Abstract
Enterococci are nosocomial pathogens that can form biofilms, which contribute to their virulence and antibiotic resistance. Although many genes involved in biofilm formation have been defined, their distribution among enterococci has not been comprehensively studied on a genome scale, and their diagnostic ability to predict biofilm phenotypes is not fully established. Here, we assessed the biofilm-forming ability of 90 enterococcal clinical isolates. Major patterns of virulence gene distribution in enterococcal genomes were identified, and the differentiating virulence genes were screened by polymerase chain reaction (PCR) in 31 of the clinical isolates. We found that detection of gelE in Enterococcus faecalis is not sufficient to predict gelatinase activity unless fsrAB, or fsrB alone, is PCR-positive (P = 0.0026 and 0.0012, respectively). We also found that agg is significantly enriched in isolates with medium and strong biofilm formation ability (P = 0.0026). Additionally, vancomycin, applied at sub minimal inhibitory concentrations, inhibited biofilm in four out of five strong biofilm-forming isolates. In conclusion, we suggest using agg and fsrB genes, together with the previously established gelE, for better prediction of biofilm strength and gelatinase activity, respectively. Future studies should explore the mechanism of biofilm inhibition by vancomycin and its possible use for antivirulence therapy.
Similar content being viewed by others
Introduction
Enterococci are Gram-positive bacteria that cause serious nosocomial infections, including urinary tract, bloodstream infections and endocarditis1. Enterococci are known for their ability to form biofilms, which are populations of cells irreversibly attached to various biotic and abiotic surfaces and encased in a hydrated matrix of exopolymeric substances, proteins, polysaccharides and nucleic acids2. Biofilms contribute to bacterial virulence in several ways. For example, adherence, an early step in biofilm formation, allows the bacteria to bind to catheters (e.g., urinary and intravascular catheters), biliary stents and silicone gastrostomy devices3. Additionally, biofilms contribute to bacterial resistance to antibiotics4 and to phagocytosis, making their eradication extremely difficult5. In a mature biofilm, the bacterial cells can tolerate antibiotics at concentrations 10–1000 times higher than those required to kill planktonic cells5.
Several enterococcal virulence proteins have been studied for their important roles in biofilm development6. These include the aggregation substance (Agg), Enterococcus f aecalis endocarditis-associated antigen A (EfaA), adhesion of collagen of E . faecalis (Ace) and biofilm on plastic operon (Bop)7. Moreover, expression of pili on the cell surface, which facilitates cell adhesion, is considered the trigger of biofilm formation8, and pili components are encoded by the endocarditis- and biofilm-associated pili genetic locus (EbpABC) and an adjacent downstream sortase-encoding gene, srt 9.
In general, biofilm formation is associated with quorum sensing, which is the regulation of bacterial gene expression in response to large cell population densities. This regulation is usually driven by molecules known as autoinducers10. In E. faecalis, biofilm formation is regulated by the well-defined quorum sensing system, fsr (faecal streptococci regulator) locus11. This locus consists of three genes, fsrA, fsrB and fsrC, immediately located next to two virulence factor-encoding genes: one encoding a gelatinase (gelE) and the other a serine protease (sprE). The expression of gelE is regulated by the fsr locus12.
Enterococcal cells do not only communicate through Fsr quorum signaling, but are also capable of communicating by peptide pheromones, secreted by recipient cells to induce the conjugative apparatus of donor cell, which mediate the transfer of pheromone-responsive plasmids13. Some of these plasmids carry genes that regulate or promote biofilm formation, such as the plasmid-encoded aggregation substance genes14. A direct link between pheromones and biofilm formation was demonstrated in Candida albicans 15. Examples of secreted enterococcal pheromones are Cpd, Cob, and Ccf16.
Because of the role of biofilms in virulence and antimicrobial resistance, different methods have been developed for biofilm prevention and removal17. One such method is the use of sub-inhibitory concentrations of antibiotics, which was shown to modify the physicochemical properties and the architecture of the outer surface of enterococcal cells, thus affecting their overall virulence1. Sub-inhibitory antibiotic concentrations have also been used to inhibit bacterial initial adhesion to abiotic substrates18.
Additionally, despite a growing body of literature about the aforementioned genetic determinants involved in biofilm formation and regulation in enterococci, no clear genotype–phenotype correlation has been established for these determinants. Associations between some of these genes and biofilm formation have been recognized (e.g., gelE 11, 19); yet, no study has systematically investigated whether the complete or partial presence of this gene set is a predictor of the biofilm formation phenotype. Such correlation would be of great diagnostic value, but is hindered by poor metadata available about published enterococcal genomes, which impede attempts for comparative genomics between biofilm and non-biofilm formers.
For the above reasons we performed a comprehensive genome survey for biofilm-associated genes, with emphasis on those that could consistently predict the strength of biofilm-forming phenotype in E. faecalis. We examined biofilm formation and antibiotic resistance phenotypes in enterococcal clinical isolates; then we screened representative E. faecalis isolates for their biofilm-regulating quorum sensing genes, as well as other virulence genetic determinants. Finally, we investigated the effect of certain antibiotics applied at sub-inhibitory concentrations on biofilm formation in strong biofilm-forming isolates. We found that agg is a good predictor of biofilm strength, that the fsrA and fsrB genes have a high predictive value of biofilm-associated gelatinase activity, and that sub-minimal inhibitory concentrations of vancomycin were able to inhibit biofilm formation.
