Abstract
We asked if twin birth influences the DNA methylation of subsequent siblings. We measured whole blood methylation using the HumanMethylation450 array for siblings from two twin and family studies in Australia and Korea. We compared the means and correlations in methylation between pairs of siblings born before a twin birth (BT siblings), born on either side of a twin birth (B/AT pairs) and born after a twin birth (AT siblings). For the genome-wide average DNA methylation, the correlation for AT pairs (rAT) was larger than the correlation for BT pairs (rBT) in both studies, and from the meta-analysis, rAT = 0.46 (95% CI: 0.26, 0.63) and rBT = −0.003 (95% CI: −0.30, 0.29) (P = 0.02). B/AT pairs were not correlated (from the meta-analysis rBAT = 0.08; 95% CI: −0.31, 0.45). Similar results were found for the average methylation of several genomic regions, e.g., CpG shelf and gene body. BT and AT pairs were differentially correlated in methylation for 15 probes (all P < 10−7), and the top 152 differentially correlated probes (at P < 10−4) were enriched in cell signalling and breast cancer regulation pathways. Our observations are consistent with a twin birth changing the intrauterine environment such that siblings both born after a twin birth are correlated in DNA methylation.
Similar content being viewed by others
Introduction
DNA methylation mainly occurs at cytosine-guanine dinucleotides (CpG) sites through the conversion of cytosine to 5-methylcytosine. This is one of several epigenetic modifications known to be involved in transcription regulation without changing the DNA sequence. DNA methylation has been proposed to play a critical role in the etiology of complex traits and diseases1.
Enlarged by pregnancy and then undergoing involution, the uterus potentially acquires specific changes as a consequence of previous births. Multiple pregnancy, an atypical pregnancy, causes even greater changes to the intrauterine environment as the uterus is enlarged far more than with a singleton pregnancy. For example, using the fundal height of women with normal singleton pregnancy as a measure of uterine distension, 99% women with twin pregnancy had fundal height > 2 standard deviations larger than the average of women with singleton pregnancy at the same gestation week2. Multiple pregnancy is also known to change maternal physiology3, 4 and increase the risks of several maternal complications4, 5, such as gestational diabetes and preeclampsia. In fact, many studies have found evidence that prenatal factors or intrauterine environment play a key role in shaping the DNA methylation of offspring6,7,8,9,10,11, with further evidence that some of these effects tend to be long-lasting and persist into later life10,11,12,13. In light of this we asked whether twin birth can alter the intrauterine environment in such a manner that influences the methylation status of subsequent siblings such that persons born after a twin birth have different methylation features compared with those born before a twin birth.
To answer this question, we studied the different methylation measures, including the global methylation, the average methylation of genomic regions and methylation at individual site, of middle-aged siblings from two twin and family studies: the Australian Mammographic Density Twins and Sisters Study (AMDTSS) and the Korean Healthy Twin Study (KHTS). We compared the means of different methylation measures between siblings born before a twin birth (denoted as BT siblings) and siblings born after a twin birth (denoted as AT siblings). We also compared the correlations across three types of sibling pairs: BT pairs, AT pairs and pairs born either side of a twin birth (B/AT pairs).
Results
Characteristics of included subjects
Table 1 shows the characteristics of included subjects from the AMDTSS and KHTS. The mean age (standard deviation) of subjects was 56.6 (8.0) and 44.9 (5.6) years, respectively. BT siblings were on average 7 years older than AT siblings in the AMTDSS (P < 0.01). There was no evidence of the BT and AT siblings differed in any other characteristics.
Discovery analysis
For the AMDTSS, from the comparison of means there was no evidence of a difference in GWAM, or in the average methylation of any genomic region, between BT and AT siblings (all P > 0.1; Table 2). From the probe-by-probe analysis, no probe differed between BT and AT siblings at the genome-wide level of significance (P = 10−7).
