Abstract
The bactericidal/permeability-increasing (BPI) fold-containing (BPIF) superfamily of genes expressed in the brain are purportedly involved in modulating brain function in response to stress, such as inflammation. Kisspeptin, encoded by kiss, is affected by inflammation in the brain; therefore, BPIF family genes might be involved in the modulation of kisspeptin in the brain. In this study, we investigated the expression of BPIF family C, like (bpifcl) in zebrafish brain and its involvement in kiss2 regulation. The identified, full-length sequence of a bpifcl isoform expressed in the zebrafish brain contained the BPI fold shared by BPIF family members. bpifcl mRNA expression in female zebrafish brains was significantly higher than that in males. Exposure of female zebrafish to 11-ketotestosterone decreased bpifcl and kiss2 mRNA expression. bpifcl knockdown by bpifcl-specific small interfering RNA administration to female zebrafish brain decreased kiss2 mRNA expression. bpifcl expression was widely distributed in the brain, including in the dorsal zone of the periventricular hypothalamus (Hd). Furthermore, bpifcl was also expressed in KISS2 neurons in the Hd. These results suggest that the Bpifcl modulates kiss2 mRNA expression under the influence of testosterone in the Hd of female zebrafish.
Similar content being viewed by others
Introduction
The bactericidal/permeability-increasing (BPI) fold-containing (BPIF) superfamily of genes is functionally classified into five groups: palate, lung and nasal epithelium clone (PLUNC); lipopolysaccharide-binding protein (LBP); BPI protein; phospholipid transfer protein (PLTP); cholesterol ester transfer protein (CETP)1, 2. All BPIF family members share the BPI fold, which has an elongated, boomerang shape consisting of two distinct N- and C-terminal domain barrels with a highly similar secondary structure3. Both domains consist of two twisted, anti-parallel β-sheets and two α-helices, with both domains connected to each other by a β-sheet4.
BPIF family of genes are expressed in a wide range of vertebrates5 and mainly involved in the innate immune system and lipoprotein metabolism. BPI and LBP are plasma proteins that play an important role in the innate immune system against invading pathogens owing to a high affinity for the lipopolysaccharides of Gram-negative bacteria3, 6. PLUNC proteins are suggested to be host defence molecules because their expression alters in response to inflammation caused by smoking, chemical irritants, and infection7, 8. CETP and PLTP are both plasma proteins that play an important role in lipoprotein metabolism9, 10. Recently, the expression of BPIF family of genes has been reported in the brain11. We have also reported the expression of BPIF family of genes, such as bpifa1, gm1006 and rya3 in the mouse brain, particularly in the preoptic area12. The expression of these genes decreases with age. Because ageing causes oxidative stress and inflammation in the brain13, the identified BPIF family of genes might respond to oxidative stress and inflammation similar to the PLUNC protein group in the brain.
Kisspeptin is encoded by kiss, expressed in various vertebrates14, and involved in various physiologies in the brain, such as reproduction15, memory16 and metabolism17. Kisspeptin also interacts with the adrenergic, serotonergic and GABAergic systems16; therefore, kisspeptin is a key modulator of brain function. Kisspeptin is a molecule known to be affected by inflammation in the brain18, 19. Since BPIF family genes, particularly BPI, LBP and PLUNC groups, respond to inflammatory agents, BPIF family of genes may interact with kisspeptin to modulate brain functions. Because the regulatory mechanisms of kisspeptin have not been fully identified, the BPIF family-kisspeptin interaction could be a novel kisspeptin regulatory mechanism in the brain.
In zebrafish, three BPIF family of genes, pltp (ENSDARG00000104495), cetp (ENSDARG00000030872) and BPIF family C, like (bpifcl; ENSDARG00000099980) are predicted. The zebrafish CETP protein has similar functionality to human CETP20, while zebrafish PLTP is annotated as part of the PLTP group owing to its sequence similarity with mammals21. Meanwhile, bpifcl has not been classified into a sub-group of the BPIF family and has unknown function. bpifcl is located on chromosome 9 and predicted to have three isoforms. In the current study, we investigated the involvement of bpifcl in the regulation of kiss2 in the zebrafish brain. bpifcl isoforms expressed and localised in zebrafish brain were initially identified. Correlations between bpifcl and kiss2 mRNA expression were investigated by comparing males and females and different developmental stages. Furthermore, the effect of bpifcl knockdown was evaluated to investigate the involvement of bpifcl in the regulation of kiss2 expression.
Results
Full-length bpifcl mRNA sequence
The identified bpifcl isoform (Genbank accession no. KX459407) consisted of 1974 nucleotides spanning 15 exons when compared to the published zebrafish genome. The coding region starts at the 65th base of exon 1 and extends to the 57th base of exon 15. Only this isoform was identified in the current study. The coding sequence was identical to the predicted bpifcl (Accession no. ENSDARG00000099980). Simple Modular Architecture Research Tool (SMART) prediction indicated that the N- and C-terminal domains of BPIF family proteins consisted of amino acids (aa) 24-243 and 258-461, respectively (Supplementary Fig. 1A). Phylogenetic tree analysis revealed that the estimated aa sequence of Bpifcl is closer to Pltp than Cetp (Supplementary Fig. 1B). The aa identity between zebrafish Bpifcl and human BPIFC (ENSG00000184459) was 19.5%.