Results
Bacterial isolate identification and relatedness
Out of 140 clinical isolates, collected from Egyptian hospitals over a period of seven years and suspected to be enterococci, 90 isolates were identified as Enterococcus spp (Table S1). These showed small brownish-black colonies surrounded by a black zone when streaked on Enterococcosel agar. Under the microscope, they were Gram positive cocci or coccobacilli arranged in pairs and short chains. Further identification by catalase test and 6.5% NaCl tolerance showed that these enterococcal isolates were catalase negative and able to grow in high concentration of NaCl. Their cultivation on Chromogenic UTI agar showed blue colonies.
The isolates were further identified by the VITEK 2 identification system, and their majority were E. faecalis (65 isolates or 72.2%), with a good number of E. faecium (22 isolates or 24.4%). Two of the remaining isolates belonged to the species E. casseliflavus and only one was identified as E. gallinarum (Table S1).
The divergent nature of the isolates was confirmed by enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR), which showed a wide variety of patterns, most of which representing distinct enterococcal clades (Fig. 1 and S1). Only a few isolates appeared to be clonal or highly related (e.g., E02/E4/E64/En33 and E10/ En23/En25, Fig. 1).
ERIC-PCR analysis of the isolates. An nweighted pair group method using arithmetic overage algorithm (UPGMA) dendrogram in a radial format representing the ERIC-PCR pattern relatedness of all tested isolates. Isolate names are color coded according to their biofilm strength (Purple, strong; Red, medium; Green, weak; Black, non-biofilm-forming).
Phenotypic assessment of the biofilm-forming ability of the entercococcal clinical isolates
Congo-Red agar biofilm assay differentiated the 90 enterococcal isolates into (i) five strong biofilm-producing isolates; (ii) eighty-one isolates that varied in their biofilm formation strength between moderate and weak; and (iii) four non-biofilm-producing isolates. This assay could not persuasively differentiate between moderate and weak biofilm formation ability. On the other hand, the Crystal Violet biofilm assay differentiated isolates into strong, moderate, weak, and non-biofilm-forming according to the O.D. values at 545 nm. Again, only five of the isolates (5.5%) were classified as strong biofilm-formers; 38 isolates (42%) were moderate; 43 were weak biofilm-formers (48%); and four (4.5%) could not form any detectable biofilm.
Gelatinase activity has been described as one of the first steps in the process of biofilm formation11. Here, all isolates were screened for their in vitro gelatinase activity, but only 27% were gelatinase positive.
Of note, neither biofilm formation strength nor gelatinase activity correlated with ERIC-PCR types, except that three strong biofilm-forming strains fell in the same clade (E01, E03 and E28, Fig. 1). The ERIC patterns somehow correlated with the species of different isolates; however, given that only two major species were represented (E. faecalis and E. faecium), strain-to-strain variations remained important to determine by ERIC.
Biofilm formation strength was not significantly associated with the bacterial species (Chi-square P value = 0.2738), but there was obvious association (Chi-square P value ≤ 0.0001) with the sample source—urine and stool samples being enriched with biofilm-forming strains with higher average strength (Table 1).
Comparative genomic analysis
Biofilm formation in enterococci is a complex trait encoded by complementary, overlapping, and possibly redundant pathways/gene clusters; yet a clear biofilm genotype–phenotype correlation remains to be established. Here, a set of 17 protein-coding genes was selected after thorough literature survey (summarized in Introduction).
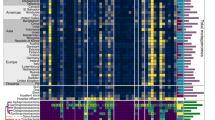
First, we screened for the presence/absence pattern of these 17 protein-coding genes in five fully sequenced E. faecalis genomes (with a BlastX cutoff of either 80% amino acid identity or an E-value of 10−80). The genomes represented different biofilm phenotypes ranging from strong biofilm formers (V583) to the non-biofilm-forming probiotic strain Symbioflor1 (Fig. 2). Apart from the highly variable ace gene and the mobile agg gene, the variation was represented by the presence/absence of fsr locus as well as an inactivating mutation in the Cpd-encoding gene (Fig. 2). A more extensive analysis on the fsr/gelE locus was conducted by the Subsystems approach on the SEED server20, and showed many variants most of which are characterized by a missing fsrAB, or—less frequently—fsrA, whereas gelE and fsrC seemed to be the most conserved genes of the locus (Table S2 and http://pubseed.theseed.org/?page=SubsystemSelect; subsystem deposited under the name: “Biofilm Formation in Enterococci”).
Comparative genomics screen for the 17 biofilm-associated protein-coding genes. Comparative genomics in a tabular format, representing patterns of gene presence and absence, analyzed by blastX against the 17 biofilm-related proteins and confirmed by blastN against primer pairs, in five representative E. faecalis strains. +: orthologous gene present; ±: paralogous gene present; −: gene absent; X: pseudogene. For Agg, more than one paralogous gene have been identified in some genomes. We differentiate the chromosomal and plasmid (p) copy of these genes.
We confirmed the aforementioned results by in silico PCR primer screening, using BLASTN searches for the primer pairs of the 17 genes mentioned above (Table 2) in the five representative genomes, and confirmed that Symbioflor1 lacked all members of the fsr/gelE operon, and that strain 62 lacked fsrA and fsrB but had fsrC.
