Abstract
Klebsiella pneumoniae, an Enterobacteriaceae that mostly causes hospital-acquired infections, belongs to the recently published WHO’s list of antibiotic-resistant pathogens that pose the greatest threat to human health. Indeed, K. pneumoniae is the enterobacterial species most concerned by both resistance to extended-spectrum cephalosporins, due to extended-spectrum β-lactamase (ESBL) production, and resistance to carbapenems, i.e. the β-lactams with the broadest activity. Carbapenem resistance is related not only to carbapenemase production, but also the production of ESBL or AmpC and the loss of general porins. Here, we characterized the mechanisms that deprived a urinary ESBL-producing, porin-deficient K. pneumoniae isolate, isolated 13 days after the end of a 40-day course of imipenem treatment, of its carbapenem resistance. These mechanisms were observed in two in-vivo derivatives of this isolate and consisted of mutations in genes encoding molecules that participate in the downregulation of the synthesis of PhoE, a porin specialized in phosphate transport. We obtained three new derivatives from one of the two original derivatives, following in-vitro antibiotic pressure, in which the carbapenem resistance was restored because of mutations in genes encoding molecules that participate in the upregulation of PhoE synthesis. Thus, we uncovered novel mechanisms of carbapenem resistance/susceptibility switching in K. pneumoniae.
Similar content being viewed by others
Introduction
Klebsiella pneumoniae is a Gram-negative pathogen responsible for infections that are mainly hospital-acquired1, 2. It was included in the recently published WHO’s list of antibiotic-resistant pathogens that pose the greatest threat to human health (http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/), because of its rapid and efficient capacity to develop both multidrug resistance (MDR)3 and extensive drug-resistance (XDR)4. MDR K. pneumoniae isolates consist of those resistant to extended-spectrum cephalosporins, due to extended-spectrum β-lactamase (ESBL) production. XDR isolates consist of those that are also resistant to carbapenems i.e. the latest generation of β-lactam molecules that possess the broadest spectrum of activity. Two distinct mechanisms that cause resistance to carbapenems have been identified: acquisition of specific enzymes that hydrolyze carbapenems, called carbapenemases, such as KPC, OXA-48, and NDM5, and the combination of ESBL and/or AmpC production and alterations of porin synthesis6, 7. The latter is currently the dominant resistance mechanism in various countries8,9,10. However, alterations of porin synthesis have also been described in carbapenemase-producing K. pneumoniae isolates11, 12, suggesting that the changes in the composition of outer membrane porins which control membrane permeation and the ß-lactam accumulation in the periplasmic space13 also plays a key role in the β-lactam resistance displayed by XDR K. pneumoniae isolates.
Three consecutive isolates that differed from each other by three carbapenem-susceptibility phenotypes were isolated from a single patient in our hospital during treatment. The first isolate displayed non-carbapenemase-related carbapenem resistance following 40 days of imipenem treatment. The second was susceptible to all carbapenems following 7 days of tigecycline treatment. The third displayed heterogeneous susceptibility to carbapenems following 7-days of tigecycline-colistin treatment. The aim of this study was to identify and characterize the genetic events that occurred in these three urinary isolates responsible for their carbapenem susceptibility. We showed that (i) the three isolates were isogenic strains, each possessing the mechanisms responsible for carbapenem resistance displayed by the first isolate and (ii) the second and third isolates displayed mutations in the pstSCAB-phoU genes that control the Pho regulon. We demonstrated that these mutations were involved in the upregulation of the synthesis of the porin PhoE and the complete and partial susceptibility to carbapenems displayed by the second and third isolates, respectively. We subjected the second cefoxitin and carbapenem-susceptible isolate to in-vitro cefoxitin and carbapenem selection pressure and showed that resistance to these two antibiotics was restored in the selected mutants due to mutations in the phoB-phoR operon, leading to the downregulation of PhoE synthesis. The involvement of phoE expression in carbapenem susceptibility had been suggested by Kaczmarek et al. following the concomitant isolation of two K. pneumoniae strains producing plasmid-mediated AmpC, devoid of general porins, and either susceptible or resistant to carbapenems14. However, the authors did not identify the genetic cascades and molecular mechanisms involved in the carbapenem resistance/susceptibility switches.
Materials and Methods
Patients and bacterial strains
The patient was a 63-year-old woman of Cambodian origin, who had been living in France for 20 years. She was hospitalized at Beaujon Hospital (AP-HP, Clichy, France) on April 2009 for a fifth recurrent urinary tract infection (UTI). She had been under treatment for disseminated (pulmonary, urinary, and bone) tuberculosis for the previous eight months. Bilateral ureteral damage necessitated the insertion of double J stents in January 2009, which were changed in March 2009. All episodes of recurrent UTI, including that of April 2009, were caused by ESBL-producing K. pneumoniae isolates. These infections required iterative treatment with carbapenems. For the fifth episode, the patient received imipenem from April 15 to May 25, 2009, including 10 days after ablation of the double J stents, believed to be responsible for the recurrent UTIs. In June 2009, the patient presented three other recurrences of UTI, demonstrated in the absence of symptoms by a systemic inflammatory response (C-reactive protein = 100 mg/L) and positive urine cultures from systematically collected samples on June 7, 14, and 21 (Fig. 1). The three isolates, called strains BJ-STC-a, BJ-STC-b, and BJ-STC-c, were tested by the agar-diffusion method and interpreted following the recommendations of the French Antibiogram Committee applicable in 2009. They were found to be resistant to extended-spectrum cephalosporins due to ESBL production, demonstrated by the double-disk synergy test15, and gentamicin, amikacin, chloramphenicol, cotrimoxazole, and fluoroquinolones (Table 1). They differed from each other by susceptibility to four antibiotics (Table 1): strain BJ-STC-a was resistant to cefoxitin and ertapenem, had intermediate susceptibility to imipenem, and was susceptible to tigecycline, whereas strain BJ-STC-b was susceptible to the four antibiotics, and strain BJ-STC-c was susceptible to imipenem and resistant to cefoxitin, ertapenem, and tigecycline. Following the isolation of strain BJ-STC-a, the patient received tigecycline (Fig. 1). After seven days of this treatment, strain BJ-STC-b, susceptible to tigecycline, was isolated. Therefore, colistin (MIC: 1 mg/l determined by E test) was added to the treatment (Fig. 1), with doses adapted to patient renal function (1 MU in IM injection every two days). Although strain BJ-STC-c was isolated under the combined treatment, this treatment was maintained until July 16, 2009. A urine analysis performed on July 27, 2009 showed only the presence of vaginal flora and the absence of K. pneumoniae. Strains BJ-STC-a, BJ-STC-b, and BJ-STC-c were studied with strain ATCC 13883T used as a control.