Localisation of bpifcl in zebrafish brain
Distributions of bpifcl mRNA in the 6 months old female zebrafish, 6 months old male fish and 60 days post-fertilization (dpf) fish were identified using digoxigenin (DIG)- in situ hybridisation (ISH). The general distribution pattern of bpifcl mRNA is illustrated in Fig. 1. Strong signals were detected in select regions of the brain: the lateral, medial and posterior zone of the dorsal telencephalic area (Fig. 1B,C, Ga, Gb, Ha, Hb, Ia and Ib), dorsal and ventral nucleus of the ventral telencephalic area (Fig. 1B, Gc, Hc and Ic), anterior and posterior parvocellular preoptic nucleus (Fig. 1C,D, Gd, Ge, Hd, He, Id and Ie), dorsal and ventral habenular nucleus (Fig. 1D, Gf, Hf and If), anterior thalamic nucleus (Fig. 1D, Gf, Hf and If), dorsal (Hd) and ventral zone of periventricular hypothalamus (Fig. 1D,E, Ge, Gg, He, Hg, Ie and Ig), posterior tuberal nucleus (Fig. 1E, Gg, Hg and Ig) and central nucleus of torus semicircularis (Fig. 1F, Gh, Hh and Ih). There were no distribution differences among 6 months old female, 6 months old male and 60 dpf fish.
Localisation of bpifcl mRNA expression in zebrafish brain. (A) Schematic sagittal drawing of the zebrafish brain. (B–F) Lines in A indicate levels of coronal sections. Schematic coronal brain drawing of zebrafish showing the distribution of bpifcl (red dots) mRNA-containing cells in the brain. (Ga–Gh) Photomicrographs of bpifcl-expressing cells in 6 months old female zebrafish brain. (Ha–Hh) Photomicrographs of bpifcl-expressing cells in 6 months old male zebrafish brain. (Ia–Ih) Photomicrographs of bpifcl-expressing cells in 60 days post-fertilisation (dpf) fish brain. For abbreviations, see Supplementary Abbreviation. Scale bar: 50 μm.
Association between bpifcl mRNA expression and kiss2 mRNA expression
There was no significant difference in bpifcl mRNA expression between 45, 60 and 120 dpf zebrafish brains (Fig. 2A and Supplementary Table 2A). Meanwhile, bpifcl mRNA expression was significantly high in the 6 months old females compared with the 6 months old males (Fig. 2B and Supplementary Table 2B). On the other hand, kiss2 mRNA was significantly low in the 6 months females (Fig. 2C and Supplementary Table 2C).
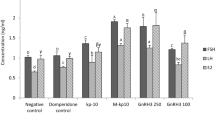
Association between bpifcl mRNA and kiss2 mRNA expression. bpifcl mRNA expression in zebrafish brain during development (A) and between 6 months old males and females (B). kiss2 mRNA expression between 6 months old males and females (C). Effect of 11-KT exposure on bpifcl (D) and kiss2 (E) mRNA expression in 6 months old male zebrafish brain. Effect of E2 exposure on bpifcl (F) and kiss2 (G) mRNA expression in 6 months old male zebrafish brain. Expression levels of 45 days post-fertilisation (dpf) (A), males (B,C) and 0 μg/L exposure (D–G) were defined as 1.0. All data are presented as the mean ± standard error of the mean and were analysed by a Student’s t-test for sex differences and one-way analysis of variance and Tukey’s post-hoc test for multiple comparisons for the others. **p < 0.01.
Effect of 11-ketotestosterone (11-KT) and β-estradiol (E2) on bpifcl and kiss2 mRNA expression
After 48 h exposure with E2 and 11-KT to the 6 months old males, bpifcl and kiss2 mRNA expressions were not significantly different, regardless of concentration (Fig. 2D–G and Supplementary Table 2D–G). gnrh3 mRNA expression was significantly different at 0.1 and 1.0 μg/L of E2 (Supplementary Fig. 2B and Supplementary Table 2O).
After 48 h exposure with 11-KT to the 6 months old females, bpifcl and kiss2 mRNA expression were significantly lower at 0.1 and 1.0 μg/L, respectively (Fig. 3A,B, Supplementary Table 2H and I), however, gnrh3 mRNA expression was not significantly different (Supplementary Fig. 2A and Supplementary Table 2N). bpifcl and kiss2 mRNA expressions were not significantly different after 6 h and 24 h exposure to 11-KT, regardless of concentration (Fig. 3A,B, Supplementary Table 2H and I).
Effect of 11-KT on 6 months old female zebrafish. Effect of 11-KT on bpifcl (A) and kiss2 (B) mRNA expression in 6 months old female zebrafish brains. Expression levels of 0 μg/L exposure at each time point (A,B) were defined as 1.0. All data are presented as the mean ± standard error of the mean and were analysed by a one-way analysis of variance and Tukey’s post-hoc test for multiple comparisons. *p < 0.05, **p < 0.01.
