Abstract
Bees are considered to be threatened globally, with severe overwinter losses of the most important commercial pollinator, the Western honeybee, a major concern in the Northern Hemisphere. Emerging infectious diseases have risen to prominence due to their temporal correlation with colony losses. Among these is Deformed wing virus (DWV), which has been frequently linked to colony mortality. We now provide evidence of a strong statistical association between overwintering colony decline in the field and the presence of DWV genotype-B (DWV-B), a genetic variant of DWV that has recently been shown to be more virulent than the original DWV genotype-A. We link the prevalence of DWV-B directly to a quantitative measure of overwinter decline (workforce mortality) of honeybee colonies in the field. We demonstrate that increased prevalence of virus infection in individual bees is associated with higher overwinter mortality. We also observed a substantial reduction of infected colonies in the spring, suggesting that virus-infected individuals had died during the winter. Our findings demonstrate that DWV-B, plus possible A/B recombinants exhibiting DWV-B at PCR primer binding sites, may be a major cause of elevated overwinter honeybee loss. Its potential emergence in naïve populations of bees may have far-reaching ecological and economic impacts.
Similar content being viewed by others
Introduction
Insect pollination, which is carried out mostly by bees, is required for 75% of all food crops1; the global value of insect pollination in 2005 has been estimated at €153 billion2, although this is likely to be an underestimate3. In addition, 85% of wild plants are pollinated by animals4, chief among these being bees. Bees are therefore of considerable economic and ecological importance. The honeybee Apis mellifera is by far the most important commercial pollinator and is relied upon heavily for the successful pollination of many food crops5. However, over the last decade severe yearly losses of honeybees in the Northern Hemisphere6,7,8,9 have raised concerns about food security10.
The emergence of several infectious diseases has coincided with elevated honeybee colony losses11,12,13. Notable among these are positive single stranded RNA (positive ssRNA) viruses that have risen to prominence since the arrival of a novel biological vector from Asia, the ectoparasitic mite Varroa destructor. The mite feeds on the hemolymph of honey bee pupae and adults14. Its ability to act as a viral vector and potential incubator of several honeybee RNA viruses has given rise to a new viral transmission route, thereby aiding the spread and re-emergence of several bee viruses15,16,17.
A major source of honey bee decline is colony depopulation during the winter. Colonies suffering elevated winter worker losses are more prone to collapse, resulting in overwinter colony losses (OCL)18. High rates of OCL are a continuing concern in temperate zones, both in Europe and North America, where 15–20% and 25–30% of colonies respectively have been consistently lost overwinter in recent years19,20,21,22. Pathogens may be major contributors to OCL. Among those that have been implicated in colony decline are several positive ssRNA viruses: Kashmir bee virus (KBV)12; Slow bee paralysis virus (SBPV)23; Israeli acute paralysis virus (IAPV)11; Acute bee paralysis virus (ABPV)24, 25; Deformed wing virus (DWV)12, 24,25,26; microsporidia, particularly Nosema ceranae, an emerging gut parasite from Asia13, 27; and V. destructor, due to its association with several of the viruses28.
One virus in particular, DWV, is considered an important predictor of colony failure25, 26, 28,29,30. Varroa destructor has altered the dynamics of DWV dramatically, reducing within-host genetic DWV strain diversity, and leading to the global spread and dominance of genetic variants related to DWV genotype A (DWV-A)17. We have recently shown that another recently described genotypic variant of DWV, genotype B (DWV-B, also known as Varroa destructor virus-1 or VDV-117, 31, 32, is more virulent than classic DWV-A33. Although DWV-B has been recorded in Europe, Africa and Asia34,35,36,37 and is widespread in UK33, its role in OCL is currently unknown.
To date, field studies on the role of pathogens in OCL have been largely based on correlations between pathogens discovered at the colony-level and qualitative assessments of colony health (i.e. stable or collapsed colony). Quantitative field measures of intra-colony disease dynamics provide a better insight into their role in OCL. For example, measurements of pathogen prevalence and workforce mortality within colonies would permit a more precise understanding of the change in disease and overall health status of overwintering colonies. We address this shortfall by assessing the pathogen landscape in 28 colonies in two apiaries before and after the winter season while experimentally controlling V. destructor infestation levels. We then determined whether there is an explicit association between DWV-B and overwinter reduction of the honeybee colony workforce.