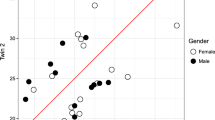
Neither BT nor B/AT pairs were correlated in GWAM (rBT = −0.01, 95% CI: −0.34, 0.31; rBAT = 0.14; 95% CI: −0.31, 0.53) (Table 3; Fig. 1). After combining rBT and rBAT, the estimate was 0.04 (95% CI: −0.24, 0.32). The correlation for AT pairs, however, was 0.48 (95% CI: 0.23, 0.67), larger than the correlation for BT pairs (P = 0.06). We did not find evidence that these correlations depended on the twins’ zygosity (P = 0.18), or any of the examined factors (all P > 0.1).
Correlations in GWAM for different types of sibling pairs in the AMDTSS and KHTS (a) Pairs of BT siblings in the AMDTSS. (b) Pairs of B/AT siblings in the AMDTSS. (c) Pairs of AT siblings in the AMDTSS. (d) Pairs of BT siblings in the KHTS. (e) Pairs of B/AT siblings in the KHTS. (f) Pairs of AT siblings in the KHTS.
For the average methylation of genomic regions, BT pairs were correlated in TSS200 (rBT = 0.36; 95% CI: 0.04, 0.61) while AT pairs were not correlated (rAT = 0.17; 95% CI: −0.19, 0.49); however, there was no difference between the two correlations (P = 0.45; Table 3). BT pairs were not correlated in any other regions (all P > 0.19). AT pairs were correlated in CpG shelf, non-CGI region, gene body, intergenic region and 3′UTR (all estimates > 0.5; all P < 6.0 × 10−6). For these five regions, the correlations were also different between BT and AT pairs (all P < 0.04). The number of statistically significant (at P < 0.05) comparisons, 5 out of 12, was more than the 0.6 that would have been expected by chance (P = 1.8 × 10−4). B/AT pairs were not correlated in any region (all P > 0.12).
From the probe analyses, for 306 the algorithm failed to converge, leaving results for 468,100 probes. There were more probes with positive correlation estimates for AT pairs compared with BT pairs (45% vs 40%, P < 10−15). The mean correlation estimate was −0.03 for BT pairs, smaller than −0.01 for AT pairs (P < 10−15). From comparing the correlations, 15 probes were identified to be differentially correlated between BT and AT pairs at the genome-wide level of significance (P = 10−7) (Table 4). Among them, two probes were more correlated for BT pairs and 13 probes were more correlated for AT pairs. This inconsistency in direction was more extreme than a 50:50 split expected under the null hypothesis that the difference is random (P < 0.01).
Replication analysis
For the KHTS, neither BT nor B/AT pairs were correlated in GWAM (rBT = 0.04, 95% CI: −0.58, 0.64; rBAT = −0.14; 95% CI: −0.77, 0.64) (Table 3; Fig. 1). The correlation was 0.43 (95% CI: 0.05, 0.70) for AT pairs, numerically larger than rBT (P = 0.35). We did not find evidence of sex heterogeneity in the correlations (P = 0.28).
For the average methylation of genomic regions, we consistently found that the AT pairs were more correlated than BT pairs between the discovery and replication analyses (P = 0.04; Table 3). BT pairs were not correlated in any region (all P > 0.16). AT pairs were correlated in all regions (all P < 0.05) except CpG island. The correlations for AT pairs were all numerically larger than those for BT pairs, and the difference for TSS200 was significant (P = 0.03). B/AT siblings were not correlated in any region except being negatively correlated in the non-CGI region (rBAT = −0.39; 95% CI: −0.58, −0.17).
Meta-analysis
From combining the AMDTSS and KHTS results, neither BT nor B/AT pairs were correlated in GWAM (rBT = −0.003, 95% CI: −0.30, 0.29; rBAT = 0.08; 95% CI: −0.31, 0.45). The correlation was 0.46 (95% CI: 0.26, 0.63) for AT pairs, larger than the correlation for BT pairs (P = 0.02; Table 3).
Neither BT nor B/AT pairs were correlated in any region, except that B/AT pairs were negatively correlated in the non-CGI region (rBAT = −0.34; 95% CI: −0.53, −0.12) (Table 3). AT pairs were correlated in all regions (P < 0.03), except that the correlation in CpG island was marginally significant (P = 0.06). The correlations were different between BT and AT pairs in each of the CpG shelf, CpG shore, non-CGI region, gene body, intergenic region, TSS200 and 3′UTR regions (all P < 0.04).