Antibiotic treatment schedule, strain isolation dates, and in-vitro strain selection. Phenotypic and genotypic characteristics of the strains are indicated, as well as the selection frequency of the in-vitro mutants. Red indicates carbapenem-resistant strains and green indicates strains susceptible to at least one carbapenem. R: resistant; S: susceptible; Carba: carbapenems; FOX: cefoxitin; ERT: ertapenem; IMI: imipenem; TIG: tigecycline.
Genome sequencing and analysis
The complete genomic sequence was determined for strains BJ-STC-a, BJ-STC-b, and BJ-STC-c. Total DNA was extracted using the Qiagen Blood & Cell Culture DNA Mini Kit (Qiagen, Courtaboeuf, France). Libraries were constructed using Nextera technology and sequenced on an Illumina HiSeq-2000 using a 2 × 100 nucleotide (nt) paired-end strategy. All reads were processed to remove low quality or artefactual nucleotides, using sequentially sickle (github.com/najoshi/sickle), AlienTrimmer16, and fqDuplicate (ftp.pasteur.fr/pub/gensoft/projects/fqtools). Read pairs were assembled using clc assembler from the CLC Genomics Workbench analysis package (www.clcbio.com/products/clc-genomics-workbench). All contigs of ≥500 nt were reordered and reoriented, using the genomic sequence of strain NTUH-K2044 as reference, with Mauve Contig Mover17. The reordered contigs were subsequently imported into the Bacterial Isolate Genome Sequence Database (BIGSdb) platform18 and analyzed for the presence of known resistance-associated genes, as well as MLST and cgMLST profiles19. Resistance genes were also sought using ResFinder with default parameters (https://cge.cbs.dtu.dk/services/ResFinder)20. The putative IS identified in the genome were characterized using ISFinder (www-is.biotoul.fr). Plasmid content was determined using PlasmidFinder (https://cge.cbs.dtu.dk/services/PlasmidFinder) and plasmid type by using pMLST (https://cge.cbs.dtu.dk/services/pMLST)21. The reads corresponding to strains BJ-STC-b and BJ-STC-c were mapped on the contigs of strain BJ-STC-a, to search for differences between the three genomes, using the Quality-based Variant Detection tool of CLC Genomics Workbench 7.0.3 with default parameters.
Mutant selection
The mutants BJ-STC-b-MFOX, BJ-STC-b-MERT, and BJ-STC-b-MIMP were obtained from strain BJ- STC-b by in vitro selection using cefoxitin, ertapenem, and imipenem, respectively, following the procedure described previously22.
Sequence studies
We used Sanger sequencing, with the primers listed in Table S1 and previously described PCR and sequencing procedures22 to (i) confirm the sequences of the ompK35, ompK36, pstS, phoU, and oqxR genes, and the rarA and oqxA inter-region, and (ii) determine the sequences of the phoB and phoR genes.
Analysis of gene expression by real-time RT-PCR
The expression of the genes encoding porins (ompK35, ompK36, and phoE), as well as that of genes encoding regulators and molecules involved in the efflux pumps OqxAB (rarA and oqxB) and AcrAB (ramA and acrB) was determined by real-time RT-PCR, as described previously22, 23, except for the determination of total RNA concentrations, which was performed using the Qubit RNA BR Assay Kit (Thermo Fischer Scientific, Illkirch, France). All primers used are listed in Table S1.
Cloning of the oqxR, pstS, phoU, phoR, and phoB genes and complementation experiments
The oqxR gene from strain ATCC 13883T and the pstS, phoU, phoR, and phoB genes of strain BJ-STC-a were cloned into the pCR™8/GW/TOPO® vector (encoding resistance to spectinomycin) following the manufacturer’s instructions (Life Technologies SAS, Saint-Aubin, France). The empty plasmid pCR™8/GW/TOPO®, used as a negative control, was generated from pCR™8/GW/TOPO®-pstS after digestion with EcoRI enzyme (Ozyme, Saint-Quentin-en-Yvelines, France) and ligation using the LigaFast Rapid DNA Ligation System (Promega, Charbonnières-les-Bains, France). After verification of the constructs by sequencing, the recombinant plasmids were electroporated into competent bacterial cells of strains BJ-STC-a for oqxR, BJ-STC-b for pstS, BJ-STC-c for phoU, BJ-STC-b-MFOX for phoR, and BJ-STC-b-MERT and BJ-STC-b-MIMP for phoB. Transformants were selected on LB agar plates containing 100 mg/L spectinomycin.
Antibiotic susceptibility testing
Minimum inhibitory concentrations (MICs) were determined in triplicate by the broth microdilution method according to the guidelines of EUCAST24 and interpreted according to the 2016 CASFM-EUCAST recommendations (www.sfm-microbiologie.org). The following antibiotics were tested in addition to ertapenem and imipenem: cefoxitin, temocillin, tigecycline, chloramphenicol, and colistin. These antibiotics were tested because resistance to them can be associated with defined resistance mechanisms (e.g. overexpression of efflux pumps for cefoxitin, chloramphenicol, and tigecycline resistance), because they were not tested (temocillin), or were tested using an inappropriate method (E-test for colistin) in 2009 when the three strains were isolated. A difference between MICs was considered to be significant after two dilutions (i.e. a 4-fold difference in MIC).
