for rs53576 and rs2254298. Although CBCL-anxiety was significantly associated with SCQ in the overall model, results regarding the effects of OXTR on SCQ scores were otherwise unchanged [rs53576 in ASD: OR = 1.4 (1.1–1.7), rs53576 in ADHD: OR = 0.8 (0.6–0.9), rs53576*dx: p < 0.0001; rs2254298 in ASD: OR = 1.3 (1.1–1.5), rs2254298 in ADHD: OR = 0.7 (0.6–0.8), rs2254298*dx: p < 0.001]. Overall, data suggest that OXTR genotype may affect anxiety to some extent, but a reciprocal pattern between diagnostic groups as seen on social measures was not observed, and anxiety did not explain the differential effects of genotype on social outcomes by diagnosis.
Discussion
In this study, we examined how common polymorphisms in neuropeptide receptor genes affected social abilities in a large group of children and youth with ASD or ADHD. This is the first study comparing genotype effects across two neurodevelopmental disorders, with the largest ADHD cohort, and one of the largest ASD cohorts for this type of analysis to date. Other strengths of this study include: participants from two clinical groups who were recruited and characterized using the same research protocol; detailed phenotyping on both participant and caregiver completed metrics; and examination for potential ancestral differences in genotype effects. Our main findings were: 1) confirmation of several previous associations regarding neuropeptide receptor genotype and social abilities in our large group of children with ASD, 2) a lack of replication of previous associations between OXTR SNPs and social abilities seen in smaller ADHD cohorts, although several new associations were detected in this larger sample, and 3) significant diagnostic differences in the association between OXTR genotype and social abilities (i.e. a genetic modifier allele associated with more social deficits in ASD had either no effect, or conferred a benefit with respect to social abilities in ADHD).
Our findings are in line with direction of effects described in previous ASD cohorts and family studies. For example, Parker et al. examined genotypic differences in social abilities in a group of 79 children with ASD, 53 unaffected siblings, and 62 controls, of varied ethnicities6. They found that carriers of the A-allele of OXTR rs2254298 had higher levels of social impairment, and carriers of the G-allele of OXTR rs53576 performed worse on an affect recognition task6. Skuse et al. tested for associations between 60 OXTR SNPs and 31 AVPR1a SNPs and social abilities in an ASD cohort of 198 families from the United Kingdom and Finland, of mostly Caucasian ancestry29. Only OXTR rs237887 genotype affected face recognition memory, where AA carriers performed significantly worse. Length of AVPR1a RS3 microsatellite promoter region did not impact social abilities in their sample29, nor in ours. A recent meta-analysis found both the OXTR rs237887 A-allele and the rs2254298 A-allele were also risk-conferring with respect to an ASD diagnosis21. It would follow that in certain vulnerable families, specific OXTR common variants may push a small subset of those at risk over the diagnostic threshold.
Few studies have examined the impact of OXTR on social functioning in ADHD27, 28. Park et al. found that rs53576 AA allele carriers had fewer social deficits in their sample of 119 participants. However, in our ADHD sample, OXTR rs53576 GG allele carriers and rs2254298 A allele carriers had fewer social deficits, opposite to the direction of effects seen in our ASD group. For OXTR rs53576, this finding may be more in keeping with patterns observed in control populations. In a recent meta-analysis of mostly typically developing adults, rs53576 G-allele homozygotes had better social abilities on a variety of metrics22. Similarly, a previous study using the RMET found that healthy adolescents who had the OXTR rs53576 GG genotype had higher accuracy overall18. Our data suggest that the direction of impact of common variants in these receptors vary by disorder- a protective variant in one condition may increase risk in another. Overall, this would indicate that common polymorphisms in OXTR represent a specific vulnerability factor, and not a universal risk factor with respect to social functioning.
Mechanistically, it may be that these non-coding intronic SNPs co-localize with specific receptor haplotypes, other important exons, or certain rare variants44, that affect the structure or function of OXTR. Alternatively, they may be markers of functional differences in the non-coding regulatory elements of neuropeptide receptor genes, indirectly affecting receptor expression levels. SNPs in non-coding regulatory elements were recently shown to exert a robust effect on oxytocin receptor expression levels within specific brain regions in prairie voles45. Similar investigations in humans are lacking and are greatly needed. At the same time, large and rare genetic variants may be more common in ASD, and could be potentially associated with more significant disruptions on oxytocin signaling46, 47. For example, contactin-associated protein 2 (CNTNAP2) mutations have been associated with ASD and developmental delay46. A recent mouse model of ASD using CNTNAP2 knockout showed disrupted oxytocin expression and social deficits, which were corrected with exogenous oxytocin administration47. While usually we conceptualize rare variants as acting on a background of common variants, the implication of our study is genetic background may also play a critical role in modifying the impact of common ‘risk’ variants on behavioural phenotypes.