Results
Pathogen detection
To identify pathogens that may be responsible for overwinter honeybee mortality, we conducted an apiary study to assess the overwinter change in pathogen composition in 28 managed honeybee colonies. Sampling was performed once in autumn, at the end of brood rearing, and once the following spring, before commencement of major brood rearing, allowing us to exclude birth rate and directly to estimate loss of workers per colony. We initially used multiple ligation probe amplification (MLPA) to target a wide range of RNA viruses and RT-PCR to identify two microsporidian gut pathogens: Nosema apis and Nosema ceranae. For these analyses, we pooled 30 honeybee workers per colony and analysed one total RNA extract per colony.
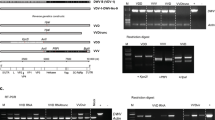
Apart from chronic bee paralysis virus (CBPV), which occurred rarely in both autumn and spring, only viruses belonging to the DWV complex and Black queen cell virus (BQCV) could be detected by MLPA (Supplementary Table S1). We subsequently employed qRT-PCR on the same pooled samples to gain a more accurate understanding of the two prominent viruses: BQCV and DWV. In addition to BQCV and the original DWV-A genotype, we screened for DWV-B which shares 84% nucleotide identity with DWV-A14. DWV-A and -B were screened for by genotype-specific qRT-PCR amplification of Leader polypeptide (Lp) and RNA-dependent RNA polymerase (RdRp) genes; these are located at the 5′ and 3′ parts of the viral genome, respectively14.
We found that 96% (N = 27) of colonies in the autumn contained significant quantities of DWV-B (Supplementary Table S2). Compared with the autumn, prevalence of DWV-B in spring colonies was significantly reduced, with 21% (N = 6) of colonies containing DWV-B (χ2 1 = 19.1, p < 0.0001; Fig. 1). Some colonies in both autumn and spring contained very low viral loads of the DWV-A genotype, but these fell below an a priori detection threshold, and were only detected using the RdRp primer pair (Supplementary Table S2). This indicated that DWV-A was not (or at best, very rarely) a cause of infection in these colonies. DWV-B titres were also significantly reduced in spring colonies compared to autumn levels (paired Wilcoxon signed rank test V = 378, p < 0.00001) (Fig. 2). Potential caveats are that we may have missed very low titres of DWV-A because of pooling 30 bees per extract and that the qPCR primers for DWV-A could have been of lower sensitivity than those for DWV-B. To address the first caveat, we subsequently sampled individual honeybees (15 individuals per colony) but still did not detect DWV-A (see ‘Overwinter mortality and pathogen prevalence’). In relation to the second caveat, primer sensitivity was similar for DWV-A and DWV-B primers (Supplementary Table S4).
Interestingly, no recombinant viruses were detected (i.e. samples containing a mix of DWV-A and DWV-B fragments, Supplementary Table S2). However, a few samples positive for DWV-B in the RdRp region (N = 3 in autumn and N = 5 in spring) could not be amplified in the Lp region (Supplementary Table S2). This was attributed to the low amount of DWV-B detected coupled with the lower sensitivity of the DWV-B primer in this region (Supplementary Table S4).
The prevalence of BQCV was also high in autumn colonies (82%, N = 22; Supplementary Table S2), and then significantly reduced in spring to 18% (N = 5; χ2 1 = 15.1, p = 0.0001; Fig. 1). BQCV titres were significantly lower in spring compared to autumn colonies (V = 272, p < 0.00001), also following the same pattern as for DWV-B. However, DWV-B titres in autumn sampling were significantly higher than those of BQCV (V = 377, p < 0.00001; see Fig. 2).
The Microsporidia N. ceranae and N. apis were detected in 54% and 25% of colonies in the autumn, respectively. Prevalences were also significantly lower in the spring, with 18% (N = 5) and 0% of colonies carrying each pathogen, respectively (χ2 1 = 8.1, 5.1; p = 0.004, 0.023 respectively; Fig. 1).
