Abstract
In this study Surface Enhanced Raman Spectroscopy (SERS) data recorded from mouse mammary glands cancer cells (4T1 cell line) was used to assess information regarding differences between control, death and viable cells after Photodynamic Therapy (PDT) treatment. The treatment used nanoemulsions (NE/PS) loaded with different chloroaluminumphthalocyanine (ClAlP) photosensitizer (PS) contents (5 and 10 µmol × L−1) and illumination (660 nm wavelength) at 10 J × cm−2 (10 minutes). The SERS data revealed significant molecular alterations in proteins and lipids due to the PDT treatment. Principal Component Analysis (PCA) was applied to analyze the data recorded. Three-dimensional and well reproductive PCA scatter plots were obtained, revealing that two clusters of dead cells were well separated from one another and from control cluster. Overlap between two clusters of viable cells was observed, though well separated from control cluster. Moreover, the data analysis also pointed out necrosis as the main cell death mechanism induced by the PDT, in agreement with the literature. Finally, Raman modes peaking at 608 cm−1 (proteins) and 1231 cm−1 (lipids) can be selected for follow up of survival rate of neoplastic cells after PDT. We envisage that this finding is key to contribute to a quick development of quantitative infrared thermography imaging.
Similar content being viewed by others
Introduction
Worldwide, breast cancer is the most common cancer among women. In 2012, 1.67 million new cases were recorded, representing 25% of women diagnosed cancer1. Treatments most commonly adopted for breast cancer are surgery, radiotherapy, chemotherapy, or a combination of these therapies. Photodynamic therapy (PDT) emerges as a new and less aggressive approach that can replace or support conventional treatments2,3,4,5,6. Currently, the PDT is used for superficial cancer treatments such as mouth and skin, including breast cancer in Stage III while spread to chest wall or breast skin3, 6. However, emerging nanostructured materials, new photosensitizers or even deeply penetrating lasers can successfully extend the use of the PDT to breast cancer7. In situ production of oxidative species by a photosensitizer (PS) while optically excited is the basis of the PDT, as the PS is capable of convert light into chemical energy. In aerobic cells, the PS converts triplet O2 into singlet O2, the latter being a strong oxidant agent (reactive oxygen species) and thus able to cause enormous oxidative stress while generated in high amounts7. Such oxidative stress can be high enough to promote cellular death or generate significant responses to it. Living cells can react in different ways under stressing stimuli. It may activate survival mechanisms or undergo death when survival mechanisms fail to restore cell homeostasis. Initially, the cells’ response to stressing stimuli is focused on defense and/or recovery from damages8. However, if stress is too severe or prolonged cells will not be able to defend themselves, leading to activation of death mechanisms. How do cells respond to stressing situation depends on the cell type and level of stress. In case of cell death, the activation of a death pathway depends on cell’s ability to deal with exposed conditions, while surviving depends on cell’s ability to create an adequate response to stimulus8. Reactive oxygen species (ROS) are among the most potent threats faced by living cells, as they can damage major classes of biological macromolecules, including proteins, lipids, nucleic acids, and carbohydrates8. Therefore, when antioxidant cells defenses are overloaded, ROS can induce cell death, and the imbalance between oxidants and antioxidants determines not only cell fate, but also the death mechanism. Several studies show that ROS, such as superoxide anion (O 2 −), hydrogen peroxide (H2O2) and nitric oxide (NO) are capable of triggering cell death by apoptosis. However, ROS can also interfere with the apoptotic death program causing cells death by necrosis8. The anticancer mechanisms related to the PDT include direct induction of cancer cell death9,10,11, vascular shutdown with subsequent tumor ischemia12, and activation/stimulation of immune responses against tumor antigens3, 7. However, it is known that the PDT can also induce generation of survivor clones with more aggressive behavior, increasing the progression and aggressiveness of the tumor cells who survived the treatment3. In this regard it is of high interest to develop a robust protocol that quantitatively assess the fraction of generated resistant clones in the short follow up term of the PDT treatment.
Raman spectroscopy has demonstrated to be a powerful tool for identification and quantification of chemical species present in a sample, since the set of vibrational modes of a particular molecule or crystal is uniquely related to its chemical identity13, 14. Raman spectroscopy is extremely useful for a wide range of applications and it has been used in qualitative and quantitative analytical studies in the fields of chemistry, biology, geology, pharmacology, solid state physics among others15,16,17. In biology, Raman spectroscopy has been widely used for identification and quantification of biomolecules18,19,20. For example, the technique was successfully used to distinguish between cancerous and non-cancerous cells and tissues21, 22. However, conventional Raman spectroscopy presents limitations in the investigation of biosystems as many biomolecules have a low cross-section and intense fluorescence effects. Therefore, in recent years more advanced experimental approaches were introduced to overcome those limitations and, in this context, Surface Enhanced Raman Spectroscopy (SERS) emerged as a powerful and innovative analytical tool for the study of biological systems23, 24. The SERS effect may enhance the Raman signal by a factor that can exceed 106, occurring when the target molecule is absorbed or placed nearby nanostructured metal surfaces25, 26. Therefore, the SERS technique can be very much useful as a tool to investigate and discriminate cancer cells subjected to different therapies, in particular the PDT27,28,29. Despite the huge sensitivity provided by the SERS, spectral differences between cancerous and non-cancerous cells are very subtle, often hard to notice by direct analysis of the spectra, thus requiring a more robust approach that can be provided by statistical analysis. In this context, Raman spectroscopy combined with multivariate analysis, such as Principal Component Analysis (PCA), has been successfully used to distinguish different types of cancer cells and even different stages of cancer30,31,32.