How exactly similar changes in receptor expression or function could result in disparate behavioural consequences in different diagnostic groups is unknown. Possible explanations include: 1) specific neurocircuits contributing to social deficits may vary between the disorders, 2) other diagnostic differences in environmental, hormonal, or epigenetic processes may be differentially accentuated through oxytocin signaling or 3) measurement factors or confounding.
With respect to neurocircuitry, emerging literature indicates that social and fear perception involve complex networks centred on the amygdala and reward centres42, 48, and that oxytocin may potentially enhance transmission in these circuits through its effects on interneurons41, 49. It may be that the brain circuits, regions, or underlying behavioural constructs contributing to differences in SCQ scores may vary between diagnostic groups. While our exploratory analyses involving anxiety measures suggested that OXTR genotype may also impact on anxiety levels to some extent, a reduction in anxiety alone was insufficient to explain the differential OXTR effects across diagnostic groups.
Alternatively, it is possible that other diagnostic differences in environmental, hormonal, or epigenetic processes may be differentially accentuated through oxytocin signaling. For example, several studies in typically developing cohorts have shown that the OXTR rs53576 G-allele was associated with increased environmental sensitivity, which led to more adverse outcomes under conditions of early childhood adversity or exclusion13,14,15,16,17. G-allele carriers were also shown to experience a more beneficial response to receiving positive social support as indicated by salivary cortisol levels50, and were more likely to seek social support under stress51. In this context, perhaps children with ASD are less likely to experience the stress buffering effects of social contact given their underlying social deficits, putting the more ‘sensitive’ allele group at greater risk. At the same time, there is evidence to suggest that oxytocin may play a role in suppressing the cortisol stress response52. Given evidence of divergent cortisol patterns in ASD vs. ADHD (i.e. enhanced cortisol responses to stress in ASD53, and attenuated cortisol profiles in ADHD54, 55), it would follow that an OXTR variant leading to increased cortisol activity would be disadvantageous in ASD, but potentially advantageous in ADHD. Differential epigenetic regulation of OXTR gene expression between the two disorders could also potentially explain diagnostic differences in SNP effects (for a review see ref. 56). While there is evidence to suggest that increased methylation of certain OXTR sites may be associated with increased social deficits and autistic traits in ASD57, 58, and callous/ unemotional traits in ADHD59, 60, to our knowledge, epigenetic profiles of OXTR have not been compared between the two disorders.
Measurement factors and confounding could also potentially contribute to observed diagnostics differences. While testing protocols were identical across diagnostic groups, it is possible that our social measures may differentially capture certain aspects of sociality in ASD vs. ADHD (e.g. attachment security, social reward, or other more complex social tasks). While common confounding factors were adjusted for in our analyses, the role of clinical severity, adaptive functioning, and parental stress or psychopathology were not explored, and could potentially affect response patterns on either social metric. The impact of different traits and measurement factors on SCQ scores in ASD and ADHD in particular merits further exploration.
In terms of other limitations, despite our sample size, we may have been underpowered to detect more subtle differences, including interactions with ancestry or analyses on the smaller group with RMET and IQ scores. Lack of statistical significance would therefore not necessarily rule out a possible biological effect of genotype on social behaviour for these measures. Analyses including a comorbid ASD/ADHD group, a control group, or a group of unaffected family members, would have proven interesting comparisons but were not feasible with the existing dataset. While results were statistically significant, differences between genotype groups were generally small and not meant to imply clinical utility. Human molecular/ expression data is greatly needed to clarify the mechanisms through which these variants may influence receptor function and downstream behaviours.
In summary, our findings indicate that common polymorphisms in neuropeptide receptor genes act as specific (as opposed to general) modifier alleles regarding social phenotypes in children with neurodevelopmental disorders, with effects that vary by diagnosis. Our findings therefore contribute to the often-conflicting literature regarding these SNPs. We are the first to implicate potential diagnostic differences as one explanation for heterogeneity in findings. Future studies comparing OXTR genotype effects across these disorders ought to examine the role of the hypothalamic pituitary axis social neurocircuitry, environmental social stress, and rare genetic, or epigenetic differences as possible mechanisms. Molecular studies investigating the functional impacts of these SNPs in humans are also greatly needed.