Pathway analysis
A total of 152 differential correlated probes (at P < 10−4), annotated to 148 unique genes, were included. The top pathways in which these probes overrepresented included “GPCR-mediated nutrient sensing in enteroendocrine cells” (4 of 85 molecules: ADCY9, CASR, ITPR1 and PRKCZ), “Phospholipase C signalling” (6 of 240 molecules: ADCY9, ARHGEF16, ARHGEF17, ITPR1, RAP1A and PRKCZ), “Corticotropin releasing hormone signalling” (4 of 111 molecules: ADCY9, ITPR1, RAP1A and PRKCZ), “Clathrin-mediated endocytosis signaling” (5 of 197 molecules: CSNK2A1, AP2M1, FGF8, DAB2 and FGF1), “Asparagine biosynthesis I” (1 of 1 molecules: ASNS) and “Breast cancer regulation by stathmin1” (5 of 203 molecules: ADCY9, ARHGEF16, ARHGEF17, ITPR1 and PRKCZ; all P < 0.01). The top diseases and disorders included “Connective tissue disorders”, “Developmental disorder”, “Gastrointestinal disease”, “Organismal injury and abnormalities” and “Skeletal and muscular disorders” (all P < 10−4). The top molecular and cellular functions included “Cellular movement”, “Cellular assembly and organization”, “Cellular function and maintenance”, “Cell death and survival” and “Cell-to-cell signalling and interaction” (all P < 5 × 10−4). The top physiological system development and function included “Digestive system development and function”, “Organismal development”, “Tissue Morphology”, “Nervous system development and function” and “Tissue development” (all P < 5 × 10−4).
Discussion
We have found, from conducting two twin and family studies that compared the blood DNA methylation measures between middle-aged BT and AT siblings, evidence consistent with twin birth changing the methylation of siblings born after a twin birth. This change appears to cause the DNA methylation of sibling pairs born after a twin birth to be similar, which is in contrast to the DNA methylation of sibling pairs both before a twin birth, or either side of a twin birth, which is not correlated.
To the best of our knowledge, our studies are the first to report on the potential for a twin birth to influence the methylation of subsequent siblings. The potential for maternal factors to change the methylation of subsequent offspring comes from two studies with similar design that have found that siblings born before and after maternal bariatric surgery have different methylation levels at several genes14, 15.
From a previous study that combined data from seven twin and family studies across the lifespan we found evidence that GWAM, a measure of global methylation16, is initially determined by intrauterine environmental effects which decrease over the life course (Li, et al. Under review). In that study, we did not find evidence that middle-aged sibling pairs overall (including BT, B/AT, AT and twin-sibling pairs) were correlated in GWAM. Here we have found that, on a closer examination, only the subgroup of AT sibling pairs was correlated. This observation indicates that twin birth can influence the covariance in GWAM for sibling pairs born after a twin birth, even when they are in middle age. It is likely that these pairs were even more highly correlated in earlier life, given that we observed this for twin pairs when we pooled of studies across the life span.
It is unlikely that our observation is explained by factors other than twin birth. Given that both types of siblings are first-degree relatives who share on average 50% of germline genetic information, this observation cannot be explained by such genetic factors. Furthermore, we previously did not find evidence of germline genetic factors explaining variation in GWAM to a detectable extent at any stage of the lifespan (Li, et al. Under review). We applied the ComBat or a mixed-effects model to minimise batch effects, and we observed similar phenomenon in two independent studies, therefore the observation is unlikely to be due to batch effects, especially those specific to any one study. We did not find evidence that the correlations depended on age, sex or any other factors we examined. It is noteworthy that the mean ages of B/AT pairs were 54.1 and 44.4 years in the AMDTSS and KHTS, respectively, similar to that of AT pairs, yet B/AT pairs were not correlated. We note that our lack of information on maternal age means that we could not examine the influence of this factor on our results. However, given that maternal age is typically negatively correlated with offspring’s age, any impact of maternal age would have been reduced by adjusting for age before estimating the correlations.