Immunodetection of outer membrane proteins
To prepare outer membrane protein as previously described25, each strain was grown in three different broth media: Mueller Hinton II (MH II), nutrient broth (NB) with low osmotic strength, increasing ompK35 gene expression, and nutrient broth containing sorbitol (NBS) with high osmotic strength, increasing ompK36 gene expression. The same amounts of protein of the final preparations were separated on SDS-PAGE gels. Briefly, samples were solubilized in loading buffer at 96 °C to totally denature the various protein complexes and samples (corresponding to an optical density of 0.02 at 600 nm), loaded on SDS-PAGE gels (10% polyacrylamide, 0.1% SDS) and run at 160 V for 90 mn. Electrotransfer of the resulting bands to nitrocellulose membranes was carried out in 0.05% SDS. After an initial saturating step in Tris-buffered saline (TBS: 50 mM Tris-HCl, 150 mM NaCl, pH 8) containing 10% skimmed milk powder overnight at 4 °C, nitrocellulose membranes were incubated in TBS containing skimmed milk powder and 0.2% Triton X-100 for 2 h at room temperature in the presence of polyclonal antibodies directed against OmpA, OmpC, or OmpF proteins. Our previous studies demonstrated strong cross-immunoreactivity between E. coli and K. pneumoniae porins26. Polyclonal antibodies directed against E. coli OmpC and OmpF denatured porins have been previously used for the efficient detection of K. pneumoniae porins in various clinical and laboratory strains26, 27. The detection of resulting antigen-antibody complexes was performed with horseradish peroxidase secondary antibody conjugated goat anti-rabbit IgG antibodies28 and revelation was performed using a Chemidoc XRS+ (BioRad, Marnes La Coquette, France).
Maximal growth rate
For comparative growth assays, strain ATCC 13883T and the various BJ-STC strains were grown overnight (O/N) at 37 °C in Luria-Bertani (LB: 10 g tryptone, 5 g yeast extract, 10 g/L NaCl) in flasks with constant stirring at 280 rpm. Then, O/N cultures were prediluted in LB to obtain an OD600 = 0.5 and the strains inoculated into two different wells each at a dilution of 1/10,000 in a Costar® 96 flat-bottomed well plate. The plates were incubated at 37 °C with stirring and growth was recorded by an Infinite 200 Tecan®, which measured the OD600 in each well every 5 min, for 24 h. Growth assays were repeated three times. The maximum growth rate (MGR) was calculated from the resulting growth curves. The OD600 values in nm were collected and Log-transformed. Curves were calculated from a smoothed spline function. The MGR was defined as the time point at which the maximum value of the derivative of the smoothed function was observed. The MGRs of strains BJ-STC-b, BJ-STC-c, and ATCC 13883T were compared to that of strain BJ-STC-a, allowing determination of the relative growth rate (RGR). Tukey’s test was used for intergroup comparisons and R software for statistical analyses. P values < 0.05 were considered to be statistically significant.
Nucleotide sequence accession number
The annotated genomic sequences of strains BJ-STC-a, BJ-STC-b, and BJ-STC-c were submitted to the ENA public sequence repository and are available under project accession number PRJEB14454.
Results
Genome analysis and clonal relatedness of the BJ-STC-a, BJ-STC-b, and BJ-STC-c strains
The genome sequences of strains BJ-STC-a, BJ-STC-b, and BJ-STC-c were assembled into 103, 134, and 106 contigs, respectively. MLST analysis performed on whole genome sequence data showed that the three strains belonged to sequence type (ST) 340. Pairwise gene-by-gene comparisons following the cgMLST scheme revealed no differences between strains BJ-STC-a, BJ-STC-b, and BJ-STC-c for the 694 cgMLST genes except for one (the folE gene). The folE gene was incomplete in the genome assembly of strain BJ-STC-c and therefore no allele was called. Thus, the three strains were isogenic.
Phenotype and genotype of antibiotic resistance
Strains BJ-STC-a, BJ-STC-b, and BJ-STC-c displayed a similar phenotype of resistance to various antibiotic families (Tables 1 and 2), including extended-spectrum cephalosporins, temocillin, aminoglycosides (gentamicin and amikacin), tetracycline, chloramphenicol, sulphonamide, trimethoprim, and fluoroquinolones. The three strains were susceptible to colistin. Finally, they differed from each other with respect to resistance to carbapenems and cefoxitin (ertapenem and cefoxitin resistance in strains BJ-STC-a and BJ-STC-c and susceptibility to these antimicrobial agents in strain BJ-STC-b; non-susceptibility to imipenem in strain BJ-STC-a and susceptibility in strains BJ-STC-b and BJ-STC-c). They also differed with respect to tigecycline susceptibility, as we observed resistance only in strain BJ-STC-c. Whole genome analysis using ResFinder and BIGSdb revealed the following resistance encoding genes in the three strains: bla SHV-11, bla TEM-1, bla OXA-1, bla CTX-M-15 (β-lactam resistance), aph(3″)-Ib and aph(6)-Id (streptomycin resistance), aac(3)-IId (gentamicin and tobramycin resistance), aac(6′)Ib-cr (amikacin and fluoroquinolone resistance), tet(D) (tetracycline resistance), a truncated chloramphenicol acetyl-transferase-encoding catB3 gene, sul2 (sulphonamide resistance), and dfrA14 (trimethoprim resistance). Mutations in the quinolone resistance determining region (QRDR) were present in genes gyrA (S83I) and parC (S80I) for the three strains. We detected no carbapenemase-encoding genes in the whole genome sequences of the three strains. We also did not detect the mutation leading to the Val57Leu amino acid substitution in the S10–30S ribosomal protein, considered to be involved in tygecycline resistance, in strain BJ-STC-c, which displayed resistance to tygecycline29.