In the present study, we report on results of SERS and PCA combined in the investigation of mouse adenocarcinoma breast cancer cells (4T1 line) treated with the PDT. A nanoemulsion, comprising monodisperse nanodroplets of castor oil containing the PS chloroaluminumphthalocyanine (NE/ClAlPc) and stabilized by Cremophor ELP® 33, was fabricated and used in the PDT protocol to treat breast cancer cells (4T1 line) cultured in vitro. For the PDT treatment (see Fig. 1a) the nanoemulsion was prepared with two different PS concentrations, namely 5 and 10 µmol × L−1 of ClAlPc and labeled NE/PS5 and NE/PS10 complexes, respectively. After the PDT treatment the cells were separated into two groups (see Fig. 1b: (i) viable cells, which were kept attached to the bottom flask and (ii) dead cells, considered here as the cells in suspension within the culture medium. Untreated cells were used as control group and the five cell groups were frozen in liquid Nitrogen for further SERS measurements). Electrodeposited Silver films were used for the SERS evaluation whereas the multivariate analysis was performed by PCA (see Fig. 1c).
Results
Figure 2 shows the average SERS spectra recorded from the 4T1 cells after the PDT treatment using both NE/ClAlPc formulations (NE/PS5 and NE/PS10). The average SERS spectrum of the control group is also presented in Fig. 2 (lower spectrum). Thirty spectra were recorded from samples collected from each cell group and were normalized to their integrated area in the range of 400–1800 cm−1. All spectra in Fig. 2 show signatures of well-defined patterns, displaying cellular fingerprints consisting of proteins, lipids, and nucleic acid complexes34.
Average SERS spectra of 4T1 breast cancer cells for control group and cells treated with the NE/PS5 and NE/PS10 complexes. Each spectrum represents an average of 30 spectra recorded from cellular samples. In the low energy region (400–1100 cm−1) the band intensities were multiplied by 10. The inserted pictures emphasize the vibrational modes related to the protein disulfide bonds (-S-S-) at ~525 cm−1 and DNA fragmentation responsible for the SERS intensity variation associated with phosphodiester bonds (~813 cm−1) and guanine/adenine bases (~1565 cm−1).
Overall, the SERS spectra shown in Fig. 2 are very much similar, with slight variations in intensity and peak position. In general, for vibrational modes around 765 cm−1 (Trp), 1122 cm−1 (CN peptide), 1173 cm−1 (Tyr), 1250 cm−1 (amide III), 1337 (CH proteins) and 1623 cm−1 (Trp/Tyr), the SERS spectra of the cells treated with the PDT protocol (viable and dead) show Raman shifts to higher energy when compared to the control group. The assignments of the vibrational modes were based on a search in the literature34,35,36,37 and are summarized in Table 1. It is worth mentioning that we found higher Raman shifts for vibrational modes associated with the dead cells groups and lower Raman shifts associated with the viable cells groups. In addition, most of the bands revealing Raman shifts to higher energy are associated with protein vibrational modes.
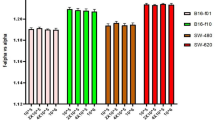
Important to note in the SERS spectra of the dead cells a new Raman band peaking at 813 cm−1 in addition to intensity reduction of the 1565 cm−1 Raman band associated with DNA vibration (phosphodiester bonds O-P-O in the backbone of DNA) and adenine/guanine bases, respectively (see light-blue strip lines in Fig. 2)38. Interestingly, the new 813 cm−1 Raman band is more intense in the spectra of the cell group treated with the NE/PS5 complex compared to the NE/PS10 complex. Moreover, in comparison with the control group the SERS spectra of all PDT treated groups showed increased intensity of the Raman features related to proteins, namely at 456 cm−1 (Phe), 608 cm−1 (Phe), 846 cm−1 (Tyr), 1003 cm−1 (Phe), 1030 cm−1 (Phe), and 1500 cm−1 (Phy). Actually, we found this intensity increase is even higher for the viable cells groups. However, no intensity increase pattern was observed for the Raman bands peaking at 520 and 549 cm−1, both associated with protein disulfide bonds (-S-S-) vibrations. In this case (520 and 549 cm−1 features), the intensity of the Raman bands remained nearly constant for both viable cells groups and revealed a strong intensity reduction for both dead cells groups (~40% and 60% for dead cells groups treated with the NE/PS5 and NE/PS10 complexes, respectively). Finally, intensity reduction of vibrational modes associated with lipids and peaking at 593 cm−1 (PtdIns), 718 cm−1 (CN fat), 973 cm−1 (CH lipids), 1051 cm−1 (CC lipids), 1231 cm−1 (CH lipids) and 1434 cm−1 (CH lipids) were also observed. For comparison, Fig. 3(a,b) collect the relative Raman peak intensity data of the bands discussed above and related to proteins and lipids, respectively. Likewise, Fig. 3(c,d) show spectral details related to selected vibrational modes of proteins and lipids, respectively.