Expression of bpifcl in KISS2 neurons and effect of knocked-down bpifcl mRNA expression
bpifcl mRNA expression in KISS2 neurons of the Hd was examined using double-label fluorescence ISH. All KISS2 neurons expressed bpifcl in the Hd (Fig. 4A–C). Furthermore, the effect of knocked-down bpifcl expression on kiss2 mRNA expression was also examined. All siRNA administrated fish survived. No localisation difference of KISS2 neurons in 6 months old female fish was exhibited by bpifcl specific siRNA administration (Fig. 4D and E). bpifcl mRNA expression was confirmed as significantly decreased in bpifcl small interfering RNA (siRNA)-injected female (Fig. 4F and Supplementary Table 2J) and male (Fig. 4H and Supplementary Table 2L) zebrafish compared to green fluorescent protein (GFP) siRNA-injected fish. Since suppression of bpifcl mRNA expression was confirmed, kiss2 mRNA expression was also examined. kiss2 mRNA expression was also significantly lower in bpifcl siRNA-injected female zebrafish (Fig. 4G and Supplementary Table 2K), but was significantly higher in bpifcl siRNA-injected males (Fig. 4I and Supplementary Table 2M) compared to GFP siRNA-injected fish.
Photomicrographs of double-label fluorescence ISH of bpifcl and kiss2 mRNA, and effect of knocked-down bpifcl mRNA expression. Anti-sense riboprobe-labelled bpifcl (A) and kiss2 (B) mRNA was co-expressed in the dorsal zone of the periventricular hypothalamus (Hd) (C). Scale bars: 20 μm. Localisation of kiss2 mRNA in the Hd of GFP siRNA injected female (D) and bpifcl siRNA injected female (E). Scale bars: 20 μm. bpifcl (F) and kiss2 (G) mRNA expression in bpifcl siRNA-injected female zebrafish, and bpifcl (H) and kiss2 (I) mRNA expression in bpifcl siRNA-injected males. Expression levels of GFP siRNA injected fish were defined as 1.0. All data are presented as the mean ± standard error of the mean and analysed by a Student’s t-test. *p < 0.05.
Discussion
In the current study, an isoform of bpifcl expressed in the brain of zebrafish was identified. Functional domain analysis using SMART showed the presence of N- and C-terminal domain of the BPIF family. Furthermore, the three-dimensional structure of the BPIFCL protein predicted by SWISS-MODEL (http://swissmodel.expasy.org) using human BPI as a template revealed a boomerang-shaped molecule with two twisted, anti-parallel β-sheets and two α-helices at both the N- and C-terminals, as well as the presence of two distinct barrel-shaped domains (data not shown). These results indicate that the identified bpifcl isoform belongs to the BPIF family of genes expressed in the brain.
bpifcl mRNA expression in mature female zebrafish brains was significantly higher than in mature males. On the other hand, kiss2 mRNA expression in mature females was significantly lower than in mature males. This suggests that sex steroids affect both of bpifcl and kiss2 mRNA expressions. During development, particularly around puberty (45 and 60 dpf), and matured fish (120 dpf), no significant difference of bpifcl mRNA expression was revealed. kiss2 mRNA expression is also remained at high levels during this period22, and localisation of bpifcl mRNA in the 60 dpf fish brain including the Hd where KISS2 neurons are present22 was not significantly different from mature fish. bpifcl mRNA expression could be associated with kiss2 mRNA expression.
Since sex steroids suggest to affect bpifcl and kiss2 mRNA expressions, fish were exposed to E2 and 11-KT, and bpifcl and kiss2 expressions were examined. Sex steroids affect sexual behaviour and spawning when male zebrafish are exposed to E2 and female fish are exposed to 11-KT23, therefore, bpifcl mRNA expression was expected to be altered by these exposure. Male fish exposed to E2 revealed no significant difference in kiss2 or bpifcl expression. On the other hand, females exposed to 11-KT revealed significantly lower levels of kiss2 and bpifcl mRNA expression after 48 h exposure, however, 11-KT exposure to male did not affect bpifcl and kiss2 mRNA expression. Since both of bpifcl and kiss2 mRNA expressions were decreased by 11-KT exposure, we administered bpifcl siRNA to evaluate involvement of bpifcl in the regulation of kiss2 mRNA expression. In both male and female zebrafish brains, bpifcl mRNA expression was successfully suppressed by bpifcl siRNA compared to GFP siRNA-injected fish. Generally, siRNA transfection in vitro leads to more than 70% suppression of the target gene24, 25, whereas in vivo administration commonly leads to 30–50% suppression26. Suppression of about 30% in the current study was comparable to in vivo siRNA administration reported in other species. bpifcl siRNA administration herein decreased kiss2 mRNA expression in female fish but not the number of KISS2 neurons. Androgen receptors are widely expressed in the Hd of zebrafish27, comparable to bpifcl expression identified in this study; this strongly suggests that testosterone could regulate Bpifcl modulation of kiss2 mRNA expression in the Hd of females. The BPIF family is known as a secreted protein3, 5,6,7,8,9,10, however, several genes such as CETP28 and PLTP29 are also expressed in cytoplasm and nucleus. Especially, PLTP is suggested to be a modulator of signal transduction of the activation of the phosphatidylinositol-3 kinase/Akt pathway29. In this study, localisation of Bpifcl in the cell was not identified, however, the phylogenetic tree analysis indicated that Bpifcl is closer to Pltp than Cetp suggesting that Bpifcl could also play a modulatory role in signal transduction pathways. A signal transduction pathway of androgen to kiss2 gene has not been identified, however, Bpifcl could be involved in modulation of the pathway. In this study, kiss2 mRNA expression in male fish exposed to 11-KT was not affected. Male zebrafish exposed to 11-KT do not change sexual behaviour23. This suggests that male fish which high levels of testosterone are endogenously expressed30 are not affected by external exposure, therefore, kiss2 mRNA expression might be unaffected by 11-KT exposure.