Plasmid content
Genome sequence analysis showed three plasmid replicons: IncFIIk, IncFIA, and IncR in the three strains of BJ-STC. The pMLST profile displayed by the IncF replicons was K1:A13:B-.
OmpF and OmpC porin profiles (RT-PCR, sequence, immunodetection)
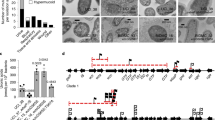
Carbapenem resistance can be due to porin alteration and ESBL production. Thus, we assessed the expression level of the general porins. The three BJ-STC strains expressed the ompK36 and ompK35 genes at approximately the same level, which was much lower than in strain ATCC 13883T (Table 2). Comparison of genomic sequence data with the genome of reference strain MGH78578 (GenBank: CP000647.1) showed that the ompK36 gene was similarly mutated in strains BJ-STC-a, BJ-STC-b, and BJ-STC-c. An insertion of a thymidine at position 4 of this gene resulted in a stop codon directly after the start codon, preventing the synthesis of a functional OmpK36 protein. The ompK35 gene presented a deletion of one of the three G nucleotides located at position 871 (out of 927 bps) in the three strains, leading to a shift in the reading frame. Moreover, an IS of the IS1 family was located seven nucleotides upstream from the start codon of the ompk35 gene in the three strains. The immunodetection experiments (Fig. 2 and Figure S1) showed that neither OmpK35 nor OmpK36 were produced by strain BJ-STC-a, regardless of the broth used for bacterial growth. We detected a band which migrated faster than that of wildtype OmpK36 in strains BJ-STC-b and BJ-STC-c, for which the synthesis of OmpK35 and OmpK36 was also altered. It was detected in strain BJ-STC-b when grown in MH II and NB broths (Fig. 2 and Figure S1) and in strain BJ-STC-c when grown in NB broth (Fig. 2). These results show the expression of another antigenically-related porin recognized by anti-porin antibodies in strains BJ-STC-b and BJ-STC-c.
Immunodetection of OmpK35 and OmpK36 synthesis. This detection was performed using polyclonal antibodies directed against denatured OmpC (OmpK 36 in Klebsiella pneumoniae) porin. Anti-OmpC antibodies are also able to detect denatured OmpF (OmpK35 in K. pneumoniae) porin due to cross-recognition. The tested strains, BJ-STC-a, BJ-STC-b, BJ-STC-c, BJ-STC-b MFOX, and BJ-STC-b MFOX T phoRwt, were grown in Mueller Hinton II (MH2), nutrient broth (NB), or nutrient broth containing sorbitol (NBS) media. Only the relevant part of the blots is shown.
Genome comparison and phoE expression
The three genomic sequences were compared by performing read mapping analysis to define the genetic basis of imipenem-susceptibility in strains BJ-STC-b and BJ-STC-c, and imipenem-resistance in strain BJ-STC-a. We detected potentially meaningful differences in the pstS and phoU genes, which regulate the expression of genes encoding phosphate transporters30, 31. We identified the mutation G825A (Fig. 3), resulting in the replacement of Trp at position 275 by a stop codon, in the pstS gene of strain BJ-STC-b, whereas the phoU gene of strain BJ-STC-c harbored mutation G78A (Fig. 3), resulting in a Met26Ile substitution. We subsequently verified the two mutations identified by genomic sequence analysis by Sanger sequencing. We measured the level of phoE transcripts, as the pstS and phoU genes regulate the expression of the phoE gene. The expression of phoE in strain BJ- STC-a was comparable to that of the reference strain ATCC 13883T, whereas it was 36-fold and almost 5-fold overexpressed in BJ-STC-b and BJ-STC-c, respectively (Table 2 and Fig. 2).
Pho regulon and location of the mutations detected in the various strains. The various molecules encoded by the pstSCAB and phoU-B genes involved in the control of Pho regulon expression are indicated (adapted from reference Gardner 43). The green and red stars represent the mutations identified that activate or repress the phoE expression, respectively.
In vitro reproduction of the carbapenem-resistant phenotype in derivatives of strain BJ-STC-b
Growth of the cefoxitin- and carbapenem-susceptible strain BJ-STC-b in the presence of cefoxitin resulted in the selection (3 × 10−7) of a mutant, BJ-STC-b-MFOX, with a β-lactam susceptibility pattern identical to that of strain BJ-STC-a: an eight-fold increase in the MIC of cefoxitin and imipenem and a 512-fold increase in the MIC of ertapenem (Table 2). The synthesis of PhoE was downregulated in this mutant, shown by RT-PCR and immunodetection (Table 2, Fig. 2). Sanger sequencing of the phoB and phoR genes, which are involved in the regulation of phoE expression, revealed a G742T mutation (Fig. 3) responsible for a premature stop codon (Glu248Stop) in the phoR gene of strain BJ-STC-b-MFOX. We used ertapenem and imipenem as selectors to obtain derivatives from strain BJ-STC-b, called BJ-STC-b-MERT (2 × 10−4) and BJ-STC-b-MIMP (4 × 10−6), respectively, that exhibit the same β-lactam susceptibility profile as strain BJ-STC-b-MFOX (Table 2). In these mutants, the expression of the phoE gene was also downregulated (Table 2) and we identified mutations within the regulator phoB gene: stop codons replaced the amino acid Gln at position 41 (C121T) in strain BJ-STC-b-MERT, and amino acid Glu at position 130 (G388T)) in strain BJ-STC-b-MIMP (Fig. 3).