Methods
Participants and study protocol
Participants were recruited via the Province of Ontario Neurodevelopmental Disorders (POND) Network, a coordinated multi-centre research initiative examining the neurobiology of neurodevelopmental disorders. Hospital Research Ethics Boards approved the study protocol at each participating institution (Holland Bloorview Kids Rehabilitation Hospital, Toronto; The Hospital for Sick Children, Toronto; McMaster Children’s Hospital, Hamilton; and Lawson Health Research Institute, London); all experiments were performed in accordance with relevant guidelines and regulations. Informed consent was obtained from parents of all participants; further details of the protocol are described elsewhere25. All participants (n = 614) were between 4 and 21 years of age, and had a clinical diagnosis of ASD or ADHD (see Table 1). Clinical diagnoses were confirmed via in-depth diagnostic assessments using established measures. Ancestry was determined by parent/caregiver self-report on the ethnic origins of the participants’ biological four grandparents. Where all four grandparents were identified as Caucasian (including Scandinavian, Ashkenazi Jewish, French Canadian, Mediterranean, Eastern European, Russian, Western European), the participant was classified as ‘Caucasian.’ Participants with one or more grandparents of non-Caucasian ancestry were classified as ‘non-Caucasian.’
Quantitative behavioural phenotype
The Social Communication Questionnaire (SCQ) is a validated measure completed by a parent/caregiver to screen for ASD35, 61, developed based on the items and factors in the Autism Diagnostic Interview-Revised62. Higher SCQ scores (out of 39) indicate a greater risk, with a cut-off of > 15 proposed to distinguish between those with and without ASD35. For context, typically developing children usually score under 10 on the SCQ, while those with ASD have mean scores closer to 20 and above63, 64. Previous research suggests that mean scores on the SCQ are comparable in children with ASD across IQ strata, and it is valid for use in children with a mental age above 2.061. For this analysis, we examined the total prorated scores on the 28 social interaction and communication items only, excluding the items pertaining to restricted/ repetitive behaviours. We did not adjust for IQ for this measure (see statistical analyses). The Reading the Mind in the Eyes Test (RMET) is a standardized test of social perception abilities65. Higher scores on the RMET are indicative of more accurate social perception. The child version of the RMET was used in this study, containing a total of 28 items34. Typically developing children tend to score between 15–20 out of 28 on this measure, while children with ASD tend to score 2–4 points lower than control subjects25, 34. RMET scores are known to vary with IQ25, therefore IQ was included as a covariate for these analyses. To more easily facilitate comparison of genotype effects across both measures, we report RMET scores with respect to the number of incorrect items. Therefore, throughout the manuscript, higher scores on either metric suggest greater social deficits. Other behavioural traits were quantified using the Child Behavior Checklist (CBCL) subscales66,67,68, and the Revised Children Anxiety and Depression Scale (RCADS)69.
Selection of genetic markers
Four SNPs in OXTR from the third intronic region (rs53576, rs2254298, rs237887, rs13316193) were selected for genotyping based on existing literature at the time of study design suggesting an association between these markers and social abilities in participants with ASD or ADHD (Table S1). These SNPs, either alone or contributing to a haplotype, have also been associated with ASD risk21, 31, 70,71,72. Analysis for linkage disequilibrium (LD) was completed using Ldlink33, and the 1000 Genomes Project database of Caucasian samples (see Supplementary Table S3). Although rs13316193 and rs237887 showed moderate linkage, all four SNPs were sequenced to facilitate replication of previous findings within specific diagnostic groups. AVPR1a RS3 microsatellite length was sequenced as well, given initial findings suggesting an association between social abilities and genotype as well as receptor expression levels in typically developing subjects36,37,38, despite no association with social abilities in ASD in one study29.
Genotyping
SNPs were analyzed on the MassARRAY Analyzer 4 system using iPLEX Gold chemistry (both Agena Biosciences, San Diego, CA, USA) using the primers in Supplementary Table S11 and the recommended manufacturer’s protocol. Genotypes were called using Typer 4.0 (Agena Biosciences). The AVPR1A RS3 polymorphism primers and methods are also listed in Table S11. Primers were adapted from that used by Tansey et al. to match the Human Feb. 2009 (GRCh37/hg19) assembly and to elevate the Tm but otherwise maintain the amplified position73. Results were analyzed using the software GeneMapper v. 3.7 (Life Technologies).