Our observations are also unlikely to be explained by the ‘epigenetic drift’, given that the participants were middle-aged. In the seminal paper that first reported this phenomenon, there was little evidence for epigenetic drift in middle age or beyond: middle-aged and elderly pairs of monozygotic twins had similar Euclidean squared distance in 5-methylcytosine content regardless of age17. We previously found evidence suggesting that the correlation in GWAM for twin pairs remains constant after early adulthood (Li, et al. Under review), which is also consistent with epigenetic drift not being manifest in middle age and beyond.
We found evidence that AT pairs were more correlated in the average methylation of several genomic regions, as well as in methylation at several methylation sites. This suggests that twin birth’s influence is not only detectable at the genome-wide level, but also at specific regions and methylation sites.
Our results are epidemiological observations. Blood methylation has also been observed to be associated with risks of complex traits and diseases; for example, GWAM or similar measures in whole blood have been found to be associated with risks of breast cancer18, 19, urothelial cell carcinoma16 and mature B-cell neoplasms20. The hypothesis of developmental origins of health and disease (DOHaD) considers that epigenome reprogramming during the fetal development period is one possible biological mechanism for the prenatal origins of diseases at later ages21, 22. Therefore, our study suggests that siblings born after a twin birth potentially have different disease (e.g. cancer) risks from those born before a twin birth. To our knowledge, no study has yet reported on different disease risks between the two types of siblings defined by a twin birth. Our study also implies that twin birth might mostly influence cell signalling and breast cancer regulation pathways. The overrepresentation of these pathways is mainly based on molecules including ADCY9, ITPR1 and PRKCZ. We also find evidence that several disorders/diseases, such as those in connective tissue, development and digestive system, and such cellular functions as cellular movement, assembly and organization involving in tumour cell migration and endocytosis of liposome are potentially influenced by twin birth. The links between these pathways/functions and diseases remain to be investigated.
We hope that our study will provide new insights on the shaping of human blood methylome, especially on the role of intrauterine environment. In addition, our study findings suggest that more research about the features of different types of siblings defined by a twin birth are justified, and have the potential for providing a better understanding the aetiology of complex diseases.
We hypothesize that twin birth influences the DNA methylation of subsequent siblings through changing the intrauterine environment. While our observations suggest that twin birth influences the DNA methylation of subsequent siblings we did not have, and our study could not provide, direct evidence that twin birth changes the intrauterine environment. The mechanisms underlying these epidemiological observations need to be further studied.
A strength of our study is that we have used two independent yet comparable studies for discovery and replication, and found similar results. The inclusion of two studies minimises the possibility for any bias due to factors specific to one study. A limitation of our study is that we were unable to investigate the influence on our results of factors other than those we had measured.
We conclude that twin birth can change the similarity in whole blood DNA methylation of siblings both born after a twin birth, without discernibly changing the methylation level of such siblings.
Methods
Subjects
Subjects were from the Australian Mammographic Density Twins and Sisters Study (AMDTSS)23 and the Korean Healthy Twin Study (KHTS)24. The AMDTSS was used for discovery and the KHTS was used for replication.
The AMDTSS is a twin and family study conducted in Australia originally designed to study mammographic density as a risk factor for breast cancer, in which 479 women comprising 132 twin pairs and their 215 sisters from 130 families were selected for methylation research. The 215 sisters including 112 BT siblings and 103 AT siblings were included in this study. Of the 130 families, 28 included at least two BT siblings, 27 included at least two AT siblings, and 23 included at least one BT sibling and one AT sibling.
The KHTS is a study of twin families conducted in South Korea designed to examine genetic and environmental factors underlying complex human diseases and traits24, in which 390 participants comprising monozygotic twins and their first-degree relatives from 97 families were selected for methylation research. 42 siblings including 27 BT siblings and 15 AT siblings were included in this study.