Complementation experiments
We assessed the role of the mutations located in the pstS, phoU, phoR, and phoB genes in the modification of phoE gene expression by cloning the corresponding genes amplified from the parental BJ-STC-a strain and using them to transform strains BJ-STC-b, BJ-STC-c, and the derivatives of BJ-STC-b. Complementation had an impact on both phoE expression and susceptibility to three of the seven antibiotics tested, namely ertapenem, imipenem, and cefoxitin (Table 2). In strains BJ-STC-b T pstSwt and BJ-STC-c T phoUwt, phoE expression was downregulated and the MICs of ertapenem, imipenem, and cefoxitin increased to reach the values observed in strain BJ-STC-a. Conversely, phoE expression was upregulated and the MICs of cefoxitin, imipenem, and ertapenem decreased in strains BJ-STC-b-MFOX T phoRwt, BJ-STC-b-METP T phoBwt, and BJ-STC-b-MIMP T phoBwt. However, the increase of phoE expression was less marked in strains BJ-STC-b-METP T phoBwt and BJ-STC-b-MIMP T phoBwt than in strain BJ-STC-b-MFOX T phoRwt (Table 2).
OqxAB and AcrAB efflux pump expression
The resistance to chloramphenicol observed in the three strains, in the absence of an intact cat gene, and the marked increase in the tigecycline MIC in strain BJ-STC-c, in the absence of modification of the S10–30S ribosomal protein, led us to analyze the genes encoding the OqxAB and AcrAB efflux pumps, for which overexpression is known to contribute to the resistance to these antibiotics23. Transcript levels of the oqxB gene and that of the rarA gene, encoding a positive regulator of the expression of the oqxB gene, were higher in strains BJ-STC-a, BJ-STC-b, and BJ-STC-c than in the reference strain ATCC 13883T (Table 2). The Val130Ala substitution in the oqxR gene, i.e. the repressor of both oqxAB and rarA gene expression, was present in all three strains. However, in strain BJ-STC-a T oqxRwt , i.e. strain BJ-STC-a complemented with the wild type gene oqxR of strain ATCC 13883T, the expression of the oqxB and rarA genes was only partially downregulated. This suggests that an additional mechanism, other than the Val130Ala substitution, was involved in both the overexpression of the oqxB and rarA genes in strain BJ-STC-a and the partial downregulation of these two genes in strain BJ-STC-a T oqxRwt (Table 2). We identified another genetic event in the three strains, namely the insertion of an additional G in the seven-G motif located between the rarA and oqxR genes at positions −123 to −129 upstream of the start codon of the rarA gene, corresponding to the DNA-binding domain of the winged helix-turn-helix of OqxR32. As a result, the MIC of chloramphenicol, a substrate of the OqxAB efflux pump, was slightly lower in strain BJ-STC-a T oqxRwt than in strain BJ-STC-a (Table 2).
Although each BJ-STC strain harbored an additional lysine (related to a T580A mutation) at the very end of RamR, the repressor of acrB and ramA expression, expression of these two genes was appreciably higher in strain BJ-STC-c only (Table 2). This overexpression was due to the insertion of ISKpn18, belonging to the IS3 family, in the ramR gene. Therefore, the MIC of tigecycline, which is a substrate of the AcrB efflux pump and not of the OqxAB efflux pump, was significantly higher in strain BJ-STC-c (4 mg/L) than in strains BJ-STC-a and BJ-STC-b (0.5 mg/L) (Table 2). In contrast, the MIC of chloramphenicol, which is a substrate of both the AcrAB and OqxAB efflux pumps, was not significantly higher in strain BJ-STC-c than in strains BJ-STC-a and BJ-STC-b.
Relative maximal growth rate (MGR)
The MGR of the three BJ-STC strains were significantly lower than that of strain ATCC 13883T (p < 0.001) (Fig. 4). However, the relative MGR of strain BJ-STC-b was non-significantly higher than that of strains BJ-STC-a and BJ-STC-c. The partial decrease in the OqxAB pump activity in strain BJ-STC-a T oqxRwt, and the repression of the PhoE expression in the complemented strains BJ-STC-b T pstSwt and BJ-STC-c T phoUwt, as well as the BJ-STC-b derivatives (BJ-STC-b-MFOX T phoRwt, BJ-STC-b-METP T phoBwt and BJ-STC-b-MIMP T phoBwt), did not result in relative MGRs significantly different from those displayed by the respective parental strains.
Relative maximal growth rate. The relative growth rate is expressed as the average of the three growth determinations ± standard deviation. a: strain BJ-STC-a; a-ToqxRwt: strain BJ-STC-a complemented with the wild type oqxR gene; b: strain BJ-STC-b; b-TpstSwt: strain BJ-STC-b complemented with the wild type pstS gene; b-MFOX: BJ-STC-b mutant selected with cefoxitin; b-MFOX-TphoRwt: BJ-STC-b mutant selected with cefoxitin and complemented with the wild type phoR gene; b-METP: strain BJ-STC-b mutant selected with ertapenem; b-METP-TphoBwt: strain BJ-STC-b mutant selected with ertapenem and complemented with the wild type phoB gene; b-MIMP: BJ-STC-b mutant selected with imipenem; b-MIMP-TphoBwt: BJ-STC-b mutant selected with imipenem and complemented with the wild type phoB gene; c: strain BJ-STC-c; c-TphoUwt: strain BJ-STC-c complemented with the wild type phoU gene.