While comparing with the control group, the SERS spectral variations of viable cells groups (treated with the NE/PS5 and NE/PS10 complexes) are higher for proteins and lower for lipids (see Fig. 3), indicating some kind of cellular stress after contact between lipid nanodroplets and cell membrane. With respect to the control group, the average total integrated areas of vibrational modes related to proteins increased 42% for the dead cells groups (35% and 50% for treatments using the NE/PS5 and NE/PS10 complexes, respectively) and 80% for the viable cells groups (85% and 75% for the NE/PS5 and NE/PS10 complexes, respectively). On the other hand, the reduction of the total integrated areas related to vibrational modes of lipids decreased on average 35% (32% and 37% for treatments using the NE/PS5 and NE/PS10 complexes, respectively) and 20% (18% and 22% for the NE/PS5 and NE/PS10 complexes, respectively) for the dead and viable cells groups, respectively. The SERS spectral variation related to vibrational modes of nucleic acids were also lower for the viable cells groups compared with the dead cells groups. The key difference we found is the Raman peak at 813 cm−1 observed only in the SERS spectra of the dead cells groups and assigned to DNA vibration.
To determine whether SERS can be used or not to distinguish from the control group the cells treated (viable and dead cells groups) with the PDT protocol (using the NE/PS5 and NE/PS10 complexes), Principal Component Analysis (PCA) was employed to analyze the recorded spectra. The covariance matrix was constructed from 150 SERS spectra (all spectral points - 30 spectra for each cell group) to determine the variance between different cells groups. Thus, as in Fig. 2, the principal components (PCs) in the range of 400–1800 cm−1 were extracted after baseline correction and normalization with respect to the spectral areas. A typical example of the variance spectrum is shown in Fig. 4 for the control group and the PDT treated cells groups (viable and dead), where solid lines represent spectral averages.
Figure 5(a,b) collect the ratio of Raman peak areas (SERS) related to viable/dead cells (A viable /A dead ) for the vibrational modes explored in Fig. 3 and related to proteins and lipids, respectively. Note that the data in Fig. 5(a,b) were organized for cells treated using both complexes NE/PS5 and NE/PS10.
Figure 6 shows the dispersion points using the first three PCs, which represent variances exceeding 41%. Notice the formation of well-defined sets in the PC space, corresponding to control cells group (black squares), dead cells groups (open and solid triangles for the NE/PS5 and NE/PS10 complexes, respectively), and viable cells groups (open and solid circles for the NE/PS5 and NE/PS10 complexes, respectively). In comparison with the control cells group Fig. 6(a,b) show the first three PCs, representing 45% (dead cells groups) and 43% (viable cells groups) of data variance, respectively. Worth mentioning that while the clusters of each dead cells groups are well separated there is an overlap between the two groups of viable cells treated with different PS doses (NE/PS5 and NE/PS10 complexes). Comparison between the control group and the cells groups (dead and viable) treated with the NE/PS5 and NE/PS10 complexes are shown in Fig. 6(c,d), respectively. Finally, Fig. 6e collects the data regarding all the studied cells groups. Important to stress that while the two clusters describing viable and dead cells show a slight overlap for the PDT treatment using the NE/PS5 complex the two clusters describing viable and dead cells treated with the NE/PS10 complex are quite separated (see Fig. 6e).
The PCA scatter plots of the 4T1 breast cancer cells showed in Fig. 6 demonstrate that the SERS spectra of all cells groups are reproductive even among spectra recorded from different groups or among spectra within the same cell group. This finding reinforces the need of a statistical approach as shown in Fig. 2, which incorporates robustness in the analysis of variation of integrated areas related to the vibrational modes of proteins and lipids.
Discussion
Raman spectroscopy has been employed in identification of death mechanisms of various types of cells, such as leukemia cells39, T lymphocytes40, 41 and melanoma cells (MEL-28)42, among others35, 43. In all these reports physical effects, chemotherapeutic agents or other pharmaceutical substances were used to induce cell death events variants that showed different impacts onto cell molecular constitution. All those studies have found significant differences between Raman spectra of treated cells compared with untreated ones (control). In a recent report, Brauche et al.43 observed intensity increase in Raman peaks associated with DNA (at 795 and 1375 cm−1) in cells that underwent apoptosis and necrosis induced by exposing at different temperatures. However, the vibrational modes the authors found at 786 and 1315 cm−1, also assigned to DNA, showed opposite behavior, i.e. intensity reduction in both cases43. The authors43 also found that the Raman peak at 1375 cm−1 was practically absent in spectra recorded from control cells and enhanced in spectra of dead cells due to cellular nucleus condensation, which is associated with both cell death mechanisms. This conclusion was based on observations reported in other publications40, 41. Additionally, it was observed that the intensity of the 1003 cm−1 Raman peak, which is associated with phenylalanine, was increased in necrotic cells while being reduced in apoptotic cells. Moreover, the intensity of the 1437 cm−1 Raman peak, associated with lipids, was increased in apoptotic cells and reduced in necrotic ones. Notingher et al.34 and Verrier et al.35 also observed similar results while studying cell death via apoptosis. Both studies observed intensity reduction of Raman peaks associated with DNA (~788 cm−1) and proteins (1005, 1322, and 1342 cm−1) and intensity increase of Raman peaks associated with lipids (1301, 1449, and 1660 cm−1). Authors in refs 42 and 43 reported similar findings while investigating necrotic cells; reduction of Raman intensity peaks associated with lipids (935, 1085, and 1448 cm−1) and increase of Raman intensity peaks associated with proteins (642, 875, 1230, 1395, and 1425 cm−1). However, Raman peaks related to vibrational modes of DNA/RNA did not show similar behavior, i.e. intensity increase of Raman features peaking at 781, 985 and 1575 cm−1 and intensity reduction of Raman peaks at 667, 990 and 1305 cm−1. Despite the divergence of behavior observed for Raman bands of DNA/RNA it is possible to conclude that apoptosis is characterized by intensity decrease of Raman modes associated with proteins and intensity increase of Raman modes related to lipids. Moreover, necrosis leads to intensity increase of Raman modes associated with proteins and intensity decrease of bands related to lipids34, 35, 43.