The AMDTSS was approved by the Human Research Ethics Committee of the University of Melbourne. The KHTS was approved by the Institutional Review Board of Samsung Medical Centre and Busan Paik Hospital. Both studies were conducted in accordance with the Helsinki Declaration. All participants from both studies provided written informed consent.
DNA methylation measurement
Each study measured DNA methylation using the Infinium HumanMethylation450 BeadChip (HM450) array and performed data pre-processing independently.
In the AMDTSS, DNA was extracted from dried blood spots stored on Guthrie cards25. DNA was sodium bisulfite converted using the EZ DNA Methylation-Gold protocol as per manufacturers’ instructions (Zymo Research, Irvine, CA) and eluted in 20 µl elution buffer. DNA samples extracted from the same family were assayed on the same chip. Raw intensity data was processed by Bioconductor package minfi 26 which included normalization of data using Illumina’s reference factor-based normalization methods (preprocessIllumina) and subset-quantile within array normalization (SWAN)27 for type I and II probe bias correction. An empirical Bayes batch-effects removal method ComBat 28 was applied to minimise the technical variation across batches. All samples passed quality control. Probes with missing value (detection P-value > 0.01) in one or more samples, 65 control probes and probes mapping to X-chromosome were excluded, leaving 468,406 autosomal probes. See Li et al.29 for more details.
In the KHTS, DNA was extracted from peripheral blood lymphocytes. The measurement was conducted in two separated experiments (experiment I and II), with individuals from the same family included in the same experiment. For each experiment, quality control and data pre-processing were performed separately, while the same analytic tools and methods were applied. The R package RnBeads 30 was applied to extract methylation values. In the quality control, a series of probe and sample filtering steps were followed: probes mapping to sex chromosomes, associated with SNPs and/or out of CpG context were removed, and CpG probes and samples were filtered at detection P-value of 0.01. The beta mixture quantile dilation (BMIQ) method31 was used for normalization.
Statistical methods
Discovery analysis
We studied a global methylation measure, genome-wide average DNA methylation (GWAM) defined as the average beta-value across autosomal probes. We compared the means in GWAM between BT and AT siblings using a linear mixed-effects model, in which GWAM was the outcome and sister type was the predictor. The model was adjusted for age and blood cell type composition (fixed effects), and for family identification number (random effect). The blood cell type composition was estimated from the methylation data using the Houseman method32 from the Bioconductor package minfi.
We estimated correlations in GWAM for BT pairs (rBT), B/AT pairs (rBAT) and AT pairs (rAT). The correlation was estimated based on computationally maximizing the likelihood of a multivariate normal model for pedigree analysis33,34,35. The correlation in GWAM was the covariance in GWAM between relatives divided by the variance of GWAM. GWAM was firstly adjusted for age and the blood cell type composition using a linear regression. Residuals from the regression were used to estimate correlations. Correlations between BT and AT pairs were compared using the likelihood ratio test (LRT), that is, a nested model in which the two correlations set to equal was fitted, and a P-value was calculated according to that twice the difference in the log likelihoods between the full and nested model approximately follows the chi-squared distribution with one degree of freedom.
To investigate if the correlations depend on the twins’ zygosity, we stratified each type of sibling pairs by the twins’ zygosity and estimated the correlation in each stratum. The model fit was compared with that of the model above using the LRT. To investigate if the correlations depend on other factors, we modelled the correlations as α + β *factor where the factor was a characteristic of the sibling pair: (1) the average age; (2) the absolute value of age difference; (3) the average birth order; (4) the absolute value of birth order difference; (5) the average year between the sibling’s birth and the twins’ birth; (6) the absolute value of the difference in year between the sibling’s birth and the twins’ birth; and (7) the twins’ age. The model fit was compared with that of the model above using the LRT.
We studied the average methylation of genomic regions. According to Illumina’s annotation file, probes were grouped according to their genomic positions (gene body, intergenic region, promoter, TSS1500, TSS200, 5′UTR, 1stExon or 3′UTR) or to their positions relative to a CpG island (CGI) (CpG island, CpG shelf, CpG shore or non-CGI region). For each region, the average beta-value across autosomal probes was calculated. The means and correlations of these genomic regions were compared using the same methods as those for GWAM. The binomial test was used to test whether the observed number of statistically significant (at P < 0.05) comparisons was more than would be expected by chance.