Statistical analyses
All analyses were two-tailed, with the alpha set at 0.05, performed in SAS 9.3 (2002–2010, SAS Institute Cary, NC, USA). We used a logistic regression analysis to examine the proportion of items selected as ‘yes’ (for the 28 SCQ social communication items) or incorrect (for the 28 RMET items), using the PROC LOGISTIC function in SAS, which allows us to specify both the total score and the number of trials that this score is based on in the model outcome. This approach permitted adjustments for total number of items (e.g. decreasing the denominator for participants who answered >75% of items but did not complete the entire metric, n = 3 total). We adjusted for the contribution of the following predictor variables: genotype group, age, sex, ancestry (and IQ for analyses involving the RMET)25. Logistic regression was chosen for the main analyses given that scores on both metrics are sums of binary variables and therefore, follow a binomial distribution. Initial hypotheses were examined in the ASD group (for OXTR rs53576, rs2254298, and rs237887) and ADHD group (for rs53576, and rs1331619), including Caucasian participants only. We selected one specific categorical genotype risk pattern per SNP (i.e. dominant, recessive, or comparison of homozygous groups) for statistical testing based on the positive findings described in previous papers (Table S1). We subsequently combined data for both the ASD and ADHD groups (Caucasian only) and assessed the SNP*diagnosis interaction term to determine whether the magnitude of the genotype effect on social abilities varied by diagnosis. We present the odds ratios (ORs) of increasing SCQ/RMET scores for the hypothesized ‘high-risk’ vs. ‘low-risk’ genotype groups, based on the literature. That is, the odds of obtaining a positive/incorrect score on a single SCQ/RMET item for a participant in the ‘high-risk’ group divided by the odds of obtaining a positive/incorrect score for a participant in the ‘low-risk’ group. Thus, an odds ratio of 1 represents no risk group differences, while odds greater than 1 suggest increased risk. For presentation purposes, we also provide the predicted SCQ/ RMET scores by genotype group (by multiplying the predicted proportion of items selected or incorrect by the total number of items) adjusted to age 11.0 years and mean IQ scores. Details on covariates are provided in Supplementary Tables S8 and S9. Unadjusted p-values are reported; a Bonferroni correction for multiple comparisons is described alongside results. Finally, we combined data across ancestry groups and tested for genotype* ancestry interactions. Where non-significant (suggesting the effect of neuropeptide receptor genotype on social abilities does not vary by ethnicity), we pooled data across groups, added ancestry as a covariate (along with genotype, age, sex +/− IQ) and calculated the effect size (OR) in the larger varied ancestry group. For exploratory analyses involving anxiety, we compared genotype groups using ANOVAs; we adjusted for age and sex using ANCOVAs.
Data availability
The datasets generated and analyzed during the current study are publicly available through Brain-CODE. http://www.braininstitute.ca/braincode/.
References
Ebstein, R. P., Israel, S., Chew, S. H., Zhong, S. & Knafo, A. Genetics of human social behavior. Neuron 65, 831–844, doi:10.1016/j.neuron.2010.02.020 (2010).
Scourfield, J., Martin, N., Lewis, G. & McGuffin, P. Heritability of social cognitive skills in children and adolescents. Br J Psychiatry 175, 559–564 (1999).
Modi, M. E. & Young, L. J. The oxytocin system in drug discovery for autism: animal models and novel therapeutic strategies. Horm Behav 61, 340–350, doi:10.1016/j.yhbeh.2011.12.010 S0018-506X(11)00287-X [pii] (2012).
Donaldson, Z. R. & Young, L. J. Oxytocin, vasopressin, and the neurogenetics of sociality. Science 322, 900–904, doi:10.1126/science.1158668 (2008).
Shamay-Tsoory, S. G. & Abu-Akel, A. The Social Salience Hypothesis of Oxytocin. Biological psychiatry 79, 194–202, doi:10.1016/j.biopsych.2015.07.020 (2016).
Parker, K. J. et al. Plasma oxytocin concentrations and OXTR polymorphisms predict social impairments in children with and without autism spectrum disorder. Proceedings of the National Academy of Sciences of the United States of America 111, 12258-12263, doi:10.1073/pnas.1402236111 1402236111 [pii] (2014).
Domes, G., Steiner, A., Porges, S. W. & Heinrichs, M. Oxytocin differentially modulates eye gaze to naturalistic social signals of happiness and anger. Psychoneuroendocrinology, doi:10.1016/j.psyneuen.2012.10.002 (2012).
Anagnostou, E. et al. Intranasal oxytocin versus placebo in the treatment of adults with autism spectrum disorders: a randomized controlled trial. Molecular autism 3, 16, doi:10.1186/2040-2392-3-16 2040-2392-3-16 [pii] (2012).
Wu, N., Li, Z. & Su, Y. The association between oxytocin receptor gene polymorphism (OXTR) and trait empathy. J Affect Disord 138, 468-472, doi:10.1016/j.jad.2012.01.009 S0165-0327(12)00016-X [pii] (2012).
Uzefovsky, F. et al. Oxytocin receptor and vasopressin receptor 1a genes are respectively associated with emotional and cognitive empathy. Horm Behav 67, 60-65, doi:10.1016/j.yhbeh.2014.11.007 S0018-506X(14)00222-0 [pii] (2015).
Smith, K. E., Porges, E. C., Norman, G. J., Connelly, J. J. & Decety, J. Oxytocin receptor gene variation predicts empathic concern and autonomic arousal while perceiving harm to others. Soc Neurosci 9, 1–9, doi:10.1080/17470919.2013.863223 (2014).