Unfortunately, few reports discuss the possible origins of different trends related to Raman intensity variation observed in vibrational modes of lipids and proteins within cells. Verrier et al.35 argued that Raman intensity decrease associated with proteins, and observed during apoptosis, is due to reduction of protein content in apoptotic cells caused by activation of caspases. Worth mentioning that Brauche et al.43 proposed a similar explanation. In this line of argument, Raman intensity increase observed in the vibrational modes of proteins during necrosis is likely due to the release of proteins from necrotic nuclei into the cytosol. Also, according to those authors, the intensity increase observed in the 1375 cm−1 Raman signal may indicate changes in the architecture of chromatin triggered by HMGB1 release (chromatin binding protein that is relocated in necrotic cells). On the other hand, Kunapareddy et al.42 suggested that Raman intensity decrease associated with lipids would be consistent with coagulation necrosis. In necrosis, destruction of cytoplasmic structures and degradation of cell membranes occur in the first 24 hours and may be responsible for the observed reduced lipid content42. Thus, Raman intensity increase associated with proteins is directly correlated to the spectral analysis. In this regard all the recorded spectra were normalized, thereby decrease observed in one spectral component (e.g. lipids) leads to increase of another one (e.g. proteins). Despite this observation, the authors mentioned that there are spectral evidences indicating changes in protein conformation or composition.
Based on the above-presented background and taking into account the results shown in Fig. 2, revealing SERS intensity increase related to proteins and intensity decrease associated with lipids, it is possible to argue that necrosis is the main process responsible for cell death of the 4T1 breast cancer cells groups treated with PDT using the NE/PS5 and NE/PS10 complexes. This finding is in agreement with results reported in previous study10, where the efficacy of the PDT treatment of cancer cells (MCF7 and 4T1) and non-cancer cells (NIH/3T3 and MCF-10A) mediated by ClAlPc associated with polymeric nanoparticles (poly(methyl vinyl ether-co-maleic anhydride)) was investigated. Moreover, the authors reported that the employed PDT treatment induced cell death mainly by apoptosis when cells were irradiated with low energy density (0.48 J × cm−2) and necrosis when irradiated with high energy density (1.38 J × cm−2)10. For tumor cells following necrosis the loss of cell membrane integrity was confirmed by ethidium bromide staining10, a result in agreement with lipid reduction. Worth mentioning at this point that the energy density used in the present study was 10 J × cm−2, thus supporting the necrosis hypothesis as the dominant cell death mechanism in our case.
Though vibrational modes related to DNA/RNA do not exhibit an unique behavior while probing cell death by apoptosis or necrosis intensity reduction of vibrational modes associated with phosphodiester bonds (O-P-O) of DNA structure was recently assigned to cellular apoptosis38. In this report, the authors used Gold nanoparticles to enhance Rayleigh and Raman signal in human oral cancer cells (SH-3) to investigate cellular apoptosis induced by hydrogen peroxide treatment. During apoptosis the intensity of the vibrational mode peaking at 838 cm−1 (O-P-O) decreased while the intensity of the Raman feature peaking at 1585 cm−1 (N7-H in guanine/adenine bases) increased. According to the authors this trend is associated with fragmentation of DNA structure in the phosphodiester bonds, thus exposing DNA bases to the Gold nanoparticles plasmonic field. Our findings differ from those reported in ref. 38. as we found the 813 cm−1 peak intensity increasing while reducing the 1565 cm−1 band intensity, suggesting that DNA fragmentation mainly occurs by breakage of base pairs rather than due to the breaking of phosphodiester bond. Separation of base pairs exposes the backbone of the DNA dorsal to the Silver substrate, resulting in enhancement of the Raman signal (SERS effect). The highest intensification of the SERS signal for the 813 cm−1 band was observed in the dead cells group treated with the NE/PS5 complex, which is assumed to be due to the highest degree of DNA fragmentation in this cellular group. In this case, in addition to separation of the DNA base pairs, breakage of the phosphodiester bonds may also occurred. Moreover, besides the spectral changes associated with DNA fragmentation, in the SERS spectra of the dead cells groups it was also observed a intensity decrease of the Raman peaks at 520 and 549 cm−1, associated with vibrations of disulfide bonds (-S-S-) of proteins. This behavior can be explained considering the breakdown of intramolecular disulfide bonds responsible for proteins tertiary/quaternary folding38.
It is well known from the literature that the key biological catalyst of disulfide bonds is ERO-α (endoplasmic reticulum oxidoreductase alpha) enzyme, which is produced in endoplasmic reticulum44. A recent study conducted with breast cancer patients reported a significant increase of the ERO-α enzyme activity in 4T1 breast cancer cells while compared to non-cancerous cells/tissues45. Therefore, it is expected that cancer cells show high concentration of biomolecules presenting disulfide bonds. Additionally, it was noticed a drastic reduction of tumor growth in 4T1 breast cancer cells when they are deprived of the ERO-α46. The authors concluded that the ERO-α enzyme contributes to the suppression of anti-tumor immunity by regulating G-CSF cytokines and CXCL1/2 via facilitating proteins folding, suggesting that inhibition of the ERO-α enzyme may difficult cancer progression.