To more systematically determine which genes could serve as biomarkers for biofilm formation in E. faecalis in an unbiased way, we conducted genome-wide comparative analysis between biofilm-forming strains and the non-biofilm-forming Symbioflor 1. Genome-scale SEED comparative analysis confirmed BLAST results and, in addition, identified several prophage and pathogenicity island genes that differentiated the different strains (e.g., phage capsid proteins, integrases, transposases) (Table S2). Bidirectional best hit analysis indicated that the chromosomal agg gene of E. faecalis V583 is not the ortholog of the agg of E. faecalis Str. Symbioflor1, and that agg gene is more related to the agg variants carried on plasmids in E. faecalis V583 and 62 (Fig. 2 and Table S3).
Taken together, the above results indicate that agg and ace sequences are too variable to serve as good PCR biomarkers; on the other hand, gelE and the fsr locus genes could be more promising, notably fsrA and fsrB. Yet, the actual correlation of these candidate genes with biofilm phenotypes remained to be tested on clinical isolates.
Screening clinical isolates for genes involved in biofilm formation
Out of the 90 enterococcal isolates, 31 were screened by PCR for the 17 genes listed above (Table 2), and the screening results are summarized as follows (Fig. 3 and Table 3): srt, ccf, bop, efaA, and cpd genes were present in 94% of the isolates. gelE, sprE, and fsrC genes in 90% of the isolates, ebpC and pil in 84%, agg in 81%, ebpA in 77%, fsrB in 65%, ebpB in 55%, cob in 50%, and ace and fsrA in 45% of the isolates.
These data are in agreement with the genome screen (Fig. 2) and subsytems analysis (Table S2), and indicate the importance of combining patterns of the gelE/fsrA locus genes in diagnosis and gelatinase activity prediction. Additionally, patterns of agg, ace combined with gelE/fsrA could distinguish weak biofilm phenotypes (Tables 4–5).
Of note, when we tried to use the patterns of gene distribution to classify the isolates, in a similar way to that used with ERIC-PCR analysis, non-biofilm-forming isolates were clustered out, as expected. However, no obvious cluster could sepearte moderate from weak biofilm-formers, but all strong biofilm-forming isolates fell in one major clade (Fig. 4).
UPGMA analysis to cluster the isolates according to the pattern of their PCR results. A UPGMA cladogram representing the clustering relatedness of 31 isolates according to the pattern of their PCR results for biofilm-associated genes. Isolate names are color coded according to their biofilm strength (Purple, strong; Red, medium; Green, weak; Black, non-biofilm-forming).
Antibiotic resistance pattern and effect of antibiotic sub-MIC on biofilm formation
The sensitivity of the isolates to azithromycin, ciprofloxacin, vancomycin, gentamycin and tigecycline was estimated by the determination of MIC values by the broth microdilution method. MICs of vancomycin was ≤ 4 µg/ml, tigecycline ≤ 0.25 µg/ml, ciprofloxacin ≤ 1 µg/ml, gentamycin ≤ 500 µg/ml, and azithromycin ≤ 0.5 µg/ml.
The strains selected for subsequent assays were those with strong biofilm-forming phenotype, which were also gelatinase positive. The five isolates with strong biofilm formation ability were sensitive to vancomycin and tigecycline (Fig. 5). Both antibiotics were tested for their sub-MIC effect on the adherence of the five isolates. In one isolate only, the adherence was significantly decreased by both vancomycin (55% decrease, p < 0.05, one-way ANOVA) and tigecycline (42% decrease, p < 0.05, one-way ANOVA) in comparison to untreated cells. The adherence of another three isolates was only significantly decreased by vancomycin (50% decrease in adherence, p < 0.05, one-way ANOVA). The adherence of one isolate was neither affected by vancomycin nor tigecycline (Fig. 6).
The effect of sub-MIC of vancomycin and tigecycline was also assessed on gelatinase activity in the five strong biofilm-forming isolates. Vancomycin inhibited gelatinase activity in the four isolates with decreased adherence and tigecycline inhibited gelatinase activity in the same isolate in which adherence was reduced.
Discussion
The main focus of this study was to determine the biofilm-forming ability of clinical enterococcal isolates, its correlation with gelatinase activity and fsr locus genotype, as well as its inhibition by antibiotics such as vancomycin. The long-term goal is to define fewer, but more precise, genetic biomarkers to predict different aspects of biofilm formation (i.e., gelatinase activity and biofilm strength) instead of screening a full panel of virulence genes. To this end, a major step was to first establish a good genotype–phenotype correlation relying on genomic analysis to define good biomarker genes for biofilm formation and gelatinase activity, as two separate—even if interdependent—phenotypes. Once the genes were defined, they were validated experimentally.
Our work demonstrates a higher than usual frequency (~96%) of biofilm production by enterococcal clinical isolates collected from Egyptian hospitals, regardless of the strength of the biofilm formed. This frequency is much higher than other reports from developing countries. For instance, Sindhanai et al. reported that 68% of isolates in Tamilnadu, India were biofilm producers21 and 64.40% of bacterial isolates from urinary tract infection in a hospital in Dhaka, Bangladesh were biofilm producers22.