Discussion
The involvement of combined AmpC overproduction and porin loss in the resistance to imipenem following imipenem treatment has been known since 199133. Since the middle of the 2000s, not only AmpC, but also ESBL, combined with porin loss was found to be responsible for carbapenem resistance34,35,36. This is certainly because of the commercialization and laboratory testing of ertapenem, of which the activity is more strongly affected by synergistic mechanism-mediated carbapenem resistance than that of imipenem37, 38. The first urinary K. pneumoniae strain (BJ-STC-a) studied herein, which was resistant to carbapenems, resembled those described in various UK hospitals in 2009 in terms of ESBL type produced (CTX-M-15) and genetic events leading to the loss of the general porins OmpK35 and Ompk36 (IS1 or nucleotide insertion)37. However, contrary to the UK strains, strain BJ-STC-a also displayed overexpression of the OqxAB efflux pump, known to expel various antibiotics (i.e cefoxitin, chloramphenicol, and fluoroquinolones), generating low level resistance to these antibiotics23. This was the sole mechanism responsible for chloramphenicol resistance in strain BJ-STC-a, whereas other mechanisms contributed to its resistance to cefoxitin (porin loss) and fluoroquinolones (target mutations). Strain BJ-STC-a displayed an ST rarely described until recently, ST34039,40,41, which is a member of clonal group 258, to which the worldwide KPC-producing strains belong42. The originality of our study is the analysis of two urinary strains sequentially isolated after a seven-day treatment with tigecycline (strain BJ-STC-b) and a seven-day treatment with tigecycline-colistin (strain BJ-STC-c). The study showed that the BJ-STC-a, BJ-STC-b, and BJ-STC-c strains were isogenic, all equipped with the various resistance mechanisms characterized in strain BJ-STC-a, notably those causing resistance to carbapenems. However, the second and the third strains displayed a carbapenem susceptibility profile different from that of the first strain. We demonstrated that the susceptibility of the second strain, BJ-STC-b, to carbapenems, as well as cefoxitin, was caused by a mutated pstS gene, leading to constitutive expression of the phoE gene. A clinical isolate of K. pneumoniae devoid of general porins, producing ESBL, and susceptible to both carbapenems and cefoxitin because of upregulation of PhoE synthesis cannot be phenotypically distinguished from one widely susceptible to carbapenems and cefoxitin. Thus, physicians are likely to use these antibiotics to treat infections due to the former strains. This led us to grow strain BJ-STC-b in the presence of cefoxitin, ertapenem, or imipenem in vitro. We obtained a phoR mutant when cefoxitin was used whereas two different phoB mutants were obtained when ertapenem and imipenem were used. These mutations led to both downregulation of the phoE gene and restoration of cefoxitin and carbapenem resistance in the two mutant types because PhoR, an inner-membrane histidine kinase sensor protein, and PhoB, a response regulator that acts as a DNA-binding protein to active or inhibit gene transcription, are the elements of the two-component regulatory system that governs the Pho regulon (Fig. 3)30, 43. Our results resemble those obtained by Kaczmarek. et al.14 in which they grew a carbapenem-susceptible, AmpC-producing, porin-deficient K. pneumoniae isolate in the presence of various carbapenems. Indeed, they found that PhoE synthesis was downregulated in the derivatives displaying carbapenem resistance. However, contrary to our study, these authors did not identify the molecular mechanisms responsible for the downregulation of PhoE synthesis. They also did not show why the parental strain was susceptible to carbapenems, whereas it produced an AmpC enzyme and had lost OmpK35 and Ompk36.
In the third strain, BJ-STC-c, which was resistant to both ertapenem and cefoxitin, but susceptible to imipenem, we found constitutive expression of the phoE gene, but at a lower level than that observed in strain BJ-STC-b. We found a mutation in the phoU gene, of which the product negatively regulates the signaling pathway of inorganic phosphate and modulates its transport through the PstSCAB proteins (Fig. 3)43. We used complementation experiments to show that this mutation was linked to phoE overexpression, susceptibility to imipenem, and the decrease in the ertapenem MIC relative to the parental BJ-STC-a strain. We postulate that the non-susceptibility to ertapenem found in strain BJ-STC-c, relative to the ertapenem susceptibility found in strain BJ-STC-b, was related to the different amount of ertapenem that accumulated in the periplasm due to the different levels of PhoE upregulation observed in these two strains. The persistence of a high cefoxitin MIC observed for strain BJ-STC-c, but not for strain BJ-STC-b, despite higher phoE overexpression, was related to the presence of the overexpression of the AcrAB efflux pump, known to expel cefoxitin, in strain BJ-STC-c. This efflux pump, in contrast to the OqxAB efflux pump, also expels tigecycline, explaining the resistance of strain BJ-STC-c to tigecycline22, 23. Finally, we found identical high MIC values of temocillin towards the three BJ-STC strains and the BJ-STC-b derivatives, suggesting that the activity of temocillin is not influenced by the Pho regulon.
The genomic study indicates that the BJ-STC isolates are isogenic strains, suggesting that they were concomitantly present in the urinary tract of the patient, who required double J stents for five months and received repeated imipenem treatments. We postulate that the suspected biofilm on the ureteral device and the constitutive activation of the Pho regulon that, beyond its role in phosphate homeostasis, interacts with the stress response and controls the expression of virulence traits30, 44, might have protected the two imipenem-susceptible strains BJ-STC-b and BJ-STC-c from imipenem activity. We also postulate that the different variants, which co-existed as persister bacteria during the antibiotic treatments due to their different metabolisms45,46,47, arose sequentially in the urine, depending on the treatment used. The physiological advantage of the imipenem-susceptible strains, BJ-STC-b and BJ-STC-c, over the imipenem-resistant strain, BJ-STC-a, was not revealed by improved fitness of the two susceptible strains relative to the resistant one. Further studies are required to obtain more insight into the advantages provided by the upregulation of the Pho regulon in ESBL-producing K. pneumoniae isolates devoid of OmpK35 and OmpK36. Our in-vitro experiments showed that the susceptibility to carbapenems recovered by strains BJ-STC-b and BJ-STC-c is ephemeral. Indeed, variants resistant to carbapenems and cefoxitin due to additional mutations (PhoR, PhoB) located downstream of the mutated regulators (PstS and PhoU) were obtained under carbapenem and cefoxitin pressure, suggesting a risk of in-vivo selection of resistant mutants due to downregulation of the Pho regulon.