Tost, H. et al. A common allele in the oxytocin receptor gene (OXTR) impacts prosocial temperament and human hypothalamic-limbic structure and function. Proceedings of the National Academy of Sciences of the United States of America 107, 13936–13941, doi:10.1073/pnas.1003296107 (2010).
Schneider-Hassloff, H. et al. Oxytocin receptor polymorphism and childhood social experiences shape adult personality, brain structure and neural correlates of mentalizing. NeuroImage 134, 671–684, doi:10.1016/j.neuroimage.2016.04.009 (2016).
McDonald, N. M., Baker, J. K. & Messinger, D. S. Oxytocin and parent-child interaction in the development of empathy among children at risk for autism. Developmental psychology 52, 735–745, doi:10.1037/dev0000104 (2016).
Dannlowski, U. et al. Disadvantage of Social Sensitivity: Interaction of Oxytocin Receptor Genotype and Child Maltreatment on Brain Structure. Biological psychiatry, doi:10.1016/j.biopsych.2015.12.010 (2015).
Bradley, B. et al. Association between childhood maltreatment and adult emotional dysregulation in a low-income, urban, African American sample: moderation by oxytocin receptor gene. Development and psychopathology 23, 439–452, doi:10.1017/S0954579411000162 (2011).
McQuaid, R. J., McInnis, O. A., Stead, J. D., Matheson, K. & Anisman, H. A paradoxical association of an oxytocin receptor gene polymorphism: early-life adversity and vulnerability to depression. Frontiers in neuroscience 7, 128, doi:10.3389/fnins.2013.00128 (2013).
Rodrigues, S. M., Saslow, L. R., Garcia, N., John, O. P. & Keltner, D. Oxytocin receptor genetic variation relates to empathy and stress reactivity in humans. Proceedings of the National Academy of Sciences of the United States of America 106, 21437–21441, doi:10.1073/pnas.0909579106 0909579106 [pii] (2009).
Kanthak, M. K. et al. Oxytocin receptor gene polymorphism modulates the effects of social support on heart rate variability. Biol Psychol 117, 43–49, doi:10.1016/j.biopsycho.2016.02.007 (2016).
Ylisaukko-oja, T. et al. Search for autism loci by combined analysis of Autism Genetic Resource Exchange and Finnish families. Annals of neurology 59, 145–155, doi:10.1002/ana.20722 (2006).
LoParo, D. & Waldman, I. D. The oxytocin receptor gene (OXTR) is associated with autism spectrum disorder: a meta-analysis. Molecular psychiatry, doi:10.1038/mp.2014.77 mp201477 [pii] (2014).
Li, J. et al. Association of Oxytocin Receptor Gene (OXTR) rs53576 Polymorphism with Sociality: A Meta-Analysis. PLoS One 10, e0131820, doi:10.1371/journal.pone.0131820 (2015).
van der Meer, J. M. et al. Are autism spectrum disorder and attention-deficit/hyperactivity disorder different manifestations of one overarching disorder? Cognitive and symptom evidence from a clinical and population-based sample. Journal of the American Academy of Child and Adolescent Psychiatry 51, 1160–1172 e1163, doi:10.1016/j.jaac.2012.08.024 S0890-8567(12)00649-1 [pii] (2012).
Buhler, E., Bachmann, C., Goyert, H., Heinzel-Gutenbrunner, M. & Kamp-Becker, I. Differential diagnosis of autism spectrum disorder and attention deficit hyperactivity disorder by means of inhibitory control and ‘theory of mind’. Journal of autism and developmental disorders 41, 1718–1726, doi:10.1007/s10803-011-1205-1 (2011).
Baribeau, D. A. et al. Examining and comparing social perception abilities across childhood-onset neurodevelopmental disorders. Journal of the American Academy of Child and Adolescent Psychiatry 54, 479–486 e471, doi:10.1016/j.jaac.2015.03.016 (2015).
Klei, L. et al. Common genetic variants, acting additively, are a major source of risk for autism. Molecular autism 3, 9, doi:10.1186/2040-2392-3-9 (2012).
Park, J. et al. Evidence that genetic variation in the oxytocin receptor (OXTR) gene influences social cognition in ADHD. Prog Neuropsychopharmacol Biol Psychiatry 34, 697–702, doi:10.1016/j.pnpbp.2010.03.029 S0278-5846(10)00122-3 [pii] (2010).
Ayaz, A. B. et al. Oxytocin system social function impacts in children with attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet 168, 609–616, doi:10.1002/ajmg.b.32343 (2015).
Skuse, D. H. et al. Common polymorphism in the oxytocin receptor gene (OXTR) is associated with human social recognition skills. Proceedings of the National Academy of Sciences of the United States of America 111, 1987-1992, doi:10.1073/pnas.1302985111 1302985111 [pii] (2014).
Insel, T. et al. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. The American journal of psychiatry 167, 748–751, doi:10.1176/appi.ajp.2010.09091379 167/7/748 [pii] (2010).