Using already-established Crystal Violet assays, we differentiated the clinical isolates into strong (5.5%), moderate (42%), weak (48%), and non-biofilm-forming (4.5%) strains. Congo-Red agar assay could not differentiate moderate from weak biofilm forming strains, so the results of the Crystal Violet assay were considered. The superiority of Crystal Violet assays has been previously described21, 23. Sindhanai and coworkers21 reported that 34% of their isolates formed strong biofilms; 49% formed moderate biofilms; and 17% formed weak biofilms. As determined in this study, differentiation between weak and moderate biofilm formation may be associated with the aggregation substance (Agg), as the gene encoding it is enriched in moderate and strong isolates (Table 4). The extent of biofilm formation and the differentiation between weak, medium and strong biofilm-forming capacity is clinically relevant as the potential implication of biofilm formation on hospitalized patients could be serious24. Of note, we performed the biofilm assays in TSB medium with additional 0.25% glucose (i.e., a final concentration of 0.5% glucose). There are mixed reports about the effect of glucose on biofilm formation: Kristich et al. reported an inhibitory effect of increasing glucose concentrations25, yet with a reasonable biofilm formation at 0.5% glucose, whereas Baldassarri et al.26 and Pillai et al.27 reported potentiating effect at concentrations <1%. Using 0.5% seemed reasonable. Likewise, we consistently performed the assays after 48 hours incubation based on prior reports28, 29; this consistency was important to guarantee that the incubation time would not be a confounding factor.
To better understand the genotype–phenotype correlations for a complex phenotype such as biofilm formation in enterococci, we performed extensive genomic analysis of E. faecalis, first starting by screening five well-defined genomes for a set of 17 biofilm-associated genes, and then by extending the screen to ~190 genomes for agg, ace and fsr locus genes.
This genomic analysis was followed by PCR detection of the 17 genes in 31 isolates with variable biofilm formation strength and variable gelatinase activity. Although the entire set of 17 genes was not fully detected in each biofilm-forming strain, many of these genes were present in all the biofilm-forming strains, i.e., gelE, ccf, sprE, bop, efaA, cpd, and fsrC (Table 3 and Fig. 4). As indicated in Results, in addition to gelE (previously determined as a biomarker for biofilm formation), agg was found to be a predictor of weak vs. strong/medium biofilm formation (Chi-square P value = 0.0026, Table 4).
As for gelE, its presence in all biofilm-forming strains and absence in non-biofilm-formers confirmed the reported importance of gelatinase for biofilm formation process11. Hancock and Perego showed that GelE is required for the formation of biofilm, as it promotes cell aggregation in microcolonies to develop a three-dimensional structure11. Two possibilities were suggested for this function of GelE: either through the enzyme’s proteolytic activity as an initiator of bacterial attachment to surfaces or through the physical presence of the protein rather than its enzymatic activity11.
GelE is positively regulated by the quorum sensing-encoding fsr locus30. Despite the presence of gelE gene in 94% of isolates, in vitro gelatinase activity was only detected in 30% of the isolates. Other studies, involving large and diverse groups of clinical isolates, showed that 56% and 59% of isolates produced gelatinase, while 88% and 92% were gelE-positive, respectively31, 32. Among 31 endodontic Enterococcus isolates, gelE was detected in all tested isolates, whereas the gelatinase activity was found in 74% of them33. These reports, together with our results, confirm that gelE is obviously essential for gelatinase activity, but that it is not sufficient as fsrA and fsrB are required for (and highly associated with) the gelatinase phenotype. Because the genomic survey we performed on ~190 genomes (Table S2), as well as our PCR results (Table 3), indicated that fsrB is more frequent than fsrA, and that no cases were found in which fsrA was present in absence of fsrB, we suggest that fsrB is sufficient to predict the gelatinase activity (Chi-Square P value = 0.0012, Table 5).
Thus, we conclude that the absence of gelatinase activity in gelE-positive strains is due to the disruption of the fsr quorum sensing locus. As described by Nakayama et al., the most common cause for loss of gelatinase production is a 23.9 kb deletion encompassing most of the fsr locus34 (fsrA and fsrB in particular35).
In several pathogens, biofilm production is regulated by quorum sensing systems, including fsr in Enterococcus, which was shown to have a pronounced effect on biofilms11. In the current study the strong biofilm-forming strains contained the entire quorum sensing locus, and loss of any of the quorum sensing genes was correlated with decrease in biofilm strength. Mohamed et al.4 reported that loss of fsr and/or gelatinase function resulted in diminished biofilm formation, in comparison with biofilm production by progenitor, fsr-positive, gelatinase-positive E. faecalis isolate. Carniol and Gilmore discussed the role of signal transduction, quorum sensing and extracellular protease activity in biofilm formation by E. faecalis 36. Another research team found that biofilm formation was reduced in all three fsr mutants (fsrA, fsrB, fsrC) by ~28 to 32% compared to wild-type E. faecalis OG1RF4.
Another focus of this study was the mutual effects between biofilm formation and antibiotics/antibiotic resistance. Here, the antimicrobial susceptibility of Enterococcus isolates to azithromycin, ciprofloxacin, vancomycin, gentamycin and tigecycline was estimated through determination of each antibiotic’s MIC by the broth microdilution method. Although studies on the antimicrobial susceptibility patterns of enterococci have affirmed the worldwide emergence of multiple-drug resistant enterococci, particularly to vancomycin37, 96.7% of our isolates were actually sensitive to vancomycin. This is not unprecedented, e.g., Karmarkar et al. reported that 76% of the isolates they screened were sensitive to vancomycin38. Additionally, all the isolates were sensitive to tigecycline, while 60.8% and 46.7% were sensitive to ciprofloxacin and gentamycin, respectively, and only 22.8% were sensitive to azithromycin. Strong biofilm-forming strains were only sensitive to vancomycin and tigecycline.