We studied the methylation measured by each autosomal probe, comparing the means and correlations in the methylation (beta-value) measured by each probe using the same methods as those for GWAM. P-values were corrected for the genomic control inflation factor. Genome-wide level of significance was 10−7. The McNemar’s test was used to compare the proportions of positive correlation estimates, and the paired t-test was used to compare the means in correlation estimates. The binomial test was used to test whether the observed direction of the genome-wide significant associations was more extreme than would have been expected by chance.
Replication analysis
For GWAM and the average methylation of genomic regions, we estimated their correlations in the KHTS. The methylation measure was firstly adjusted for age, sex, experiment, the estimated blood cell type composition (fixed effects), array and position on the array (random effects) using a linear mixed-effects model in the whole dataset. Residuals from the model were used for correlation estimation. Sex heterogeneity in correlations was examined using the LRT. The binomial test was used to test the consistency of results between discovery and replication analyses.
Meta-analysis
Results from discovery and replication analyses were pooled using a fixed-effect meta-analysis under inverse-variance weighting. The correlations were compared between BT and AT pairs based on asymptotic theory.
Pathway analysis
We performed pathway analysis for the probes differentially correlated at P < 10−4 between BT and AT pairs. These probes were annotated to genes according to the closest transcription start site (TSS)36. The gene list was uploaded to QIAGEN Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity) for assessing overrepresentation relative to all human gene functions37.
Correlation estimation and modelling were performed using the Sequential Oligogenic Linkage Analysis Routines (SOLAR) program version 8.1.1 (http://solar-eclipse-genetics.org/). Regression analyses were performed using R version 3.2.4 (http://www.r-project.org). The mixed-effects model was fitted using the lmer function from the R package lme4. The linear regression was fitted using the lm function in R. The meta-analysis was performed using the metagen function from the R package meta. The McNemar’s test, t-test, binomial test were performed using the mcnemar. test, t. test and binom. test function in R, respectively.
Data availability
The discovery data are available at the Gene Expression Omnibus under accession number GSE100227. The replication data are available on the request to the authors. The SOLAR code could be found in the Supplementary Text.
References
Petronis, A. Epigenetics as a Unifying Principle in the Aetiology of Complex Traits and Diseases. Nature 465, 721–727 (2010).
Westin, B. Gravidogram and Fetal Growth. Comparison with Biochemical Supervision. Acta. Obstet. Gynecol. Scand. 56, 273–282 (1977).
Blickstein, I. & Keith, L. G. Multiple Pregnancy: Epidemiology, Gestation, and Perinatal Outcome. 2nd edn, 436–443 (Taylor & Francis, 2005).
Norwitz, E. R., Edusa, V. & Park, J. S. Maternal Physiology and Complications of Multiple Pregnancy. Semin. Perinatol. 29, 338–348 (2005).
Campbell, D. A Review of Maternal Complications of Multiple Pregnancy. Twin Res. 4, 146–149 (2001).
Gordon, L. et al. Neonatal DNA Methylation Profile in Human Twins Is Specified by a Complex Interplay between Intrauterine Environmental and Genetic Factors, Subject to Tissue-Specific Influence. Genome Res. 22, 1395–1406 (2012).
Ollikainen, M. et al. DNA Methylation Analysis of Multiple Tissues from Newborn Twins Reveals Both Genetic and Intrauterine Components to Variation in the Human Neonatal Epigenome. Hum. Mol. Genet. 19, 4176–4188 (2010).
Joubert, B. R. et al. Maternal Plasma Folate Impacts Differential DNA Methylation in an Epigenome-Wide Meta-Analysis of Newborns. Nat Commun. 7, 10577, doi:10.1038/ncomms10577 (2016).
Joubert, B. R. et al. DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-Wide Consortium Meta-Analysis. Am. J. Hum. Genet. 98, 680–696 (2016).