Overwinter mortality and pathogen prevalence
To understand the link between pathogens and overwinter workforce mortality in finer detail, we estimated the prevalence of pathogens within colonies by sampling 15 additional individual honeybees from each colony in autumn, using MLPA for RNA viruses and RT-PCR for Nosema spp. To verify the rarity of DWV-A across the colonies in the pooled samples (0% true prevalence from 30 pooled individuals per colony (N = 840), 95% confidence intervals (CIs): 0–0.45%), a subset of 37 individual workers found positive for DWV in MLPA (representing ca. 37% of DWV MLPA-positive samples, distributed randomly across 7 colonies) was subjected to qRT-PCR using the DWV-A specific primers. DWV-A was not present in any of the samples tested, supporting our previous analyses and suggesting that, if present, DWV-A had an extremely low titre. Therefore, DWV-positive individual honeybees screened by MLPA are treated as DWV-B, and are henceforth reported as such.
The analysis of individual honeybees confirmed the disease patterns detected in pools of 30 worker honeybees per colony (above), with DWV-B featuring most frequently among autumn bees (Supplementary Table S2). Generalized linear models (GLMs) were conducted to infer whether within-colony pathogen prevalence (including V. destructor infestation estimated from 150 honeybees per colony) were important predictors of overwinter workforce mortality. GLMs were conducted either with DWV-B prevalence (model 1) or V. destructor infestation (model 2) in addition to all other detected pathogens and apiary site as explanatory variables. Simplified final models either contained only DWV-B or only V. destructor but not BQCV as significant predictors of overwinter colony workforce mortality (Table 1).
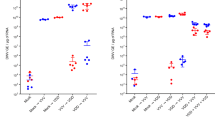
The relationships between pathogen prevalence and overwinter mortality for the DWV-B and BQCV viruses (Fig. 3), CBPV and Microsporidian pathogens (Supplementary Fig. S1), and V. destructor (Fig. 4) were tested using GLMs (Table 1) and Spearman’s correlations (Figs 3 and 4), which indicated that DWV-B and V. destructor were closely tied to overwinter colony decline but BQCV was not. In colonies suffering >50% adult worker mortality (N = 10 colonies), DWV-B prevalence and V. destructor infestation averaged (standard deviation, SD): 33% (SD 21%) and 13.3% (SD 11.3%) and in the remaining colonies (N = 18), 14.4% (SD 16.1%) and 1.4% (SD 2.5%) respectively.
(a) Relationship between V. destructor infestation and overwinter mortality within each colony. Spearman’s ρ and significance levels are shown. (b) Levels of V. destructor infestation and overwinter mortality across the two acaricide treatments (in the periodic treatment, acaricides were removed from 26th July until 18th October 2011).
Varroa destructor infestation and acaricide treatment were negatively associated (Wilcoxon signed rank test, W = 195, p < 0.0001), demonstrating that intensive acaricide treatment had worked effectively to remove V. destructor mites from colonies (Fig. 4b). Additionally, V. destructor and within-colony DWV-B prevalence were positively correlated (GLM, t = 2.486, p = 0.02), while BQCV, CBPV and Nosema spp. were not found to be affected by the mite infestation levels or acaricide treatment (p > 0.05 in all cases).
DWV composition
In order to verify the DWV-B variant detected in our study, a viral propagation derived from a DWV-B positive colony extract was subjected to ultra-deep sequencing. Results (presented in ref. 33) confirmed our qRT-PCR finding; DWV was the only virus detected in our field extract, with DWV-B being the main genetic variant detected. A very low background level of DWV-A was also present but accounted for only 0.13% of all DWV reads compared to 99.87% of DWV-B reads. A third DWV variant, DWV-C38, was not detected (Supplementary Table S3).
An important caveat is that, though we did not detect any DWV-A/B recombinants in our samples, either by qPCR or by ultra-deep sequencing, we cannot exclude the possibility that one or more colonies harboured a DWV-A/B recombinant that caused high mortality. If this is the case, then the recombinant comprises predominantly DWV-B at both 5′ (Lp gene) and 3′ (RdRp gene) parts of its genome and DWV-A somewhere in its centre.
Discussion
Colony losses are a major source of ongoing honeybee population decline in temperate zones21, 22, 39. The V. destructor mite and its association with viruses have together been implicated as a possible cause of widespread losses of honeybee colonies globally9, 11, 24, 26, 29, 40. But determining which among several potential factors of OCL are more important has remained elusive. In a field study, we now find the prevalence of DWV-B, an emerging variant of DWV17, 31, 32, and V. destructor infestation to be highly correlated with overwinter honeybee worker loss. Low virus titres in bees surviving to spring further suggested that individuals infected with high DWV-B titres had died overwinter. To our knowledge, our study is the first to provide strong correlative evidence of this distinct genotypic variant of DWV and OCL in the absence of the classical DWV-A.