Figure 4(a,b) show the ratio A viable /A dead collected for different vibrational modes associated to proteins and lipids while using the NE/PS5 and NE/PS10 complexes, respectively. Worth mentioning that even under 10 J/cm2 illumination, either using the NE/PS5 complex or the NE/PS10 complex, cells undergo necrosis as previously reported9. Moreover, the higher the PS doses the lower the viable cell population after performing the PDT. Therefore, vibrational modes peaking at 608, 846, 1003 and 1500 cm−1 for proteins and 718 and 1231 cm−1 for lipids are reasonable candidates for monitoring cell population ratio as shown in Fig. 4(a,b), respectively. Among the selected vibrational modes for monitoring viable and dead cells the highest discrimination is 608 cm−1 for proteins and 1231 cm−1 for lipids, as shown in Fig. 4(a,b), respectively. Additionally, note in Fig. 4(a,b) the pair of vibrational modes peaking at 608 cm−1 (proteins) and 1231 cm−1 (lipids) seems to be a good candidate for monitoring viable/dead cell population, given the values of A viable /A dead below 2 (NE/PS5) and 1 (NE/PS10) and above 2 (NE/PS5) and 1 (NE/PS10) for proteins and lipids, respectively. This observation fully agrees with increasing SERS line intensity for proteins and decreasing SERS line intensity for lipids contents in cells undergoing necrosis. Therefore, we claim that the corresponding pair of vibrational modes (for proteins and lipids) are potentially the best candidates for quantitative follow up of cell survival after the PDT treatment employed in the present study. This finding is a key point while using spatial infrared (IR) thermography imaging in oncology47, 48. In the context of thermographic IR imaging quantitative capability represents the forefront trend of research in this area, particularly because of the non-invasive characteristic of the technique and advances in computing and image-analysis software in the last decades47, 48. However, identification of IR bands that can be used to enhance the IR imaging performance is challenging, and the present report offers a robust approach to fulfill this requirement. In this regard, a previous screening of promising vibrational modes that can offer superior discrimination of neoplastic cells can be performed using the approach introduced in the present study. In principle, the approach herein proposed can be extended to a wide variety of neoplastic cells and tissues, thus fostering the development and transferring the spatial infrared thermography imaging technology from the bench to the clinic.
Though extremely useful for future cancer diagnosis development and follow up protocols the explanation of the observed SERS spectral variations among control cells, viable cells and dead cells groups is far from to be a simple task. However, using a robust statistical tool, we can infer that significant intensity increase in the Raman vibrational modes associated with proteins in viable cells may be related to activation of intracellular defense mechanisms. Among these mechanisms it has been highlighted alteration on protein synthesis encoded by genes stimulated or suppressed during cellular stress caused by the PDT treatment3. In this process, genes encoding inducing signals of cell proliferation promotes production of proteins (growth factors, growth factor receptors, transcription factors, and apoptosis regulators) responsible for cell proliferation. When cell proliferation is stimulated several molecular mechanisms are activated in an attempt to control the process of cell division. New proteins are synthesized to account for those mechanisms, such as the cyclin proteins. In dead cells, the production of proteins is suppressed and, therefore, dead cells show less protein content while compared to viable cell. Thus, the significant intensity increase we found in the Raman bands assigned to proteins in the viable cells, as observed in the recorded SERS spectra, are likely related to activation of defense mechanisms induced by oxidative stress due to the employed PDT treatment. A careful analysis of the SERS spectra shown in Fig. 2 reveals that it is possible to observe spectral variations among the three cells groups studied here (control, dead, and viable) and between the two cells groups treated with different PS doses (NE/PS5 and NE/PS10 complexes). Furthermore, the employed PCA tool adds robustness to our conclusions. Figure 6(a,e) show that the data points collected from the dead cells fall in well separated clusters, according to the PS dosage received (treatment with the NE/PS5 or NE/PS10 complex). Moreover, the data points from the dead cells group treated with the NE/PS10 complex (solid triangles) are located closer to the control group than the dead cells group treated with the NE/PS5 complex (open triangles). This finding may indicate that the dead cells group treated with the NE/PS10 complex undergoes less biochemical changes before the death mechanism is triggered. On the other hand, in the viable cells groups it is observed a superposition of data points on the PCs space, as shown in Fig. 6(b,e). However, compared with the viable cells group treated with the NE/PS5 complex we found that the data points related to the viable cells group treated with the NE/PS10 complex is farther from the data points related to the control group. As far as the employed PDT protocol is concerned this finding may indicate that the viable cells group presents higher biochemical change when treated with the highest PS doses (NE/PS10 complex).