Richmond, R. C. et al. Prenatal Exposure to Maternal Smoking and Offspring DNA Methylation across the Lifecourse: Findings from the Avon Longitudinal Study of Parents and Children (Alspac). Hum. Mol. Genet. 24, 2201–2217 (2015).
Tobi, E. W. et al. DNA Methylation Signatures Link Prenatal Famine Exposure to Growth and Metabolism. Nat. Commun. 5, 5592, doi:10.1038/ncomms6592 (2014).
Lee, K. W. et al. Prenatal Exposure to Maternal Cigarette Smoking and DNA Methylation: Epigenome-Wide Association in a Discovery Sample of Adolescents and Replication in an Independent Cohort at Birth through 17 Years of Age. Environ. Health Perspect. 123, 193–199 (2015).
Breton, C. V. et al. Prenatal Tobacco Smoke Exposure Is Associated with Childhood DNA Cpg Methylation. PloS one 9, e99716, doi:10.1371/journal.pone.0099716 (2014).
Guenard, F. et al. Differential Methylation in Glucoregulatory Genes of Offspring Born before Vs. After Maternal Gastrointestinal Bypass Surgery. Proc. Natl. Acad. Sci. USA 110, 11439–11444 (2013).
Berglind, D. et al. Differential Methylation in Inflammation and Type 2 Diabetes Genes in Siblings Born before and after Maternal Bariatric Surgery. Obesity 24, 250–261 (2016).
Dugue, P. A. et al. Genome-Wide Measures of DNA Methylation in Peripheral Blood and the Risk of Urothelial Cell Carcinoma: A Prospective Nested Case-Control Study. Br. J. Cancer 115, 664–673 (2016).
Fraga, M. F. et al. Epigenetic Differences Arise During the Lifetime of Monozygotic Twins. Proc. Natl. Acad. Sci. USA 102, 10604–10609 (2005).
Severi, G. et al. Epigenome-Wide Methylation in DNA from Peripheral Blood as a Marker of Risk for Breast Cancer. Breast Cancer Res. Treat. 148, 665–673 (2014).
van Veldhoven, K. et al. Epigenome-Wide Association Study Reveals Decreased Average Methylation Levels Years before Breast Cancer Diagnosis. Clin. Epigenetics 7, 67 (2015).
Wong Doo, N. et al. Global Measures of Peripheral Blood-Derived DNA Methylation as a Risk Factor in the Development of Mature B-Cell Neoplasms. Epigenomics 8, 55–66 (2016).
Waterland, R. A. & Michels, K. B. Epigenetic Epidemiology of the Developmental Origins Hypothesis. Annu. Rev. Nutr. 27, 363–388 (2007).
Wadhwa, P. D., Buss, C., Entringer, S. & Swanson, J. M. Developmental Origins of Health and Disease: Brief History of the Approach and Current Focus on Epigenetic Mechanisms. Semin. Reprod. Med. 27, 358–368 (2009).
Odefrey, F. et al. Common Genetic Variants Associated with Breast Cancer and Mammographic Density Measures That Predict Disease. Cancer Res. 70, 1449–1458 (2010).
Sung, J. et al. Healthy Twin: A Twin-Family Study of Korea–Protocols and Current Status. Twin Res. Hum. Genet. 9, 844–848 (2006).
Joo, J. E. et al. The Use of DNA from Archival Dried Blood Spots with the Infinium Humanmethylation450 Array. BMC Biotechnol. 13, 23, doi:10.1186/1472-6750-13-23 (2013).
Aryee, M. J. et al. Minfi: A Flexible and Comprehensive Bioconductor Package for the Analysis of Infinium DNA Methylation Microarrays. Bioinformatics 30, 1363–1369 (2014).
Maksimovic, J., Gordon, L. & Oshlack, A. Swan: Subset-Quantile within Array Normalization for Illumina Infinium Humanmethylation450 Beadchips. Genome Biol. 13, R44, doi:10.1186/gb-2012-13-6-r44 (2012).