Colony losses have been linked to the presence of other positive ssRNA viruses, in particular to the ABPV-KBV-IAPV complex11, 25 and to SBPV9 that, in association with V. destructor, have been linked to high virulence and individual-level bee mortality41, 42. Our data are in line with the view that DWV, although less virulent when vectored by V. destructor than viruses such as ABPV and SBPV40, 43, may be more widespread and damaging at the colony and population level44. Unlike ABPV and SBPV, honeybee pupae (and their V. destructor ectoparasites) infected with DWV typically do not die prior to eclosion. Survival of infected pupae into adulthood permits the ongoing spread of infection by co-emerging V. destructor mites, and therefore increases the persistence of infections within colonies. Field evidence shows that, of the documented honeybee viruses, DWV occurs most frequently, and at high levels globally wherever V. destructor is also present17. In accordance, a recent study based on phylogenetic analysis of DWV strongly suggested that the dispersal of DWV around the globe followed a similar pattern to the spread of V. destructor mites45.
Although levels of V. destructor infestation in colonies were significantly correlated with the prevalence of DWV-B, making it difficult to disentangle their effects in OCL, reduced virus detection in the spring suggests that colony depopulation during winter can be attributed to the death of heavily DWV-B infected individuals. In addition, we have recently shown that, under lab conditions, DWV-B is more virulent that DWV-A, leading to significantly reduced honeybee worker survival even in the absence of V. destructor 33. Taken together, these results suggest that the variance in overwinter honeybee loss can be largely accounted for by differences among colonies in autumn DWV-B loads.
Interestingly, we only detected the DWV-B genotype among our honeybee samples, and found little evidence of either the classic DWV-A, or recombinant forms of DWV-A and -B. An increasing number of studies report the presence of recombinants between DWV-A /-B in nature46,47,48,49, and it has been suggested that recombinants may be more virulent than DWV-A or -B, at least in honeybee pupae48. In the present study, we did not detect any mixed infections in our field samples to indicate the presence of recombinants. In addition, recombinant DWV-A/B viruses so far reported by others46,47,48,49 all comprise DWV-A at their 3′ end (including the RdRp gene), whereas colonies in our study contained only DWV-B RdRp; we can therefore exclude the possibility that we oversaw previously detected recombinant variants. Yet we cannot exclude the possibility that one or more colonies in which we detected DWV-B may have harboured a novel recombinant DWV-A/B virus comprising DWV-B at its 5′ and 3′ parts, where respectively our Lp and RdRp primers bound to the DWV genome. Alternatively, for a small number of samples with low virus load, the Lp region remained uncharacterized (no amplification with either the DWV-A or DWV-B Lp primers), possibly due to the lower sensitivity of the Lp primers. Given a recent study by Dalmon et al.49, which suggests the presence of a recombination breakpoint located in the region where our Lp primers bind to the DWV genome, we cannot exclude the possibility of recombinants being present in some of our colonies.
Knowledge of the distribution of DWV genotypes across Europe and more widely across the globe is largely lacking. In a recent survey across Great Britain, we found that DWV-B is widespread across the landscape33. Preliminary data suggest that DWV-B in addition to recombinant DWV-A/-B forms are currently more prevalent in Europe than in North America46, 48, 50. Our findings highlight the significant risk posed by this emerging virus genotype to the wellbeing of managed honeybee populations; understanding the global distribution of DWV genotypes is an urgent necessity.
In conclusion, our findings support the view that DWV-B poses a particular problem in temperate zones, when queens cease egg laying during winter and the workforce is not replenished by newly emerging workers for several months. Given its putatively higher virulence, this specific genetic variant of DWV can be considered a sufficient cause of premature death in otherwise healthy adult worker bees, even in the absence of its vector the V. destructor mite. However, the extent and rate of overwinter decline of a standing population of honeybee workers in a colony, and therefore the chance of that colony’s survival into spring, will be a function of initial workforce size, virus load and prevalence, in addition to other factors such as availability of overwinter resources. Models of honeybee colony dynamics that incorporate Varroa-virus interactions explicitly (reviewed in ref. 51) seem especially appropriate in light of our data.