Jacob, S. et al. Association of the oxytocin receptor gene (OXTR) in Caucasian children and adolescents with autism. Neurosci Lett 417, 6–9, doi:10.1016/j.neulet.2007.02.001 (2007).
Chen, F. S., Barth, M. E., Johnson, S. L., Gotlib, I. H. & Johnson, S. C. Oxytocin Receptor (OXTR) Polymorphisms and Attachment in Human Infants. Frontiers in psychology 2, 200, doi:10.3389/fpsyg.2011.00200 (2011).
Machiela, M. J. & Chanock, S. J. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557, doi:10.1093/bioinformatics/btv402 (2015).
Baron-Cohen, S., Wheelwright, S., Spong, A., Scahill, V. & Lawson, J. Are intuitive physics and intuitive psychology independent? A test with children with Asperger Syndrome. Journal of Developmental and Learning Disorders 5, 47–78 (2001).
Berument, S. K., Rutter, M., Lord, C., Pickles, A. & Bailey, A. Autism screening questionnaire: diagnostic validity. Br J Psychiatry 175, 444–451 (1999).
Knafo, A. et al. Individual differences in allocation of funds in the dictator game associated with length of the arginine vasopressin 1a receptor RS3 promoter region and correlation between RS3 length and hippocampal mRNA. Genes Brain Behav 7, 266–275, doi:10.1111/j.1601-183X.2007.00341.x (2008).
Avinun, R. et al. AVPR1A variant associated with preschoolers’ lower altruistic behavior. PLoS One 6, e25274, doi:10.1371/journal.pone.0025274 PONE-D-11-12719 [pii] (2011).
Avinun, R., Ebstein, R. P. & Knafo, A. Human maternal behaviour is associated with arginine vasopressin receptor 1A gene. Biol Lett 8, 894–896, doi:10.1098/rsbl.2012.0492 rsbl.2012.0492 [pii] (2012).
Levin, R. et al. Association between arginine vasopressin 1a receptor (AVPR1a) promoter region polymorphisms and prepulse inhibition. Psychoneuroendocrinology 34, 901–908, doi:10.1016/j.psyneuen.2008.12.014 (2009).
Ophir, A. G., Campbell, P., Hanna, K. & Phelps, S. M. Field tests of cis-regulatory variation at the prairie vole avpr1a locus: association with V1aR abundance but not sexual or social fidelity. Horm Behav 54, 694–702, doi:10.1016/j.yhbeh.2008.07.009 (2008).
Knobloch, H. S. et al. Evoked axonal oxytocin release in the central amygdala attenuates fear response. Neuron 73, 553–566, doi:10.1016/j.neuron.2011.11.030 (2012).
Janak, P. H. & Tye, K. M. From circuits to behaviour in the amygdala. Nature 517, 284–292, doi:10.1038/nature14188 (2015).
van Steensel, F. J., Bogels, S. M. & Perrin, S. Anxiety disorders in children and adolescents with autistic spectrum disorders: a meta-analysis. Clin Child Fam Psychol Rev 14, 302–317, doi:10.1007/s10567-011-0097-0 (2011).
Liu, X. et al. Novel rare variations of the oxytocin receptor (OXTR) gene in autism spectrum disorder individuals. Hum Genome Var 2, 15024, doi:10.1038/hgv.2015.24 (2015).
King, L. B., Walum, H., Inoue, K., Eyrich, N. W. & Young, L. J. Variation in the Oxytocin Receptor Gene Predicts Brain Region-Specific Expression and Social Attachment. Biological psychiatry, doi:10.1016/j.biopsych.2015.12.008 (2015).
Alarcon, M. et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. American journal of human genetics 82, 150–159, doi:10.1016/j.ajhg.2007.09.005 (2008).
Penagarikano, O. et al. Exogenous and evoked oxytocin restores social behavior in the Cntnap2 mouse model of autism. Science translational medicine 7, 271ra278, doi:10.1126/scitranslmed.3010257 (2015).
Twining, R. C., Vantrease, J. E., Love, S., Padival, M. & Rosenkranz, J. A. An intra-amygdala circuit specifically regulates social fear learning. Nat Neurosci. doi:10.1038/nn.4481 (2017).
Owen, S. F. et al. Oxytocin enhances hippocampal spike transmission by modulating fast-spiking interneurons. Nature 500, 458–462, doi:10.1038/nature12330 (2013).
Chen, F. S. et al. Common oxytocin receptor gene (OXTR) polymorphism and social support interact to reduce stress in humans. Proceedings of the National Academy of Sciences of the United States of America 108, 19937–19942, doi:10.1073/pnas.1113079108 (2011).