Conclusion
Surface Enhanced Raman Spectroscopy (SERS) was successfully used to investigate breast cancer cells treated with photodynamic therapy (PDT). The photosensitizer (PS) used in the PDT treatment was formulated as nanoemulsions (NE/PS) with two different PS (chloroaluminumphthalocyanine) contents (5 and 10 µmol × L−1). Analysis of the SERS data using Principal Component Analysis (PCA) showed that the employed PDT treatment with different PS concentrations leads to significant biochemical changes related to proteins, lipids, and DNA. Changes in SERS intensity related to proteins and lipids indicated necrosis as the main cellular death mechanism after the PDT treatment. Also, compared to untreated cells (control group), the SERS spectra collected from dead cells groups showed significant changes in the vibrational modes of DNA. The increased intensity of vibrational modes of phosphodiester bond (O-P-O) plus the intensity reduction of the SERS bands related to the guanine/adenine bases suggested that after the PDT treatment DNA fragmentation mainly occurs by breakage of its base pairs. Additionally, while compared with control group, the SERS data recorded from viable cells groups showed that the significant intensity increase of the vibrational modes related to proteins are likely associated with cellular defense mechanisms induced by oxidative stress due to the PDT treatment. Moreover, the PCA analysis indicated that biochemical changes observed in dead cells treated with the NE/PS10 complex was lower than changes observed in dead cells treated with the NE/PS5 complex. This finding can be understood as long as the death cell mechanism (necrosis) in cells treated with the former (NE/PS10 complex) is triggered at earlier times compared with the treatment performed with the latter (NE/PS5 complex). Finally, we found the SERS modes 608 and 1231 cm−1, ascribed respectively to proteins and lipids, are the best candidates for follow up of 4T1 cells survival subjected to PDT treatment. We claim that this finding is a key contribution to the development of quantitative infrared thermography imaging.
Methods
Nanoemulsion of ClAlPc
Nanoemulsions containing the photosensitizer (PS: ClAlPc) were prepared from solubilization of ClAlPc in ethanol 99 °GL at 1.7 mmol × L−1. Then, 12 g of a mixture comprising 75% of Cremophor ELP® and 25% of castor oil was added to the previous ClAlPc solution, producing a mixture containing 40 µmol × L−1 of ClAlPc. Next, the ethanol was removed at 100 °C under gentle magnetic stirring (300 rpm for 15 minutes). After cooling the mixture down to room temperature 70 mL of culture medium was added under mild agitation for extra 15 minutes. Then, culture medium was added to fix the final ClAlPc concentration at 5 and 10 µmol × L−1 of AlClPc and the final complexes were labeled NE/PS5 and NE/PS10, respectively.
Culture and treatment of breast cancer cells
Mouse mammary glands cancer cells (line 4T1 ATCC® CRL-2539) were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum and 1% antibiotic solution (100 IU mg × L−1 penicillin and 100 mg × mL−1 streptomycin). The cells were maintained at 37 °C in a humidified atmosphere of 5% CO2 for 24 hours. Then, the cells were divided into two main groups: control group (kept under the same growing condition for another 24 hours) and treated groups (submitted to the PDT treatment using NE/PS5 or NE/PS10 complex). Afterwards, all cells groups were frozen in liquid Nitrogen.
For the PDT treatment, cells were separated into two groups and treated with the NE/PS5 or NE/PS10 complex. In both cases, cells were exposed to the complexes for 15 minutes in dark and washed twice with phosphate-buffered saline (PBS). Next, cells were irradiated with LED lamp (660 nm) at an energy density of 10 J × cm−2 for 10 minutes. After treatment, cells were cultured for 24 hours under the culture condition described above. After 24 hours supernatant cells (dead cells) were separated from surviving cells (viable cells) and both groups were frozen in liquid Nitrogen.
SERS substrate and sample preparation for SERS measurement
Electrodeposited films of Silver were used as SERS substrate. The Silver films were prepared by electrodeposition of Silver ions onto Silver electrodes. After cleaning and polishing the Silver electrodes were immersed within a solution of 200 mL distilled water containing 22 mg of AgNO3. Next, the as-produced electrodes were subjected to a potential difference of 3 V, electric current of approximately 1000 µA for a period of 1 hour. Then, the negative electrodes (SERS substrates) were immediately dried under Nitrogen flow.
The frozen cells were ground and stirred to obtain a liquid and homogeneous solution. Then, 10 µL of the cells were deposited onto the Silver film previously prepared. Next, the films were dried under Nitrogen flow and kept within sealed containers with plastic film until they were used for measurements. Raman measurements were performed shortly after the probing samples were prepared (up to 4 hours after preparation) to avoid film’s oxidation and cells’ degradation.
Instrumentation
For acquisition of Raman spectra a triple spectrometer Jobin Yvon model T64000 equipped with a 1800 groves/mm grating and a CCD (Charge-Coupled Device) 2048 × 512 pixels cooled Nitrogen was used. The SERS measurements were performed with the macro-Raman system, in which the laser beam was focused onto the sample using a cylindrical lens with an excitation power of 0.1 W × cm−2, to prevent sample heating and damaging cells. The 488 nm Argon laser line was used as the excitation source.
Data analysis
Analysis of the Raman data was performed with the LabSpec 4.10 software. The background signal was subtracted from the recorded Raman spectra and the resulting data were individually normalized with respect to the integrated area in the range of 400–1800 cm−1. To improve analysis of spectral differences among the five cells groups SERS measurements were carried out on 30 samples for each group. Next, Principal Component Analysis (PCA) was employed in the analysis of all 150 spectra (30 spectra for each 5 cells groups). The PCA was implemented within the MINITAB® software49.
Data Availability
All data generated or analysed during this study are included in this published article.
References
INTERNATIONAL AGENCY FOR RESEARCH ON CANCER - IARC, Cancer Incidence and Mortality Worldwide: CancerBase. Lyon, France, 2013; http://globocan.iarc.fr/, accessed: January, 2016.
Reibel, J. Prognosis of oral pre-malignant lesions: significance of clinical, hitopathological, and molecular biological characteristics. Critical Rev. Oral Biol. Med. 14, 47–60 (2003).