By evaluating the effect of sub-MIC of vancomycin and tigecycline on adherence of strong biofilm-forming isolates, we found that vancomycin decreased the adherence significantly in four isolates while tigecycline only decreased the adherence in one isolate. Tigecycline has previously been reported to inhibit biofilm formation in enterococci. Maestre and colleagues demonstrated that tigecycline decreased the adherence of 55% of the isolates that had strong biofilm-formation ability39. To the best of our knowledge, there are no published reports of biofilm inhibition by vancomycin in enterococci, but there is a precedent case reported for a clinical staphylococcal isolates16. Further studies should explore the mechanism of biofilm inhibition by vancomycin and whether it is through direct interaction with the gelatinase enzyme or with any of its regulators. Other possibilities for such inhibition could be related to alteration of gene regulator circuits in response to vancomycin-induced stress or to selection of a bacterial subpopulation that has higher tolerance for low doses of vancomycin.
Conclusion
We conclude that the biofilm-forming ability of enterococcal isolates sampled in this study, and representing different Egyptian hospitals, does not particularly correlate with their overall genetic make-up, which was highly divergent (as reflected by ERIC-PCR patterns). Several genetic determinants, including the quorum sensing locus, which encodes proteins that regulate biofilm formation, were detected in these biofilm-forming isolates, but none of them was detected in non-biofilm-formers. The genes with highest diagnostic values, as determined by in silico genome analysis and validated by in vitro PCR screens and phenotypic assays, are gelE, agg, and fsrB. Finally, we show that vancomycin, and to a lesser extent tigecycline, could inhibit biofilm formation when used at concentrations below their measured MICs. This work highlights the importance of integrating phenotypic and genotypic assays of biofilm-forming determinants in clinical enterococcal isolates. In addition, it calls for further understanding of the mechanism by which some antibiotics inhibit biofilm formation, thus affecting enterococcal virulence and possibly resistance to other antibiotics.
Methods
Bacterial isolates and culture media
Isolates were collected from Egyptian hospitals in the period from 2009 to 2015. Enterococcus colonies were isolated by surface streaking of clinical specimens on Enterococcosel agar (Difco laboratories, USA) and identified by Gram stain followed by catalase test. The identity was further confirmed by cultivation on Chromogenic UTI agar (Oxoid, UK), currently known as Brilliance UTI agar. This differential culture medium provides presumptive identification of several urinary tract pathogens. Unlike other species, Enterococcus species express beta-glucosidase but not beta-galactosidase or tryptophan deaminase. The beta-glucosidase activity targets the chromogen, x-glucoside, and produces blue colonies. Confirmed Enterococcus strains were streaked on Brain Heart agar slants and kept at 4 °C for the period of experimentation. Duplicate glycerol stocks of each isolate were stored at −80 °C.
Bacterial isolate identification by the VITEK 2 system
A number of isolated colonies (5–10) from a fresh overnight pure culture were resuspended in 3.0 mL of sterile saline solution. The turbidity was adjusted to 0.5 McFarland standard. GP (Gram Positive) identification cards were inoculated with the suspended microorganism by an integrated vacuum apparatus, and test tubes containing the bacterial suspension were placed into a special rack while the identification cards were placed in the neighboring slot. Cards were sealed and inserted into the VITEK 2 reader-incubator, and then subjected to fluorescence measurement every 15 min. Results were obtained automatically and interpreted by the ID-GPC database.
Polymerase Chain Reaction (PCR) and ERIC-PCR
DNA from prescreened Enterococcus strains was extracted as described by Soumet et al.40. Specific primer pairs were designed and used for amplification of the different genes (Table 2). PCR amplification was performed in a Thermocycler (SensoQuest, Germany) in 0.2 ml reaction tubes, each with 25 µl reaction mixtures. Reaction mixtures were made of 10 pM of each primer, 200 µM of each deoxyribonucleotide (Promega, USA), 5 X reaction buffer, 1.5 mM MgCl2, 0.5 U Taq polymerase (Promega, USA) and 0.25 µg extracted enterococcal genomic DNA (or non-template control of nuclease-free water). PCR products were analyzed in 1% agarose gels by electrophoresis, stained with ethidium bromide and visualized under UV light (Fig. S2).
A modified enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) has been developed for typing enterococci41. This method was used as previously described, with the primers ERIC-1: ATGTAAGCTCCTGGGGATTCAC and ERIC-2: AAGTAAGTGACTGGG GTGAGCG. PCR conditions were as follows: For ERIC-1, an initial denaturation at 94 °C for 5 min was followed by five cycles of 94 °C for 5 min., 35 °C for 5 min. and 72 °C for 5 min; then 30 cycles of 94 °C for 1 min., 35 °C for 1 min., 72 °C for 2 min and a final extension of 72 °C for 10 min. For ERIC-2, the initial denaturation step was followed by 35 cycles of 94 °C for 1 min., 25 °C for 1 min. then, 72 °C for 4 min, and a final extension of 72 °C for 10 min.
Unweighted pair group method using arithmetic overage algorithm (UPGMA), available at (http://genomes.urv.cat/UPGMA/) was used for ERIC-PCR gel electrophoretic pattern analysis and dendrogram generation. Trees were rendered, colored, and annotated by FigTree v.1.2.3 (obtained from http://tree.bio.ed.ac.uk/software/figtree).
Congo-red agar biofilm assay
The investigation of biofilm production by the Congo-red agar assay was proposed by Freeman et al.42. Enterococcal cell suspensions were inoculated on Congo-red agar plates. Black colonies with a dry crystalline consistency indicated a positive result. If the colonies were dark but with no dry crystalline colonial morphology, the biofilm-forming ability was considered intermediate. Non-biofilm producers grow as pink colonies (Fig. S3).