kiss2 neurons are present in the Hd, Hv and posterior tuberal nucleus (nPT)22. kiss2 mRNA was not detected in the Hv in the current study. Since Servili and coworkers31 showed no kiss2 neurons in the Hv, localisation of kiss2 neurons in the Hv is still debatable. In the nPT, bpifcl mRNA was not expressed. Therefore, kiss2 and bpifcl mRNA could be co-localised in only the Hd. Interestingly, kiss2 mRNA expression was significantly increased by bpifcl siRNA administration in males. Meanwhile, neither E2 nor 11-KT affected to kiss2 mRNA expression in male fish. These suggest that Bpifcl is involved in several regulatory mechanisms of kiss2 mRNA expression and differently involved in the regulation of kiss2 mRNA expression between male and female. We identified Bpifcl as a regulator of kiss2 mRNA expression in female. This regulatory mechanism could be involved in testosterone-related physiologies other than reproduction. Since androgens mediate inflammation in the brain32, this regulatory mechanism could respond to inflammation in the brain.
In vertebrates, the main reproductive regulatory system is the production and secretion of gonadotropin-releasing hormone (GnRH) from GnRH3 neurons in the preoptic area. GnRH regulates gonadal maturation by stimulating the synthesis and secretion of gonadotropins33. Kisspeptin, a product of kiss expression, is a critical regulator of GnRH secretion15. In the current study, exposure of female zebrafish to 11-KT did not affect gnrh3 mRNA expression. In female mummichogs, exposure to 5α-dihydrotestosterone for 21 days was shown to affect fecundity, particularly low egg production34. Trenbolone acetate, a synthetic androgen, was also shown to decrease ovarian follicles in female Japanese medaka after 14 days of exposure35. Here, female zebrafish were exposed to 11-KT for 48 h to investigate any direct correlation(s) with bpifcl. Either this exposure time is too short to affect gnrh3 mRNA expression or kisspeptin might not regulate gnrh3 in zebrafish. In fact, a previous study showed that knocking-out kiss2 in zebrafish did not affect their reproductive capability36. Here we found gnrh3 mRNA expression was significantly increased in male fish exposed to E2. gnrh3 mRNA expression is affected by E2 in teleosts37, and oestrogen receptors are expressed in the preoptic area, where GnRH neurons are present in fish38. Therefore, current E2 results suggest a direct effect on gnrh3 mRNA expression.
An orthologous gene to zebrafish bpifcl has not been identified in other species. However, BPIF genes have been estimated in other species, such as humans (NCBI gene ID: 254240), mouse (270757), rat (299685), bird (107204354), turtle (102945958) and snake (106539050). Furthermore, as we have identified that several BPIF family genes are expressed in the brain and affected by ageing and chronic selective serotonin reuptake inhibitor treatment25. Therefore, an orthologous gene to zebrafish bpifcl could be encoded in other species. Furthermore, modulation of kisspeptin by Bpifcl might be a conserved function in vertebrates since kisspeptin is conserved in vertebrates.
In the current study, we showed involvement of a BPIF family gene regulates gene expression in the zebrafish brain. Although many BPIF family genes are estimated in mammals5, most of the BPIF family genes have unknown function. Some BPIF family genes could be involved in the regulation of gene expression, like bpifcl. The current results suggest that testosterone regulates Bpifcl modulation of kiss2 expression in the Hd of zebrafish, particularly in females. Considering we found that bpifcl expression was widely distributed in the brain, it is reasonable to posit that Bpifcl plays a role in regulating other neurons, particularly those regulated by testosterone, such as KISS2 neurons.
Methods
Animals
Zebrafish (Danio rerio) were maintained in fresh water at 27.0 ± 1.0 °C under a controlled natural photo regimen (14-h light/10-h dark cycle). Fish were fed an Adult Zebrafish Diet (Zeigler Bros., PA, USA) twice daily. To obtain developing embryos, one pair of fish was placed in a tank with glass marbles overnight to allow mating. Fertilised eggs were then syphoned from the tank and maintained at 27.0 ± 1.0 °C. After hatching, larvae were fed paramecium twice daily for the first 3 weeks and brine shrimps (Zebrafish Management Ltd., Hampshire, UK) twice daily for the following 4 weeks. This study was approved by the Animal Ethics Committee of Monash University (Melbourne, Australia; Approval no. MARP/2012/147). All experimental procedures were performed in accordance with guidelines of the Animal Ethics Committee of Monash University.