In conclusion, this is the first study showing that mutations located in the pstSABC-phoU operon deprive a XDR K. pneumoniae clinical isolate (producing CTX-M-15, devoid of OmpK35 and OmpK36 and overproducing the OqxAB efflux pump) of its resistance to carbapenems and cefoxitin. It also shows that the versatility of the expression of outer membrane porins appears to play a key role in bacterial adaptation and may play an important role in persister cells during antibiotic treatment.
References
Podschun, R. & Ullmann, U. Klebsiella spp. as nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11, 589–603 (1998).
Moradigaravand, D., Martin, V., Sharon, P. & Parkhill, J. Evolution and epidemiology of multidrug-resistant Klebsiella pneumoniae in the United Kingdom and Ireland. MBio 8, e01976–01916 (2017).
Pendleton, J.-N., Gorman, S.-P. & Gilmore, B.-F. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti-Infect Ther 11, 297–308 (2013).
Du, J. et al. Molecular epidemiology of extensively drug-resistant Klebsiella pneumoniae outbreak in Wenzhou, Southern China. J Med Microbiol 65, 1111–1118 (2016).
Pitout, J.-D., Nordmann, P. & Poirel, L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother 59, 5873–5884 (2015).
Wozniak, A. et al. Porin alterations present in non-carbapenemase-producing Enterobacteriaceae with high and intermediate levels of carbapenem resistance in Chile. J Med Microbiol 61, 1270–1279 (2012).
Tsai, Y. K. et al. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob Agents Chemother 55, 1485–1493 (2011).
Huang, T. D. et al. Increasing proportion of carbapenemase-producing Enterobcareriaeae and emergence of MCR-1 producer through a multicentric study among hospital-based and private laboratories in Belgium from September to November 2015. Eurosurveillance 22 (2017).
Robert, J., Pantel, A., Merens, A., Lavigne, J. P. & Nicolas-Chanoine, M. H. Incidence rates of carbapenemase-producing Enterobacteriaceae clinical isolates in France: a prospective nationwide study in 2011–12. J Antimicrob Chemother 69, 2706–2712 (2014).
Wang, J.-T. et al. Carbapenem-nonsusceptible Enterobacteriaceae in Taiwan. PLoS ONE 10, e0121668 (2015).
Zhang, Y. et al. Contribution of β-lactamases and porin proteins OmpK35 and OmpK36 to carbapenem resistance in clinical isolates of KPC-2-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 58, 1214–1217 (2014).
Garcia-Fernandez, A. et al. Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob Agents Chemother 56, 2143–2145 (2012).
Masi, M., Réfregiers, M., Pos, K.-M. & Pagès, J.-M. Mechanisms of envelope permeability and antibiotic influx and efflux in Gram-negative bacteria. Nat Microbiol 2, 17001 (2017).
Kaczmarek, F. M., Dib-Hajj, F., Shang, W. & Gootz, T. D. High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of bla ACT-1 β-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin PhoE. Antimicrob Agents Chemother 50, 3396–3406 (2006).
Jarlier, V., Nicolas, M. H., Fournier, G. & Philippon, A. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis 10, 867–878 (1988).
Criscuolo, A. & Brisse, S. AlienTrimmer: a tool to quickly and accurately trim off multiple short contaminant sequences from high-throughput sequencing reads. Genomics 102, 500–506 (2013).
Rissman, A. I. et al. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics 25, 2071–2073 (2009).
Jolley, K. A. & Maiden, M. C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11, 595 (2010).
Bialek-Davenet, S. et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis 20, 1812–1820 (2014).
Zankari, E. et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67, 2640–2644 (2012).
Carattoli, A. et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58, 3895–3903 (2014).
Bialek-Davenet, S. et al. In vitro selection of ramR and soxR mutants overexpressing efflux systems by fluoroquinolones as well as cefoxitin in Klebsiella pneumoniae. Antimicrob Agents Chemother 55, 2795–2802 (2011).
Bialek-Davenet, S. et al. Differential contribution of AcrAB and OqxAB efflux pumps to multidrug resistance and virulence in Klebsiella pneumoniae. J Antimicrob Chemother 70, 81–88 (2015).
EUCAST & ESCMID. Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin Microbiol Infect 9, 1–7 (2003).
Gayet, S., Chollet, R., Molle, G., Pagès, J. M. & Chevalier, J. Modification of outer membrane protein profile and evidence suggesting an active drug pump in Enterobacter aerogenes clinical strains. Antimicrob Agents Chemother 47, 1555–1559 (2003).
Simonet, V., Mallea, M., Fourel, D., Bolla, J. M. & Pages, J. M. Crucial domains are conserved in Enterobacteriaceae porins. FEMS Microbiol Lett 136, 91–97 (1996).
Hasdemir, U. O., Chevalier, J., Nordmann, P. & Pages, J. M. Detection and prevalence of active drug efflux mechanism in various multidrug-resistant Klebsiella pneumoniae strains from Turkey. J Clin Microbiol 42, 2701–2706 (2004).
Philippe, N. et al. In vivo evolution of bacterial resistance in two cases of Enterobacter aerogenes infections during treatment with Imipenem. PLoS ONE 10, e0138828 (2015).
Villa, L., Feudi, C., Fortini, D., García-Fernández, A. & Carattoli, A. Genomics of KPC-producing Klebsiella pneumoniae sequence type 512 clone highlights the role of RamR and ribosomal S10 protein mutations in conferring tigecycline resistance. Antimicrob Agents Chemother 58, 1707–1712 (2014).