Agostinis, M., Tucci, P. & Melino, G. Cell death pathology: Perspective for human diseases. Biochem. Biophys. Res. Commun. 414, 451–455 (2011).
Wei, Y., Zhou, F., Zhang, D., Chen, Q. & Zing, D. A graphene oxide based smart drug delivery system for tumor mitochondria-targeting photodynamic therapy. Nanoscale 8, 3530 (2016).
Lovell, J. F., Liu, T. W. B., Chen, J. & Chen Zheng, G. Activatable Photosensitizers for Imaging and Therapy. Chem. Rev. 110, 2839–2857 (2010).
Monge-Fuentes, V., Muehlmann, L. A. & Azevedo, R. B. Perspectives on the application of nanotechnology in photodynamic therapy for the treatment of melanoma. Nano Rev. 5, 24381 (2014).
Dougherty, T. J. et al. Photodynamic Therapy. J. Natl. Cancer Inst. 12, 889–905 (1998).
Fulda, S., Gorman, A. M., Hori, O., Samali, A. Cellular Stress Responses: Cell Survival and Cell Death. Int. J. Biochem. Cell. Biol. 2010 (2010).
Muehlmann, L. A., Joanitti, G. A., Silva, J. R., Longo, J. P. F. & Azevedo, R. B. Liposomal photosensitizers: potential platforms for anticancer photodynamic therapy. Braz. J. Med. Biol. Res. 44, 729–737 (2011).
Muehlmann, L. A. et al. Aluminum–phthalocyanine chloride associated to poly(methyl vinyl ether-co-maleic anhydride) nanoparticles as a new third-generation photosensitizer for anticancer photodynamic therapy. Int. J. Nanomedicine 9, 1199–1213 (2014).
Longo, J. P. F. et al. Effects of photodynamic therapy mediated by liposomal aluminum-phthalocyanine chloride on chemically induced tongue tumors. Chemotherapy 1(103), 2 (2012).
Mijan, M. C. et al. Vascular Shutdown and Pro-inflammatory Cytokine Expression in Breast Cancer Tumors after Photodynamic Therapy Mediated by Nano-sized Liposomes Containing Aluminium-Chloride-Phthalocyanine. J. Nanomed. Nanotechnol. 5, 1–9 (2014).
Si, M. Z., Kang, Y. P. & Zhang, Z. G. Surface-enhanced Raman scattering (SERS) spectra of Methyl Orange in Ag colloids prepared by electrolysis method. Appl. Surf. Sci. 255, 6007–6010 (2009).
da Silva, S. W. et al. Effect of the Zn content in the structural and magnetic properties of ZnxMg1−xFe2O4 mixed ferrites monitored by Raman and Mössbauer spectroscopies. J. Appl Phys. 107, 09B503 (2010).
Aragon, F. H. et al. Evidences of the evolution from solid solution to surface segregation in Ni-doped SnO2 nanoparticles using Raman spectroscopy. J. Raman Spectrosc. 42, 1081–1086 (2011).
Chem, H. J., Wang, Y. L., Qu, J. Y. & Dong, S. J. Self-assembled silver nanoparticle monolayer on glassy carbon: an approach to SERS substrate. J. Raman Spectrosc. 38, 1444–1448 (2007).
The, S. K. et al. Diagnostic potential of near-infrared Raman spectroscopy in the stomach: differentiating dysplasia from normal tissue. Br. J. Cancer, BJC. 98, 457–465 (2008).
Haka, A. S. et al. Diagnosing breast cancer by using Raman spectroscopy. Proc. Natl. Acad. Sci. 102, 12371–12376 (2005).
de Gelder, J., De Gussem, K., Vandenabeele, P. & Moens, L. Reference database of Raman spectra of biological Molecules. J. Raman Spectrosc. 38, 1133–1147 (2007).
Van de Vondel, E., Mensch, C. & Johannessen, C. Direct Measurements of the Crowding Effect in Proteins by Means of Raman Optical Activity. J. Phys. Chem. B 120, 886–890 (2016).
Abramczyk, H., Brozek-Pluska, B., Surmacki, J., Jablonska, J. & Kordek, R. The label-free Raman imaging of human breast câncer. J. Mol. Liq. 164, 123–131 (2011).
Brozek-Pluska, B. et al. Raman spectroscopy and imaging: applications in human breast cancer diagnosis. Analyst. 137, 3773–3780 (2012).
da Paz, M. C. et al. Anti-CEA loaded maghemite nanoparticles as a theragnostic device for colorectal cancer. Int. J. Nanomedicine 7, 5271–5282 (2012).
Santos, C. M. B. et al. SERS as a valuable tool for detection and treatment follow-up of fungal infection in mice lungs: use of amphotericin B and its nanoencapsulation onto magnetic nanoparticles. J. Raman Spectrosc. 44, 695–702 (2013).
Albrecht, M. G. & Creighton, J. A. Anomalously intense Raman spectra of pyridine at a silver electrode. J. Am. Chem. Soc. 99, 5215–5217 (1977).
Fleischmann, M., Hendra, P. J. & Mcquillan, A. J. Raman spectra of pyridine adsorbed at a silver electrode. Chem. Phys. Lett. 26, 163–166 (1974).
Lee, S. et al. Rapid and sensitive phenotypic marker detection on breast câncer cells using surface-nhanced Raman scattering (SERS) imaging. Biosens. Bioelectron. 51, 238–243 (2014).