Cloning of full-length bpifcl cDNA
Zebrafish were anaesthetised by immersing them in a 0.01% solution of benzocaine (Sigma, MO, USA) and killed by decapitation in order to dissect the fresh brains. Total RNA was isolated from whole brain with TRIzol reagent (Invitrogen, CA, USA) and converted to cDNA with SuperScript III (Invitrogen) and oligo d(T) primers. A partial coding sequence region was amplified by polymerase chain reaction (PCR). The primers were designed based on the genomic sequence of a predicted zebrafish bpifcl gene (Accession no. ENSDARG00000099980); primer sequences were as follows: forward, 5′-ATGCAGAGGCTGATGTTCCTCCTG-3′; reverse, 5′-TTATGGAGCATTCAGCCCATCAG-3′. The PCR reaction was performed in a 20 μL reaction mixture containing 1X HotStarTaq Master Mix (Qiagen, Hilden, Germany), 0.5 μM of each primer and 1 μL of cDNA. The amplified DNA was subjected to direct sequencing.
The full-length bpifcl mRNA sequence was identified by 3′- and 5′-rapid amplification of cDNA ends (RACE). Gene-specific primers were designed based on the identified partial mRNA sequence. For 3′-RACE, 1 μg of total RNA was converted to cDNA with SuperScript III and 50 pmol of oligo d(T)-containing adapter primer in a 20 μL reaction volume. The converted cDNA was subjected to PCR in a 10 μL reaction volume containing 1X HotStarTaq Master Mix, 1 pmol of abridged universal amplification primer (Invitrogen) and 10 pmol of a gene-specific primer (5′-TGAGGTCCGGCTTTCTCTTGCCCAC-3′).
For 5′-RACE, 1 μg of the total RNA was converted to cDNA in a 20 μL reaction volume with SuperScript III and 2 pmol of a gene-specific primer (5′-ACAGTCCTGTTCCCTCCACAAACAC-3′). The polycytosine-tailed cDNA with terminal transferase (Roche Diagnostics, Mannheim, Germany) was subjected to PCR in a 20 μL reaction volume containing 1X HotStarTaq Master Mix, 1 pmol of 5′-RACE abridged anchor primer (Invitrogen) and 10 pmol of a gene-specific primer (5′-CGGCTCAGCTGTTTCGCACTGCACG-3′). Furthermore, 1 μL of the PCR reaction mixture was subjected to nested PCR in a 20 μL reaction mixture containing 1X HotStarTaq Master Mix, 1 pmol of abridged universal amplification primer and 10 pmol of a gene specific primer (5′-TATCCAGACAGGCATCATCTGAGAC-3′). The complete nucleotide sequence of bpifcl mRNA was confirmed by sequence analysis of each PCR product. The first ATG codon with a Kozak translation initiation sequence was considered the initial codon39, 40. Functional domain regions were further analysed by SMART41, 42.
Localisation of bpifcl mRNA in adult zebrafish brain using ISH
6 months old male zebrafish, 6 months old female fish and 60 dpf fish were anaesthetized by immersion in 0.01% benzocaine solution and killed by decapitation in order to dissect the fresh brains. The brains were fixed in buffered 4% paraformaldehyde for 6 h, cryoprotected in 20% sucrose and then embedded in Tissue Tek OCT compound (Sakura Finetechnical, Tokyo, Japan). Coronal sections (12 μm thick) were cut on a cryostat and thaw-mounted onto 3-aminopropylsilane-coated glass slides. The RNA probes were synthesised by in vitro transcription from a pGEM-T Easy vector (Promega, Madison, WI, USA), containing sequences amplified with a forward 5′-CGAACCGTGAACATACCCGT-3′ and reverse 5′-TCTGTCGAGCGCCTGTAGA-3′ primer. Sense and anti-sense DIG-labelled riboprobes were synthesised using MAXIscript (Ambion, Austin, TX, USA) and DIG RNA Labeling Mix (Roche Diagnostics). DIG-ISH was performed as described previously22, 43 with minor modifications. Brain regions were followed according to the neuroanatomy of the zebrafish brain44.
Double-label fluorescence ISH of bpifcl and kiss2 mRNA
Riboprobes for bpifcl and kiss2 22 mRNA were labelled with biotin and DIG, respectively, using MAXIscript with either biotin or DIG RNA Labeling Mix (Roche Diagnostics). The biotin-labelled bpifcl probe was detected using horseradish peroxidase-streptavidin and AlexaFluor 594 Tyramide (Invitrogen), whereas the DIG-labelled kiss2 probe was detected using a Tyramide Signal Amplification Plus kit (PerkinElmer/NEN Life Science Products, MA, USA). The biotin-labelled bpifcl and DIG-labelled kiss2 riboprobes were mixed in a cocktail for the hybridisation. After hybridisation, a peroxidase-conjugated anti-DIG antibody (Roche Diagnostics) diluted 1:500 in a buffer containing 0.1 M Tris (pH 7.4), 0.15 M NaCl and Tween buffer (0.05% Triton X-100) with 1% normal goat serum was applied to each slide for 2 h at room temperature, followed by incubation with Tyramide Signal Amplification Working Solution for 1 min for colour development of kiss2 probes. After blocking with 3% H2O2 in a 0.1 M Tris-HCl (pH 7.4) and 0.15 M NaCl buffer for 30 min at room temperature, a horseradish peroxidase-conjugated streptavidin antibody diluted 1:100 in the 0.1 M Tris-HCl (pH 7.4) and 0.15 M NaCl buffer buffer with 2% bovine serum albumin was applied to each slide overnight at 4 °C. The colour development reaction for bpifcl probes was initiated by adding a 1:100 dilution of reconstituted AlexaFluor 594 Tyramide to the Amplification Buffer (Invitrogen) with 0.0015% H2O2 for 15 min. Separate images were captured with the appropriate excitation wavelengths, and computer software (NIS Elements; Nikon, Tokyo, Japan) was used to superimpose the two images.