Crystal Violet biofilm assay
Biofilm formation was also assessed by the Crystal Violet assay as described by Christensen et al.43. Briefly, overnight cultures were inoculated in fresh trypticase soy broth (TSB) containing 0.25% glucose. The culture density was spectrophotometrically adjusted to approximately 0.5 McFarland standard (at absorbance = 600 nm). Each broth culture was diluted 1:100 in fresh broth, and 200 µl of each diluted culture was distributed in three wells of a 96-well microtiter plate and incubated for 48 hours at 37 °C. Negative control wells were included. Planktonic cells were removed, and wells were (i) washed with sterile phosphate-buffered saline for removal of non-adherent cells, (ii) decanted, and then (iii) left to dry. Adherent cells were stained with 200 µl of 2% (W/V) crystal violet for 15 minutes. Excess stain was gently rinsed off with water. Plates were allowed to air-dry. The dye bound to the adherent cells was re-solubilized with 200 µl of 33% (v/v) glacial acetic acid. The optical density (OD) was measured at 545 nm in a plate reader (ELx800, Biotek, USA). Readings from triplicate wells were averaged (Fig. S4).
Strains were classified into categories according to the O.D. measurement of their biofilms as follows: Those with O.D. ≤ O.D.c (O.D. of the negative control) were considered non-adherent or biofilm negative. If O.D.c < O.D. ≤ (2x O.D.c), strains were classified as weakly adherent; (2x O.D.c) < O.D. ≤ (4x O.D.c) = moderately adherent; and (4x O.D.c) < O.D. = strongly adherent.
Gelatinase assay
An inoculum from a pure culture was grown on agar plates containing 3% (w/v) gelatin and incubated for 48 hours. After incubation, plates were flooded with Frazier solution. A positive gelatinase activity was exhibited as transparent halo zones surrounding colonies44 (Fig. S5).
Antibiotics and determination MIC values
The MICs of vancomycin, tigecycline, ciprofloxacin, gentamycin and azithromycin were determined by broth microdilution performed in 96-well plates containing Mueller Hinton broth, in accordance with CLSI guidelines45.
Evaluation of the effect of antibiotic sub-MIC on biofilm formation and gelatinase activity
MICs of different antibiotics were determined by the broth microdilution method, and the antibiotics to which strong biofilm-forming E. faecalis isolates were sensitive were selected for studying the effect of their sub-MIC on biofilm production and gelatinase activity. Bacterial suspensions of the strong biofilm-forming isolates were prepared as described under the Crystal Violet assay: Wells (200 µl) were inoculated with bacterial suspensions, TSB containing 0.25% glucose and the corresponding antibiotic sub-MIC concentrations or TSB without antibiotic (antibiotic-free controls). Plates were incubated for 48 hours at 37 °C. The O.D. of isolates with and without antibiotics at sub-MIC was measured at 545 nm using the plate reader as detailed above. Results were taken as the average of triplicate reads.
The evaluation of sub-MIC antibiotic concentrations on gelatinase activity was performed in a similar way to their effect on biofilm formation. Bacterial suspensions were mixed with TSB containing the corresponding antibiotic (at sub-MICs) or supplemented TSB without antibiotic (antibiotic-free controls). A loopful of this mixture was grown on agar plates containing 3% (w/v) gelatin and incubated for 48 hr, then halos were recorded as detailed above.
Comparative genomic analysis
Five representative strains whose genomes have been previously sequenced were used in this analysis. Three of the five strains have known biofilm formation ability: E. faecalis V583 (Accession # AE016830.1), E. faecalis OG1RF (Accession # CP002621.1) and E. faecalis D32 (Accession # CP003726.1). The other two strains were E. faecalis Str. Symbioflor1 (Accession # HF558530.1), a probiotic strain with no biofilm-forming capacity, and E. faecalis 62 (Accession # CP002491.1), a commensal strain isolated from a healthy child46.
Amino acid and DNA sequences of the selected proteins and their coding genes, respectively, were obtained from NCBI based on the standard strain V583. Analysis tools for genome screens and comparative genomics were BLASTX, BLASTN47 (http://blast.ncbi.nlm.nih.gov/) and the SEED subsystems analysis and database comparative genomics tool20 (http://pubseed.theseed.org/?page = MultiGenomeCompare http://pubseed.theseed.org/comp_genomes.cgi) with reference organism set to V583 as follows: organism = 226185.1 and comparison organisms set to: 936153.3 (62), 1206105.3 (D32), 474186.5 (OG1RF) and 1261557.3 (Symbioflor 1).
Statistical Analysis
All statistical analyses, including descriptive statistics and hypothesis tests (e.g., Chi-Square, Fisher Exact Test, ANOVA and Student t-test were performed on Data Desk v. 6.3 (Data Description Inc., Ithaca, NY, USA) and GraphPad Prism (GraphPad Software Tools, Inc., La Jolla, CA, USA).
References
Richards, M. J., Edwards, J. R., Culver, D. H. & Gaynes, R. P. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect. Control Hosp. Epidemiol. 21, 510–515, doi:10.1086/501795 (2000).
Costerton, J. W. Cystic fibrosis pathogenesis and the role of biofilms in persistent infection. Trends Microbiol. 9, 50–52 (2001).