Kim, H. S. et al. Culture, distress, and oxytocin receptor polymorphism (OXTR) interact to influence emotional support seeking. Proceedings of the National Academy of Sciences of the United States of America 107, 15717–15721, doi:10.1073/pnas.1010830107 (2010).
Heinrichs, M., Baumgartner, T., Kirschbaum, C. & Ehlert, U. Social support and oxytocin interact to suppress cortisol and subjective responses to psychosocial stress. Biological psychiatry 54, 1389–1398 (2003).
Spratt, E. G. et al. Enhanced cortisol response to stress in children in autism. Journal of autism and developmental disorders 42, 75–81, doi:10.1007/s10803-011-1214-0 (2012).
Isaksson, J., Nilsson, K. W., Nyberg, F., Hogmark, A. & Lindblad, F. Cortisol levels in children with attention-deficit/hyperactivity disorder. J Psychiatr Res 46, 1398–1405, doi:10.1016/j.jpsychires.2012.08.021 (2012).
Blomqvist, M. et al. Salivary cortisol levels and dental anxiety in children with attention deficit hyperactivity disorder. Eur J Oral Sci 115, 1–6, doi:10.1111/j.1600-0722.2007.00423.x (2007).
Grafodatskaya, D., Chung, B., Szatmari, P. & Weksberg, R. Autism spectrum disorders and epigenetics. Journal of the American Academy of Child and Adolescent Psychiatry 49, 794–809, doi:10.1016/j.jaac.2010.05.005 (2010).
Rijlaarsdam, J. et al. Prenatal stress exposure, oxytocin receptor gene (OXTR) methylation and child autistic traits: The moderating role of OXTR rs53576 genotype. Autism research: official journal of the International Society for Autism Research, doi:10.1002/aur.1681 (2016).
Gregory, S. G. et al. Genomic and epigenetic evidence for oxytocin receptor deficiency in autism. BMC Med 7, 62, doi:1741-7015-7-62 [pii] 10.1186/1741-7015-7-62 (2009).
Cecil, C. A. et al. Environmental risk, Oxytocin Receptor Gene (OXTR) methylation and youth callous-unemotional traits: a 13-year longitudinal study. Molecular psychiatry 19, 1071–1077, doi:10.1038/mp.2014.95 (2014).
Dadds, M. R. et al. Methylation of the oxytocin receptor gene and oxytocin blood levels in the development of psychopathy. Development and psychopathology 26, 33–40, doi:10.1017/S0954579413000497 (2014).
Rutter, M., Bailey, A. & Lord, C. The Social Communication Questionnaire. 32 (Western Psychological Services, United States of America, 2003).
Lord, C., Rutter, M. & Le Couteur, A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. Journal of autism and developmental disorders 24, 659–685 (1994).
Chandler, S. et al. Validation of the social communication questionnaire in a population cohort of children with autism spectrum disorders. Journal of the American Academy of Child and Adolescent Psychiatry 46, 1324–1332, doi:10.1097/chi.0b013e31812f7d8d (2007).
Mulligan, A., Richardson, T., Anney, R. J. & Gill, M. The Social Communication Questionnaire in a sample of the general population of school-going children. Ir J Med Sci 178, 193–199, doi:10.1007/s11845-008-0184-5 (2009).
Baron-Cohen, S., Wheelwright, S., Hill, J., Raste, Y. & Plumb, I. The “Reading the Mind in the Eyes” Test revised version: a study with normal adults, and adults with Asperger syndrome or high-functioning autism. Journal of child psychology and psychiatry, and allied disciplines 42, 241–251 (2001).
Achenbach, T. M., Vermont, V. D. o. P. U. o. & Edelbrock, C. S. Manual for the child behavior checklist and revised child behavior profile. (Department of Psychiatry of the University of Vermont, 1983).
Achenbach, T. M. Integrative guide for the 1991 CBCL/4-18, YSR, and TRF profiles. (Department of Psychiatry, University of Vermont, 1991).
Ferdinand, R. F. Validity of the CBCL/YSR DSM-IV scales Anxiety Problems and Affective Problems. J Anxiety Disord 22, 126–134, doi:10.1016/j.janxdis.2007.01.008 (2008).
Chorpita, B. F., Yim, L., Moffitt, C., Umemoto, L. A. & Francis, S. E. Assessment of symptoms of DSM-IV anxiety and depression in children: a revised child anxiety and depression scale. Behaviour research and therapy 38, 835–855 (2000).
Wu, S. et al. Positive association of the oxytocin receptor gene (OXTR) with autism in the Chinese Han population. Biological psychiatry 58, 74-77, doi:10.1016/j.biopsych.2005.03.013 (2005).
Lerer, E. et al. Association between the oxytocin receptor (OXTR) gene and autism: relationship to Vineland Adaptive Behavior Scales and cognition. Molecular psychiatry 13, 980–988, doi:10.1038/sj.mp.4002087 (2008).