bpifcl mRNA expression analysis during development in male and female zebrafish
Zebrafish at 45 dpf (n = 13), 60 dpf (n = 12), 120 dpf (female, n = 8; male, n = 6) and 6 months old males (n = 10) and females (n = 10) were anaesthetised by immersing them in 0.01% benzocaine solution, followed by decapitation and fresh brain dissection. Standard lengths of the fish were not measured. Total RNA was isolated from whole brains with TRIzol reagent and converted to cDNA with a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Levels of β-actin, bpifcl and kiss2 mRNA were examined by real-time PCR. Real-time PCR was performed in 10 μL reaction mixtures containing 1X SensiFAST SYBR Master Mix (BioLine Reagent, London, UK), 0.2 μM of each forward and reverse primer and 1 μL of cDNA. The primers used are shown in the Supplementary Table 1. Real-time PCR was performed using a 7500 Fast PCR system (Applied Biosystems) with conditions of 95 °C for 10 min, 40 cycles of 95 °C for 15 s and 60 °C for 1 min, followed by a dissociation step. The levels of each mRNA type were normalised to β-actin mRNA using the ΔΔCt method.
Effect of E2 and 11-KT on bpifcl and kiss2 expression
6 months old male zebrafish were exposed to either 0.01% dimethyl sulfoxide (DMSO, Sigma) and 0.1 or 1.0 μg/L E2 in 0.01% DMSO for 48 h, and 6 months old female fish were exposed to 0.01% DMSO and 0.1 or 1.0 μg/L 11-KT in 0.01% DMSO. Fish were randomly selected and divided into three groups (n = 10 per group). The treatment was performed in tanks [280 mm (L) × 210 mm (W) × 180 mm (H)] containing 2 L of the above solution. During the experiment, the solution was changed after 24 h. The 0.1 μg/L dose of E2 was selected because it has been shown to affect the reproductive system of male zebrafish45, 46. The corresponding 11-KT concentrations were selected to match and compare with E2 doses. During exposure, fish were fed an Adult Zebrafish Diet (Zeigler Bros.,) twice daily. After exposure, β-actin, bpifcl, kiss2 and gnrh3 expression was examined as described above. The primers used are shown in the Supplementary Table 1.
Effect of knocked-down bpifcl expression on kiss2 mRNA expression
Custom-designed in vivo siRNA (Ambion), bpifcl-specific siRNA (sense, 5′-GGAACGAACCGUGAACAUAtt-3′; anti-sense, 5′-UAUGUUCACGGUUCGUUCCtg-3′) or GFP-specific siRNA (sense, 5′-GCAUCAAGGUGAACUUCAAtt 3′; anti-sense, 5′-UUGAAGUUCACCUUGAUGCcg-3′) was intracranially administered to the 6 months old zebrafish. bpifcl or GFP siRNA (60 pmol/µL) was mixed with the same volume of 10% glucose and 0.06 volume of Turbofect (Thermo Scientific, MA, USA) and incubated for 20 min at room temperature. Anaesthetised fish with 0.01% benzocaine solution (n = 10 per group) were placed on a sponge soaked with water, the skulls punctured with a 25 G × 1-in needle (Terumo, Tokyo, Japan) in the midline at the telencephalon-diencephalon border47 and 1.0 µL of the mixture intracranially injected. The fish were placed to recover in static water and fed an Adult Zebrafish Diet (Zeigler Bros.,) twice daily. After 48 h maintenance, β-actin, bpifcl and kiss2 expression were examined by real-time PCR as described above. GFP siRNA-injected fish were used as negative controls.
Statistical analysis
Statistical analysis of gene expression during development, by gender and for 11-KT and E2 exposure was performed using a one-way analysis of variance and Tukey’s post-hoc test for multiple comparisons using SPSS 20 software. A Student’s t-test was applied for sex differences and bpifcl knock-down experiments. p < 0.05 was considered significant.
References
Masson, D., Jiang, X. C., Lagrost, L. & Tall, A. R. The role of plasma lipid transfer proteins in lipoprotein metabolism and atherogenesis. J Lipid Res 50(Suppl), S201–206 (2009).
Bingle, L. & Bingle, C. D. Distribution of human PLUNC/BPI fold-containing (BPIF) proteins. Biochem Soc Trans 39, 1023–1027 (2011).