Methods
Field Experiment
Fourteen honeybee colonies from an apiary at Kenzingen (48°11′30″N 7°46′6″E) and 14 colonies at an apiary in Simonswald (48°6′1″N 8°3′21″E), both in Baden-Württemberg, Germany, were monitored overwinter from 2011–2012. All colonies were treated with the acaricides flumethrin (Bayvarol®) and coumaphos (CheckMite+™) except from 26th July until 18th October 2011, when acaricides was removed from half of the colonies at each site47 Worker bees from each colony were freeze-killed on dry-ice on 26th September 2011 and 9th April 2012 and subsequently preserved at −80 °C for pathogen analysis. To quantify levels of overwinter worker mortality we applied the Liebefeld method52 to estimate numbers of bees in each colony in autumn (17th October 2011), after most brood rearing had stopped, and early spring (28th February 2012), before new workers had enclosed. These data allowed us to quantify precisely overwinter mortality of each colony (Supplementary Table S2). Levels of V. destructor infestation were assessed by counting the number of mites associated with samples of 150 adult workers per colony (17th October 2011)53 (Supplementary Table S2).
RNA extraction and pathogen detection
For each colony, 30 adult worker bees from each sampling period (September 2011 and April 2012) were pooled in RNAse free mesh bags (Bioreba, Reinach, Switzerland) and crushed in 6 mL DEPC treated (RNAse free) water while snap-frozen in liquid nitrogen, 150 μl of which was used for RNA isolation. In addition, 15 adults per colony (collected during exactly the same sampling times as the pooled samples above) were individually crushed in 500 μl RLT buffer plus 1% b-mercaptoethanol, 100 μl of which was used for RNA isolation. RNA was obtained using the RNeasy mini kit in a QiaCube robot (Qiagen, Hilden, Germany). We thereby aimed to capture all RNA viruses in our extracts, including those with low titre.
We targeted the amplification of 10 widespread viruses and 2 Microsporidia (N. ceranae, N. apis). As we conducted an investigation of overwinter mortality when brood production largely ceases, we did not screen for brood diseases such as European and American Foulbrood (Paenibacillus larvae, Melissococcus plutonius respectively) or Chalkbrood (Ascosphaera apis). For virus screens of pooled and individual samples, we employed multiple ligation-dependent probe amplification (MLPA) using the RT-MLPA® kit (MRC-Holland, Netherlands) according to manufacturer’s instructions, using probes designed for the positive strand of the following RNA virus complexes: DWV-VDV-KV; CBPV; ABPV-IAPV-KBV; BQCV; SBPV; SBV, and β-actin (host housekeeping gene)54. Amplified fragments were viewed on a QIAxcel capillary fragment analyzer (Qiagen), using an acceptance threshold of 0.1 relative fluorescence units.
As MLPA probes were unable to distinguish between DWV-A and DWV-B, pooled samples were re-analyzed using qRT-PCR to gain higher resolution data on DWV and BQCV, the main RNA viruses found among colonies. Total cDNA was synthesized using M-MLV Revertase (Promega) following manufacturer’s instructions, using 800 ng of sample RNA. For absolute viral load, separate qPCRs were performed for DWV-A, DWV-B, BQCV and A. mellifera RP49 (housekeeping gene) in a Bio-Rad C1000 Thermal Cycler (Bio-Rad) using 2 x SensiMix SYBR and Fluorescein (Bioline), 0.2 µM of each primer and 1 µl of 1:10 diluted cDNA in a final volume of 10 µl. Negative controls comprising all reaction components except cDNA template were included in each run. Each reaction was performed in duplicate and the average quantification cycle (Ct) value was used (the maximum accepted Ct difference between replicates was set to one Ct). Amplification was performed using the following thermal profile: 5 min at 95 °C, followed by 40 cycles of 10 sec at 95 °C and 30 sec at 57 °C (including a read at each cycle). Post amplification melting curve analysis was used to check for non-specific amplification (50–90 °C with an increment of 0.5 °C s−1). To minimize risk of false positives, an upper threshold of 35 Ct for detection of DWV-A, DWV-B and BQCV was applied55.