Di Napoli, A., Warrier, V., Baron-Cohen, S. & Chakrabarti, B. Genetic variation in the oxytocin receptor (OXTR) gene is associated with Asperger Syndrome. Molecular autism 5, 48, doi:10.1186/2040-2392-5-48 139 [pii] (2014).
Tansey, K. E. et al. Functionality of promoter microsatellites of arginine vasopressin receptor 1A (AVPR1A): implications for autism. Molecular autism 2, 3, doi:10.1186/2040-2392-2-3 2040-2392-2-3 [pii] (2011).
Acknowledgements
This research was supported by the grant IDS-I l-02 from the Ontario Brain Institute. The authors thank the following individuals for research support and data collection: Tara Goodale, M.Sc., Reva Schachter, M.Sc., Mithula Sriskandarajah, B.Sc., Marlena Colasanto, M.Sc., Jennifer Gomez, M.A., and Laura Park, M.Sc, from The Hospital for Sick Children; Susan Day Fragiadakis, M.A., Naomi Peleg, M.Sc., and Leanne Ristic, B.A., from Holland Bloorview; Richa Mehta, B.A., Christina Sommerdyk, M.Sc., from the Lawson Health Research Institute; Carolyn Russell, B.Sc., Alessia Greco, M.A., Mike Chalupka, B.A., B.Sc., Christina Chrysler, B.A., Irene O’Connor, M.Ed. Psych., from McMaster Children’s Hospital. Weili (Liz) Li, PhD, and the statistical analysis facility at The Centre for Applied Genomics and the Hospital for Sick Children assisted with planning analyses. Dr. Baribeau was supported in part by the American Psychiatric Association Resident Psychiatric Research Scholars Program, the American Academy of Child and Adolescent Psychiatry and the University of Toronto Postgraduate Research Awards.
Author information
Authors and Affiliations
Contributions
E.A. and D.B. conceived of the study. D.B. contributed to the analyses, led the synthesis and interpretation of results, and wrote the manuscript. A.D. performed statistical analyses, aided in interpretation, and contributed to writing and editing of the manuscript. T.P. and S.S. conducted genetic analyses, and contributed to writing and interpretation of findings. R.S., P.A., P.Z., R.N., S.G., J.C., J.B., and J.L. helped with recruitment participants, interpretation of results, and contributed to editing of the manuscript. A.I. coordinated data, and aided in writing and interpretation of results.
Corresponding author
Ethics declarations
Competing Interests
Dr. Schachar has consulted to Highland Therapeutics, Eli Lilly and Co., and Purdue Pharma. He has commercial interest in a cognitive rehabilitation software company, ‘eHave.’ Dr. Arnold holds a patent for ‘SLCIAI Marker for Anxiety Disorder’ (granted May 6, 2008). Dr. Szatmari has received royalties from Guilford Press. Dr. Anagnostou has served as a consultant to Roche. She has received grant funding from SanofiCanada and SynpapDx. She holds a provisional patent for the device, ‘Anxiety Meter’. She has received royalties from APPI and Springer. Dr. Scherer is an academic scientific consultant advising or is in licensing arrangements with Athena, Diagnostics, King Abdulaziz University Lineagen, Deep Genomics and Population Diagnostics. He also holds the CIHR-GlaxoSmithKline Endowed Chair in Genome Sciences at the Hospital for Sick Children and University of Toronto. Drs Baribeau, Iaboni, Dupuis, Crosbie, McGinn, Brian, Nicolson, Georgiades, Paton and Lerch report no biomedical financial interests or potential conflicts of interest.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Baribeau, D.A., Dupuis, A., Paton, T.A. et al. Oxytocin Receptor Polymorphisms are Differentially Associated with Social Abilities across Neurodevelopmental Disorders. Sci Rep 7, 11618 (2017). https://doi.org/10.1038/s41598-017-10821-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-10821-0
This article is cited by
-
Association of verbal and non-verbal theory of mind abilities with non-coding variants of OXTR in youth with autism spectrum disorder and typically developing individuals: a case-control study
BMC Psychiatry (2024)
-
Patterns of neural activity in response to threatening faces are predictive of autistic traits: modulatory effects of oxytocin receptor genotype
Translational Psychiatry (2024)
-
Genetic Variations in Elements of the Oxytocinergic Pathway are Associated with Attention/Hyperactivity Problems and Anxiety Problems in Childhood
Child Psychiatry & Human Development (2024)
-
Oxytocin Receptor Expression in Hair Follicle Stem Cells: A Promising Model for Biological and Therapeutic Discovery in Neuropsychiatric Disorders
Stem Cell Reviews and Reports (2023)
-
Signalling pathways in autism spectrum disorder: mechanisms and therapeutic implications
Signal Transduction and Targeted Therapy (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.