Beamer, L. J., Carroll, S. F. & Eisenberg, D. The BPI/LBP family of proteins: a structural analysis of conserved regions. Protein Sci 7, 906–914 (1998).
Bruce, C., Beamer, L. J. & Tall, A. R. The implications of the structure of the bactericidal/permeability-increasing protein on the lipid-transfer function of the cholesteryl ester transfer protein. Curr Opin Struct Biol 8, 426–434 (1998).
Bingle, C. D., Seal, R. L. & Craven, C. J. Systematic nomenclature for the PLUNC/PSP/BSP30/SMGB proteins as a subfamily of the BPI fold-containing superfamily. Biochem Soc Trans 39, 977–983 (2011).
Mogensen, T. H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev 22, 240–273, Table of Contents (2009).
Ghafouri, B., Stahlbom, B., Tagesson, C. & Lindahl, M. Newly identified proteins in human nasal lavage fluid from non-smokers and smokers using two-dimensional gel electrophoresis and peptide mass fingerprinting. Proteomics 2, 112–120 (2002).
Ghafouri, B., Kihlstrom, E., Tagesson, C. & Lindahl, M. PLUNC in human nasal lavage fluid: multiple isoforms that bind to lipopolysaccharide. Biochim Biophys Acta 1699, 57–63 (2004).
Kawano, K., Qin, S. C., Lin, M., Tall, A. R. & Jiang, X. C. Cholesteryl ester transfer protein and phospholipid transfer protein have nonoverlapping functions in vivo. J Biol Chem 275, 29477–29481 (2000).
Tu, A. Y., Nishida, H. I. & Nishida, T. High density lipoprotein conversion mediated by human plasma phospholipid transfer protein. J Biol Chem 268, 23098–23105 (1993).
Tong, Y. et al. Phospholipid transfer protein (PLTP) deficiency accelerates memory dysfunction through altering amyloid precursor protein (APP) processing in a mouse model of Alzheimer’s disease. Hum Mol Genet 24, 5388–5403 (2015).
Moriya, S., Soga, T., Wong, D. W. & Parhar, I. S. Transcriptome composition of the preoptic area in mid-age and escitalopram treatment in male mice. Neurosci Lett 622, 67–71 (2016).
Hearps, A. C. et al. Aging is associated with chronic innate immune activation and dysregulation of monocyte phenotype and function. Aging Cell 11, 867–875, doi:10.1111/j.1474-9726.2012.00851.x (2012).
Pasquier, J. et al. Comparative evolutionary histories of kisspeptins and kisspeptin receptors in vertebrates reveal both parallel and divergent features. Front Endocrinol (Lausanne) 3, 173 (2012).
Thompson, E. L. et al. Central and Peripheral Administration of Kisspeptin-10 Stimulates the Hypothalamic-Pituitary-Gonadal Axis. Journal of Neuroendocrinology 16, 850–858, doi:10.1111/j.1365-2826.2004.01240.x (2004).
Telegdy, G. & Adamik, A. The action of kisspeptin-13 on passive avoidance learning in mice. Involvement of transmitters. Behav Brain Res 243, 300–305 (2013).
Castellano, J. M. & Tena-Sempere, M. Metabolic regulation of kisspeptin. Adv Exp Med Biol 784, 363–383 (2013).
Castellano, J. M. et al. Acute inflammation reduces kisspeptin immunoreactivity at the arcuate nucleus and decreases responsiveness to kisspeptin independently of its anorectic effects. Am J Physiol Endocrinol Metab 299, E54–61 (2010).
Iwasa, T. et al. Hypothalamic Kiss1 and RFRP gene expressions are changed by a high dose of lipopolysaccharide in female rats. Horm Behav 66, 309–316 (2014).
Fang, L., Liu, C. & Miller, Y. I. Zebrafish models of dyslipidemia: relevance to atherosclerosis and angiogenesis. Transl Res 163, 99–108 (2014).
Curwen, V. et al. The Ensembl automatic gene annotation system. Genome Res 14, 942–950 (2004).
Kitahashi, T., Ogawa, S. & Parhar, I. S. Cloning and expression of kiss2 in the zebrafish and medaka. Endocrinology 150, 821–831 (2009).
Pradhan, A. & Olsson, P. E. Zebrafish sexual behavior: role of sex steroid hormones and prostaglandins. Behav Brain Funct 11, 23 (2015).
Maeda, Y., Fukushima, K., Nishizaki, K. & Smith, R. J. In vitro and in vivo suppression of GJB2 expression by RNA interference. Hum Mol Genet 14, 1641–1650 (2005).
Moriya, S., Takiguchi, M. & Seki, N. Expression of the WT1 gene -KTS domain isoforms suppresses the invasive ability of human lung squamous cell carcinoma cells. Int J Oncol 32, 349–356 (2008).
Urban-Klein, B., Werth, S., Abuharbeid, S., Czubayko, F. & Aigner, A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther 12, 461–466 (2005).
Gorelick, D. A., Watson, W. & Halpern, M. E. Androgen receptor gene expression in the developing and adult zebrafish brain. Developmental Dynamics 237, 2987–2995, doi:10.1002/dvdy.21700 (2008).