Lamarche, M. G., Wanner, B. L., Crepin, S. & Harel, J. The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol. Rev. 32, 461–473 (2008).
Gardner, S.-G., Johns, K.-D., Tanner, R. & McCleary, W.-R. The PhoU protein from Escherichia coli interacts with PhoR, PstB, and metals to form a phosphate-signaling complex at the membrane. Journal of Bacteriology 196, 1741–1752 (2014).
Veleba, M., Higgins, P. G., Gonzalez, G., Seifert, H. & Schneidersa, T. Characterization of RarA, a novel AraC family multidrug resistance regulator in Klebsiella pneumoniae. Antimicrob Agents Chemother 56, 4450–4458 (2012).
Lee, E. H. et al. Association of two resistance mechanisms in a clinical isolate of Enterobacter cloacae with high-level resistance to imipenem. Antimicrob Agents Chemother 35, 1093–1098 (1991).
Bidet, P. et al. In vivo transfer of plasmid-encoded ACC-1 AmpC from Klebsiella pneumoniae to Escherichia coli in an infant and selection of impermeability to imipenem in K. pneumoniae. Antimicrob Agents Chemother 49, 3562–3565 (2005).
Skurnik, D. et al. Development of ertapenem resistance in a patient with mediastinitis caused by Klebsiella pneumoniae producing an extended-spectrum β-lactamase. J Med Microbiol 59, 115–119 (2010).
Leavitt, A. et al. Ertapenem resistance among extended-spectrum-β-lactamase-producing Klebsiella pneumoniae isolates. J Clin Microbiol 47, 969–974 (2009).
Doumith, M., Ellington, M. J., Livermore, D. M. & Woodford, N. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother 63, 659–667 (2009).
Jacoby, G. A., Mills, D. M. & Chow, N. Role of β-lactamases and porins in resistance to ertapenem and other β-lactams in Klebsiella pneumoniae. Antimicrob Agents Chemother 48, 3203–3206 (2004).
Cerdeira, L. et al. Draft genome sequence of a CTX-M-15-producing Klebsiella pneumoniae sequence type 340 (clonal complex 258) isolate from a food-producing animal. Journal of global antimicrobial resistance 7, 67–68 (2016).
Horna, G., Velasquez, J., Fernandez, N., Tamariz, J. & Ruiz, J. Characterization of the first KPC-2-producing Klebsiella pneumoniae ST340 in Peru. Journal of Global Antimicrobial Resistance 9, 36–40 (2017).
Martin, W. M. et al. Coproduction of KPC-2 and QnrB19 in Klebsiella pneumoniae ST340 isolate in Brazil. Diagn Microbiol Infect Dis 83, 375–376 (2015).
Peirano, G. et al. The importance of clonal complex 258 and IncFK2-like plasmids among a global collection of Klebsiella pneumoniae with bla KPCs. Antimicrob Agents Chemother 61, 2610–2616 (2017).
Gardner, S. G. et al. Genetic analysis, structural modeling, and direct coupling analysis suggest a mechanism for phosphate signaling in Escherichia coli. BMC Microbiol 16, S2 (2015).
Santos-Beneit, F. The Pho regulon: a huge regulatory network in bacteria. Front Microbiol 6, 1–13 (2015).
Brauner, A., Fridman, O., Gefen, O. & Balaban, N.-Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14, 320–330 (2016).
Van Acker, H. & Coenye, T. The role of efflux and physiological adaptation in biofilm tolerance and resistance. J Biol Chem 10, 12565–12572 (2016).
Van den Bergh, B. et al. Frequency of antibiotic application drives rapid evolutionary adaptation of Escherichia coli persistence. Nat Microbiol 7, 16020 (2016).
Acknowledgements
We thank Anne Davin-Regli and Muriel Masi for their fruitful discussions and Sabina Fajal Legrand for her assistance during the clinical data collection. We thank Damien Mornico for help with the data submission to sequence repositories. The research leading to these results was conducted as part of the TRANSLOCATION consortium and has received support from the Innovative Medicines Initiatives Joint Undertaking under Grant Agreement n°115525, resources which consist of financial contributions from the European Union’s seventh framework program (FP7/2007-2013) and EFPIA companies in-kind contributions. This work was also supported by Aix-Marseille University and the Service de Santé des Armées.
Author information
Authors and Affiliations
Contributions
M.H.N.C. designed the research. S.B.D. and N.M. performed the genome analysis and both the bacteriological and molecular biology assays. J.V. performed the immunodetection assays. M.D. performed the strain growth assays. S.B. supervised the whole genome sequencing and assembly. M.H.N.C. supervised the overall project with contribution from J.M.P. for the part focusing on porin regulation and synthesis. M.H.N.C. wrote the manuscript with contributions from S.D.B., J.M.P., and S.B. All authors read the manuscript and have approved it for submission.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bialek-Davenet, S., Mayer, N., Vergalli, J. et al. In-vivo loss of carbapenem resistance by extensively drug-resistant Klebsiella pneumoniae during treatment via porin expression modification. Sci Rep 7, 6722 (2017). https://doi.org/10.1038/s41598-017-06503-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-06503-6
This article is cited by
-
Differential expression of outer membrane proteins and quinolone resistance determining region mutations can lead to ciprofloxacin resistance in Salmonella Typhi
Archives of Microbiology (2023)
-
Porin-Mediated Carbapenem Resistance in Klebsiella pneumoniae: an Alarming Threat to Global Health
Current Clinical Microbiology Reports (2023)
-
Lipid A-Ara4N as an alternate pathway for (colistin) resistance in Klebsiella pneumonia isolates in Pakistan
BMC Research Notes (2021)
-
First description of antimicrobial resistance in carbapenem-susceptible Klebsiella pneumoniae after imipenem treatment, driven by outer membrane remodeling
BMC Microbiology (2020)
-
Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria
Nature Reviews Microbiology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.