Dautle, M. P., Wilkinson, T. R. & Gauderer, M. W. Isolation and identification of biofilm microorganisms from silicone gastrostomy devices. J. Pediatr. Surg. 38, 216–220, doi:10.1053/jpsu.2003.50046 (2003).
Mohamed, J. A., Huang, W., Nallapareddy, S. R., Teng, F. & Murray, B. E. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect. Immun. 72, 3658–3663, doi:10.1128/iai.72.6.3658-3663.2004 (2004).
Lewis, K. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45, 999–1007, doi:10.1128/aac.45.4.999-1007.2001 (2001).
Pires-Bouccedil, P. D., Izumi, E., Furlaneto-Maia, L., Sturion, L. & Suzart, S. Effects of environmental and nutritional factors on gelatinolytic activity by Enterococcus faecalis strains isolated from clinical sources. Afri. J. Microbiol. Res. 4, 969–976 (2010).
Nallapareddy, S. R. & Murray, B. E. Ligand-signaled upregulation of Enterococcus faecalis ace transcription, a mechanism for modulating host-E. faecalis interaction. Infect. Immun. 74, 4982–4989, doi:10.1128/iai.00476-06 (2006).
Mandlik, A., Swierczynski, A., Das, A. & Ton-That, H. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 16, 33–40, doi:10.1016/j.tim.2007.10.010 (2008).
Nallapareddy, S. R. et al. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J. Clin. Invest. 116, 2799–2807, doi:10.1172/jci29021 (2006).
Miller, M. B. & Bassler, B. L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199, doi:10.1146/annurev.micro.55.1.165 (2001).
Hancock, L. E. & Perego, M. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 186, 5629–5639, doi:10.1128/jb.186.17.5629-5639.2004 (2004).
Nakayama, J. et al. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol. Microbiol. 41, 145–154 (2001).
Frank, K. L. et al. Use of recombinase-based in vivo expression technology to characterize Enterococcus faecalis gene expression during infection identifies in vivo-expressed antisense RNAs and implicates the protease Eep in pathogenesis. Infect. Immun. 80, 539–549, doi:10.1128/iai.05964-11 (2012).
Dunny, G. M. & Berntsson, R. P. Enterococcal sex pheromones: evolutionary pathways to complex, two-signal systems. J. Bacteriol. 198, 1556–1562, doi:10.1128/jb.00128-16 (2016).
Alby, K. & Bennett, R. J. Interspecies pheromone signaling promotes biofilm formation and same-sex mating in Candida albicans. Proc. Natl Acad. Sci. USA 108, 2510–2515, doi:10.1073/pnas.1017234108 (2011).
Clewell, D. B., Francia, M. V., Flannagan, S. E. & An, F. Y. Enterococcal plasmid transfer: sex pheromones, transfer origins, relaxases, and the Staphylococcus aureus issue. Plasmid 48, 193–201 (2002).
Srinivasan, S., Harrington, G. W., Xagoraraki, I. & Goel, R. Factors affecting bulk to total bacteria ratio in drinking water distribution systems. Water Res 42, 3393–3404, doi:10.1016/j.watres.2008.04.025 (2008).
Kadry, A. A., Tawfik, A., Abu El-Asrar, A. A. & Shibl, A. M. Reduction of mucoid Staphylococcus epidermidis adherence to intraocular lenses by selected antimicrobial agents. Chemotherapy 45, 56–60 (1999).
Perez, M. et al. IS256 abolishes gelatinase activity and biofilm formation in a mutant of the nosocomial pathogen Enterococcus faecalis V583. Can. J. Microbiol. 61, 517–519, doi:10.1139/cjm-2015-0090 (2015).
Aziz, R. K. et al. SEED Servers: high-performance access to the SEED genomes, annotations, and metabolic models. PLoS ONE 7, e48053, doi:10.1371/journal.pone.0048053 (2012).
Sindhanai, V., Avanthiga, S. S. & Suresh Chander, V. C. Antibiotic susceptibility pattern of biofilm forming and biofilm non forming enterococci species. IOSR J. Dent. Med. Sci 15, 33–37 (2016).
Akhter, J., Ahmed, S., Saleh, A. A. & Anwar, S. Antimicrobial resistance and in vitro biofilm-forming ability of Enterococci spp. isolated from urinary tract infection in a tertiary care hospital in Dhaka. Bangladesh Med. Res. Counc. Bull. 40, 6–9 (2014).
Hassan, A. et al. Evaluation of different detection methods of biofilm formation in the clinical isolates. Braz. J. Infect. Dis. 15, 305–311 (2011).
Mohamed, J. A. & Huang, D. B. Biofilm formation by enterococci. J. Med. Microbiol. 56, 1581–1588, doi:10.1099/jmm.0.47331-0 (2007).
Kristich, C. J., Li, Y. H., Cvitkovitch, D. G. & Dunny, G. M. Esp-independent biofilm formation by Enterococcus faecalis. J. Bacteriol. 186, 154–163 (2004).
Baldassarri, L. et al. Enterococcus spp. produces slime and survives in rat peritoneal macrophages. Med. Microbiol. Immunol. 190, 113–120 (2001).
Pillai, S. K. et al. Effects of glucose on fsr-mediated biofilm formation in Enterococcus faecalis. J. Infect. Dis. 190, 967–970, doi:10.1086/423139 (2004).
Merritt, J. H., Kadouri, D. E. & O’Toole, G. A. Growing and analyzing static biofilms. Curr. Protoc. Microbiol. Chapter 1, Unit 1B 1, doi:10.1002/9780471729259.mc01b01s00 (2005).