Maiti, K. K. et al. Multiplex targeted in vivo cancer detection using sensitive near-infrared SERS nanotags. Nano Today. 7, 85–93 (2012).
Hoonejani, M. R., Pallaoro, A., Braun, G. B., Moskovits, M. & Meinhart, C. D. Quantitative multiplexed simulated-cell identification by SERS in microfluidic devices. Nanoscale 7, 16834 (2015).
Thakur, J. S. et al. Raman spectral signatures of mouse mammary tissue and associated lymph nodes: normal, tumor and mastitis. J. Raman Spectrosc. 38, 127–134 (2007).
Oshima, Y., Shinzawa, H., Takenaka, T., Furihata, C. & Sato, H. Discrimination analysis of human lung cancer cells associated with histological type and malignancy using Raman spectroscopy. J. Biomed. Opt. 15, 017009–08 (2010).
Su, L. et al. Raman Spectral Properties of Squamous Cell Carcinoma of Oral Tissues and Cells. Laser Physics. 22, 311–316 (2012).
Muehlmann, L. A. et al. Aluminium-phthalocyanine chloride nanoemulsions for anticancer photodynamic therapy: Development and in vitro activity against monolayers and spheroids of human mammary adenocarcinoma MCF-7 cells. J. Nanobiotechnology 13(1), 36 (2015).
Notingher, I. et al. Discrimination between ricin and sulphur mustard toxicity in vitro using Raman spectroscopy. J. R. Soc. Interface 1, 79–90 (2004).
Verrier, S., Notingher, I., Polak, J. M. & Hench, L. L. In Situ Monitoring of Cell Death Using Raman Microspectroscopy. Biopolymers 74, 157–162 (2004).
Zhou, J. et al. Surface-enhanced Raman spectroscopy investigation on human breast cancer cells. Chem Cent J. 7, 37 (2013).
Binoy, J. et al. NIR-FT. Raman and FTIR spectral studies and ab initio calculations of the anticancer drug combretastatin A4. J. Raman Spectrosc. 35, 939–946 (2004).
Kang, B., Austin, L. A. & El-Sayed, M. A. Observing Real-Time Molecular Event Dynamics of Apoptosis in Living Cancer Cells using Nuclear-Targeted Plasmonically Enhanced Raman Nanoprobes. ACS Nano 8, 4883–4892 (2014).
Chan, J. W. et al. Nondestructive Identification of Individual Leukemia Cells by Laser Trapping Raman Spectroscopy. Anal. Chem. 80, 2180–2187 (2008).
Pupples, G. J. et al. Laser Irradiation and Raman Spectroscopy of Single Living Cells Chromosomes: Sample Degradation Occurs with 514.5 nm but not with 660 nm Laser Light. Exp Cell Res. 195, 361–367 (1991).
Pully, V. V., Lenferink, A. T. M. & Otto, C. Time-lapse Raman imaging of single live lymphocytes. J. Raman Spectrosc. 42, 167–173 (2011).
Kunapareddy, N., Freyer, J. & Mourant, J. Raman spectroscopic characterization of necrotic cell death. J. Biomed. Opt. 13, 054002 (2008).
Brauche, E., Thude, S., Brucker, S. Y. & Schenke-Layland, K. Cell death stages in single apoptotic and necrotic cells monitored by Raman microspectroscopy. Sci. Rep. 4, 4698 (2014).
Araki, K. & Nagata, K. Functional in vitro analysis of the ERO1 protein and protein-disulfide isomerase pathway. J. Biol. Chem. 286, 32705–32712 (2011).
Kutomi, G. et al. Human endoplasmic reticulum oxidoreductin 1-α is a novel predictor for poor prognosis of breast cancer. Cancer Sci. 104, 1091–1096 (2013).
Tanaka, T. et al. Oxidative Protein Folding Myeloid-Derived Suppressor Cells via Drives the Production of Tumor-Promoting Cancer-Associated Oxidoreductase ERO1-α. J. Immunol. 194, 2004–2010 (2015).
Pirtini, M. C. & Herman, C. Quantification of the thermal signature of a melanoma lesion. Int. J. Therm. Sci. 50, 421–431 (2011).
Ring, E. F. J. & Ammer, K. Infrared thermal imaging in medicine. Physical Meas. 33(3), R33 (2012).
Minitab, Inc. “MINITAB statistical software.” Minitab Release, 13 (2000).
Acknowledgements
Financial support from the Brazilian agencies CNPq (Grant no. 304252/2012-3), FINEP (Grant No. PRO0745/10), CAPES Grant no. 150442/2012-2), FAPDF, and FINATEC is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
S.W.S. conceived the project and planed the experiments. A.B.V., J.P.F.L. and B.F.T. performed the experiments, L.A.M., P.E.N.S., R.B.A. fabrication of the photosensitizer and samples. S.W.S. and P.C.M. prepared the manuscript. All authors discussed the results and comment on the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Veloso, A.B., Longo, J.P.F., Muehlmann, L.A. et al. SERS Investigation of Cancer Cells Treated with PDT: Quantification of Cell Survival and Follow-up. Sci Rep 7, 7175 (2017). https://doi.org/10.1038/s41598-017-07469-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-07469-1
This article is cited by
-
Characterization of multifunctional nanocomposites assembled from Ag-decorated Fe3O4 and Eu-doped Y2O3
Journal of the Korean Physical Society (2022)
-
Surface enhanced Raman scattering artificial nose for high dimensionality fingerprinting
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.