Johnson, W. E., Li, C. & Rabinovic, A. Adjusting Batch Effects in Microarray Expression Data Using Empirical Bayes Methods. Biostatistics 8, 118–127 (2007).
Li, S. et al. Genetic and Environmental Causes of Variation in the Difference between Biological Age Based on DNA Methylation and Chronological Age for Middle-Aged Women. Twin Res. Hum. Genet. 18, 720–726 (2015).
Assenov, Y. et al. Comprehensive Analysis of DNA Methylation Data with Rnbeads. Nat. Methods 11, 1138–1140 (2014).
Teschendorff, A. E. et al. A Beta-Mixture Quantile Normalization Method for Correcting Probe Design Bias in Illumina Infinium 450 K DNA Methylation Data. Bioinformatics 29, 189–196 (2013).
Houseman, E. A. et al. DNA Methylation Arrays as Surrogate Measures of Cell Mixture Distribution. BMC bioinformatics 13, 86, doi:10.1186/1471-2105-13-86 (2012).
Lange, K., Westlake, J. & Spence, M. A. Extensions to Pedigree Analysis. Iii. Variance Components by the Scoring Method. Ann. Hum. Genet. 39, 485–491 (1976).
Hopper, J. L. & Mathews, J. D. Extensions to Multivariate Normal Models for Pedigree Analysis. Ann. Hum. Genet. 46, 373–383 (1982).
Hopper, J. L. & Mathews, J. D. A Multivariate Normal Model for Pedigree and Longitudinal Data and the Software ‘Fisher’. Aust. J. Statist. 36, 153–176 (1994).
Price, M. E. et al. Additional Annotation Enhances Potential for Biologically-Relevant Analysis of the Illumina Infinium Humanmethylation450 Beadchip Array. Epigenetics Chromatin. 6, 4, doi:10.1186/1756-8935-6-4 (2013).
Kramer, A., Green, J., Pollard, J. Jr. & Tugendreich, S. Causal Analysis Approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530 (2014).
Acknowledgements
We would like to thank the subjects participating in each study. The data analysis was facilitated by Spartan, the High Performance Computing system (HPC) of the University of Melbourne. The AMDTSS was facilitated through the Australian Twin Registry, a national research resource in part supported by a Centre for Research Excellence Grant from the National Health and Medical Research Council (NHMRC) APP 1079102. The AMDTSS was supported by NHMRC (grant numbers 1050561 and 1079102), Cancer Australia and National Breast Cancer Foundation (grant number 509307). The KHTS was supported by a fund (2014-E71004-00) by Research of Korea Centers for Disease Control, and Prevention and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (grant number NRF-2017R1A2B2002136). SL is supported by the Australian Government Research Training Program Scholarship and Richard Lovell Travelling Scholarship from the University of Melbourne. TLN is supported by a NHMRC Post-Graduate Scholarship and the Richard Lovell Travelling Scholarship from the University of Melbourne. MCS is a Senior Research Fellow of NHMRC. JLH is a Senior Principal Research Fellow of NHMRC and a Distinguished Visiting Professor at Seoul National University.
Author information
Authors and Affiliations
Contributions
Study conception and design: S.L., J.S. and J.L.H. Data collection and preparation: E.K., E.M.W., J.-H.E.J., T.L.N., J.S. Y.-M.S., G.G.G., M.C.S., J.S. and J.L.H. Data analysis: S.L. and E.K. Data interpretation: S.L., E.M.W., L.B.F., R.S., M.C.S., J.L.H. Manuscript draft: S.L. E.M.W., L.B.F, M.C.S. and J.L.H. All authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, S., Kim, E., Wong, E.M. et al. Twin birth changes DNA methylation of subsequent siblings. Sci Rep 7, 8463 (2017). https://doi.org/10.1038/s41598-017-08595-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-08595-6
This article is cited by
-
A Pregnancy and Childhood Epigenetics Consortium (PACE) meta-analysis highlights potential relationships between birth order and neonatal blood DNA methylation
Communications Biology (2024)
-
Reproductive history and blood cell DNA methylation later in life: the Young Finns Study
Clinical Epigenetics (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.