Papadimitriou-Olivgeris, M. et al. Biofilm synthesis and presence of virulence factors among enterococci isolated from patients and water samples. J. Med. Microbiol. 64, 1270–1276, doi:10.1099/jmm.0.000151 (2015).
Qin, X., Singh, K. V., Weinstock, G. M. & Murray, B. E. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 183, 3372–3382, doi:10.1128/jb.183.11.3372-3382.2001 (2001).
Roberts, J. C., Singh, K. V., Okhuysen, P. C. & Murray, B. E. Molecular epidemiology of the fsr locus and of gelatinase production among different subsets of Enterococcus faecalis isolates. J. Clin. Microbiol. 42, 2317–2320 (2004).
Mohamed, J. A. & Murray, B. E. Lack of correlation of gelatinase production and biofilm formation in a large collection of Enterococcus faecalis isolates. J. Clin. Microbiol. 43, 5405–5407, doi:10.1128/jcm.43.10.5405-5407.2005 (2005).
Sedgley, C. M. et al. Virulence, phenotype and genotype characteristics of endodontic Enterococcus spp. Oral Microbiol. Immunol. 20, 10–19, doi:10.1111/j.1399-302X.2004.00180.x (2005).
Nakayama, J., Kariyama, R. & Kumon, H. Description of a 23.9-kilobase chromosomal deletion containing a region encoding fsr genes which mainly determines the gelatinase-negative phenotype of clinical isolates of Enterococcus faecalis in urine. Appl. Environ. Microbiol. 68, 3152–3155 (2002).
Bourgogne, A. et al. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 9, R110, doi:10.1186/gb-2008-9-7-r110 (2008).
Carniol, K. & Gilmore, M. S. Signal transduction, quorum-sensing, and extracellular protease activity in Enterococcus faecalis biofilm formation. J. Bacteriol. 186, 8161–8163, doi:10.1128/jb.186.24.8161-8163.2004 (2004).
Werner, G. et al. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill. 13 (2008).
Karmarkar, M. G., Gershom, E. S. & Mehta, P. R. Enterococcal infections with special reference to phenotypic characterization & drug resistance. Indian J. Med. Res. 119(Suppl), 22–25 (2004).
Maestre, J. R. et al. In vitro interference of tigecycline at subinhibitory concentrations on biofilm development by Enterococcus faecalis. J. Antimicrob. Chemother. 67, 1155–1158, doi:10.1093/jac/dks014 (2012).
Soumet, C., Ermel, G., Fach, P. & Colin, P. Evaluation of different DNA extraction procedures for the detection of Salmonella from chicken products by polymerase chain reaction. Lett. Appl. Microbiol. 19, 294–298 (1994).
Versalovic, J., Koeuth, T. & Lupski, J. R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19, 6823–6831 (1991).
Freeman, D. J., Falkiner, F. R. & Keane, C. T. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 42, 872–874 (1989).
Christensen, G. D. et al. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22, 996–1006 (1985).
Lopes Mde, F., Simoes, A. P., Tenreiro, R., Marques, J. J. & Crespo, M. T. Activity and expression of a virulence factor, gelatinase, in dairy enterococci. Int. J. Food Microbiol. 112, 208–214, doi:10.1016/j.ijfoodmicro.2006.09.004 (2006).
CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-first Informational Supplement M100-S21., (Wayne, PA, 2011).
Brede, D. A., Snipen, L. G., Ussery, D. W., Nederbragt, A. J. & Nes, I. F. Complete genome sequence of the commensal Enterococcus faecalis 62, isolated from a healthy Norwegian infant. J. Bacteriol. 193, 2377–2378, doi:10.1128/jb.00183-11 (2011).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402 (1997).
Acknowledgements
We thank Professor AbdelGawad Hashem and Professor Magdy Amin for insightful comments and useful advice.
Author information
Authors and Affiliations
Contributions
T.M.E., A.S.Y. and R.K.A. conceived the study and designed experiments; T.M.E. and Y.A.H. provided the isolates; Y.A.H. performed all laboratory experiments; Y.A.H. and R.K.A. performed bioinformatics and genome analysis and prepared all figures and tables; Y.A.H., T.M.E., A.S.Y. and R.K.A. analyzed the data. Y.A.H., H.M.A. drafted the manuscript; A.S.Y. and R.K.A. edited the final version; all authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hashem, Y.A., Amin, H.M., Essam, T.M. et al. Biofilm formation in enterococci: genotype-phenotype correlations and inhibition by vancomycin. Sci Rep 7, 5733 (2017). https://doi.org/10.1038/s41598-017-05901-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-05901-0
This article is cited by
-
Genotype–phenotype correlation of fecal Streptococcus regulator (fsr) locus with gelatinase activity and biofilm formation intensity in clinical E. faecalis isolates
Future Journal of Pharmaceutical Sciences (2024)
-
Antimicrobial resistance patterns, virulence genes, and biofilm formation in enterococci strains collected from different sources
BMC Infectious Diseases (2024)
-
Repurposing fusidic acid as an antimicrobial against enterococci with a low probability of resistance development
International Microbiology (2024)
-
Virulence and antibiotic-resistance genes in Enterococcus faecalis associated with streptococcosis disease in fish
Scientific Reports (2023)
-
Antibiofilm activity of green synthesized silver nanoparticles against biofilm associated enterococcal urinary pathogens
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.