Izem, L., Greene, D. J., Bialkowska, K. & Morton, R. E. Overexpression of full-length cholesteryl ester transfer protein in SW872 cells reduces lipid accumulation. J Lipid Res 56, 515–525 (2015).
Dong, W., Albers, J. J. & Vuletic, S. Phospholipid transfer protein reduces phosphorylation of tau in human neuronal cells. J Neurosci Res 87, 3176–3185 (2009).
Asahina, K., Kobashi, T. & Soeda, H. Changes in Plasma Electrolyte and Hormone Concentrations during Homing Migration of Chum Salmon Oncorhynchus keta with Special Reference to the Development of Nuptial Color. Nippon Suisan Gakkaishi 57, 599–605, doi:10.2331/suisan.57.599 (1991).
Servili, A. et al. Organization of two independent kisspeptin systems derived from evolutionary-ancient kiss genes in the brain of zebrafish. Endocrinology 152, 1527–1540 (2011).
Brown, C. M., Xu, Q., Okhubo, N., Vitek, M. P. & Colton, C. A. Androgen-mediated immune function is altered by the apolipoprotein E gene. Endocrinology 148, 3383–3390 (2007).
Steven, C. et al. Molecular characterization of the GnRH system in zebrafish (Danio rerio): cloning of chicken GnRH-II, adult brain expression patterns and pituitary content of salmon GnRH and chicken GnRH-II. Gen Comp Endocrinol 133, 27–37 (2003).
Glinka, C. O. et al. The effects of model androgen 5alpha-dihydrotestosterone on mummichog (Fundulus heteroclitus) reproduction under different salinities. Aquat Toxicol 165, 266–276 (2015).
Forsgren, K. L., Qu, S., Lavado, R., Cwiertny, D. & Schlenk, D. Trenbolone acetate metabolites promote ovarian growth and development in adult Japanese medaka (Oryzias latipes). Gen Comp Endocrinol 202, 1–7 (2014).
Tang, H. et al. The kiss/kissr systems are dispensable for zebrafish reproduction: evidence from gene knockout studies. Endocrinology 156, 589–599 (2015).
Trudeau, V. L., Somoza, G. M., Nahorniak, C. S. & Peter, R. E. Interactions of estradiol with gonadotropin-releasing hormone and thyrotropin-releasing hormone in the control of growth hormone secretion in the goldfish. Neuroendocrinology 56, 483–490 (1992).
Zempo, B., Kanda, S., Okubo, K., Akazome, Y. & Oka, Y. Anatomical distribution of sex steroid hormone receptors in the brain of female medaka. J Comp Neurol 521, 1760–1780 (2013).
Kozak, M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15, 8125–8148 (1987).
Kozak, M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci USA 87, 8301–8305 (1990).
Letunic, I., Doerks, T. & Bork, P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Research 40, D302–D305, doi:10.1093/nar/gkr931 (2012).
Schultz, J., Milpetz, F., Bork, P. & Ponting, C. P. SMART, a simple modular architecture research tool: Identification of signaling domains. Proceedings of the National Academy of Sciences 95, 5857–5864 (1998).
Parhar, I. S., Ogawa, S. & Sakuma, Y. Laser-captured single digoxigenin-labeled neurons of gonadotropin-releasing hormone types reveal a novel G protein-coupled receptor (Gpr54) during maturation in cichlid fish. Endocrinology 145, 3613–3618 (2004).
Wulliman, M. F., Rupp, B. & Reichert, H. Neuroanatomy of the zebrafish brain: a topological atlas (Birkhäuser, 2012).
Fuzzen, M. L., Bernier, N. J. & Van Der Kraak, G. Differential effects of 17beta-estradiol and 11-ketotestosterone on the endocrine stress response in zebrafish (Danio rerio). Gen Comp Endocrinol 170, 365–373 (2011).
Brion, F. et al. Impacts of 17beta-estradiol, including environmentally relevant concentrations, on reproduction after exposure during embryo-larval-, juvenile- and adult-life stages in zebrafish (Danio rerio). Aquat Toxicol 68, 193–217 (2004).
Ogawa, S., Nathan, F. M. & Parhar, I. S. Habenular kisspeptin modulates fear in the zebrafish. Proceedings of the National Academy of Sciences 111, 3841–3846, doi:10.1073/pnas.1314184111 (2014).
Acknowledgements
We would like to thank to Kai We Ng, Rachel Shalini and Young Scholar Program students of Monash University Malaysia for their technical supports, and Enago (www.enago.jp) for the English language review. This work was supported by grant from ScienceFund of Ministry of Science, Technology and Innovation, Malaysia (02-02-10-SF0107).
Author information
Authors and Affiliations
Contributions
S.M. and I.S.P. designed the study. S.M. and N.T. performed the experiments and analyses. S.M. drafted the manuscript and figures. I.S.P. gave constructive comments of this work.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Moriya, S., Tahsin, N. & Parhar, I.S. Bpifcl modulates kiss2 expression under the influence of 11-ketotestosterone in female zebrafish. Sci Rep 7, 7926 (2017). https://doi.org/10.1038/s41598-017-08248-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-08248-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.