For DWV-A/-B we conducted qRT-PCR amplification in two regions at either end of the genome: the leader polypeptide (Lp, 5′ end of genome) and RNA-dependent RNA polymerase (RdRp, 3′ end of genome), in order to screen for possible recombinants. Samples positive for DWV-B in both RdRp and Lp regions that showed negative amplification (or Ct > 35) for DWV-A (Lp and RpRd region) were classified as DWV-B positive. In the case of samples (n = 3 autumn, n = 5 spring samples) that showed positive amplification only in the DWV-B RdRp region and not in the DWV-B Lp region, samples were also considered to be DWV-B positive as no amplification was detected in the DWV-A Lp or DWV-A RdRp regions to indicate the presence of a recombinant. Absolute quantification of DWV-A, DWV-B and BQCV was calculated using duplicate DNA standard curves of purified PCR products (DWV-A/-B) or plasmids (BQCV, RP49), with efficiencies between 90% to 97% and correlation coefficients (R2) from 0.985–0.999. Pooled sample virus loads are reported as average viral titre per bee, calculated by the mean viral load of 30 pooled bees and taking into account the qRT-PCR starting quantities (Supplementary Table S2). Absolute quantities and prevalence of DWV-A and -B in pooled colony samples were calculated using the RdRp primers. Additionally, we also calculated normalized viral titres to the amount of RP49 mRNA per sample to account for differences in RNA integrity across samples (Supplementary Table S2). Nosema spp. detection was performed from total cDNA by RT-PCR using species-specific primers. All (q)RT-PCR primers used in the present study are listed in Supplementary Table S4. The high specificity of DWV primers spanning in the RdRp region has been previously verified in McMahon et al.33 (see Supplementary Table S3 in ref. 33).
Virus verification
In order to verify the identity of DWV genotypes detected in our samples, one of the DWV-B positive pooled-colony samples, devoid of BQCV and CBPV, namely colony ts3, was used to produce a virus extract and was subsequently subjected to ultra-deep sequencing on an Illumina platform (GATC Biotech, Konstanz, Germany). Details of the method are available in McMahon et al.33, where exactly the same viral extract was used for experimental purposes.
Statistical analysis
All analyses were performed in R v 3.1.356. We compared pathogen presence in autumn and spring colonies using McNemar’s χ2 (repeated colony measure) and tested for differences in pathogen load in autumn and spring using paired Wilcoxon signed rank tests. The latter analysis was performed using either absolute viral titre derived from standard curves or viral titre normalized to the host housekeeping gene (RP49). As both approaches yielded the same results, reported test statistics refer to absolute viral titres. Bee mortality at apiaries was analyzed in a generalized linear model as a two-vector response variable (number of dying and surviving bees). Varroa destructor infestation and treatment were negatively associated but as V. destructor was a significantly better fit (in terms of residual deviance) than acaricide treatment, it was retained in the model. Varroa destructor infestation and DWV-B were positively associated (see results). While V. destructor was superior to DWV-B in terms of model fit, we conducted parallel models with either DWV-B (model 1) or V. destructor infestation (model 2) because the causal relationship between DWV-B or V. destructor and mortality is unclear. All quantitative predictors were standardized to a mean of zero and standard deviation of one prior to analysis.
Models containing either DWV-B or V. destructor in addition to BQCV, CBPV, N. ceranae, N. apis and apiary site were simplified by performing automated model selection using the ‘dredge’ function of the R-package ‘MuMIn’57. Inspection of residual deviance indicated overdispersion, which was accommodated by using a quasibinomial error structure. We used the Akaike information criterion adjusted for overdispersion and corrected for small sample sizes (QAICc58) calculated by the ‘dredge’ function for all plausible subsets of model, to select explanatory variables driving overwinter worker mortality. Model residuals were explored graphically, while Shapiro-Wilk’s and White’s Tests were used to examine normality (‘shapiro.test’ function) and heteroskedasticity (‘het.test’ function), respectively. McFadden’s pseudo r 2 values are reported. The Spearman’s rank correlation test was applied to assess the relationship between the prevalence of each pathogen (%) and overwinter worker mortality (%).
Compliance with ethical standards
All experiments conducted and presented in the manuscript comply with the laws and rules of the institution and country in which they were performed.
Data availability statement
The datasets generated during the current study are included in this published article (and its Supplementary Information files).
References
Klein, A.-M. et al. Importance of pollinators in changing landscapes for world crops. Proc. R. Soc. B Biol. Sci. 274, 303–313 (2007).
Gallai, N., Salles, J. M., Settele, J. & Vaissière, B. E. Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol Econ 68 (2009).
Potts, S. G. et al. Safeguarding pollinators and their values to human well-being. Nature. 540, 220–229 (2016).
Ollerton, J., Winfree, R. & Tarrant, S. How many flowering plants are pollinated by animals? Oikos. 120, 321–326 (2011).
Morse, R. A. & Calderone, N. W. The value of honey bees as pollinators of U.S. crops in 2000. Bee Cult 1–15 (2000).
Oldroyd, B. P. What’s killing American honey bees? PLoS Biol. 5, 1195–1199 (2007).
Stokstad, E. The case of the empty hives. Science. 316, 970–972 (2007).
Moritz, R. F. A. et al. Research strategies to improve honeybee health in Europe *. Apidologie. 41, 227–242 (2010).
Neumann, P. & Carreck, N. L. Honey bee colony losses. J. Apic. Res. 49, 1–6 (2010).
Potts, S. G. et al. Global pollinator declines: trends, impacts and drivers. Trends Ecol. Evol. 25, 345–53 (2010).
Cox-Foster, D. L. et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 318, 283–287 (2007).
Johnson, R. M., Evans, J. D., Robinson, G. E. & Berenbaum, M. R. Changes in transcript abundance relating to colony collapse disorder in honey bees (Apis mellifera). Proc. Natl. Acad. Sci. USA 106, 14790–5 (2009).
Paxton, R. Does infection by Nosema ceranae cause ‘Colony Collapse Disorder’ in honey bees (Apis mellifera)? J. Apic. Res. 49, 80 (2010).
de Miranda, J. R. & Genersch, E. Deformed wing virus. J. Invertebr. Pathol. 103, S48–S61 (2010).
Chen, Y. P. & Siede, R. Honey bee viruses. Adv. Virus Res. 70, 33–80 (2007).
Genersch, E. & Aubert, M. Emerging and re-emerging viruses of the honey bee (Apis mellifera L.). Vet. Res. 41, 54 (2010).
Martin, S. J. et al. Global honey bee viral landscape altered by a parasitic mite. Science. 336, 1304–1306 (2012).
Russell, S., Barron, A. B. & Harris, D. Dynamic modelling of honey bee (Apis mellifera) colony growth and failure. Ecol. Modell. 265, 158–169 (2013).
van der Zee, R. et al. Managed honey bee colony losses in Canada, China, Europe, Israel and Turkey, for the winters of 2008–9 and 2009–10. J. Apic. Res. 51, 100–114 (2012).
Spleen, A. M. et al. A national survey of managed honey bee 2011-12 winter colony losses in the United States: results from the Bee Informed Partnership. J. Apic. Res. 52, 44–53 (2013).
Seitz, N. et al. A national survey of managed honey bee 2014–2015 annual colony losses in the USA. J. Apic. Res. 54, 292–304 (2015).
Laurent, M., Hendrikx, P., Ribiere-Chabert, M. & Chauzat, M.-P. A pan-European epidemiological study on honeybee colony losses 2012–2014. 2013, (2014).
Carreck, N. & Neumann, P. Honey bee colony losses. J. Apic. Res. 49, 1 (2010).
Genersch, E. et al. The German bee monitoring project: a long term study to understand periodically high winter losses of honey bee colonies. Apidologie. 41, 332–352 (2010).
Francis, R. M., Nielsen, S. L. & Kryger, P. Varroa-Virus interaction in collapsing honey bee colonies. PLoS One 8 (2013).
Highfield, A. C. et al. Deformed Wing Virus implicated in overwintering honeybee colony losses. Appl. Environ. Microbiol. 75, 7212–7220 (2009).
Higes, M. et al. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 10, 2659–2669 (2008).
Rosenkranz, P., Aumeier, P. & Ziegelmann, B. Biology and control of Varroa destructor. J. Invertebr. Pathol. 103, S96–S119 (2010).
Dainat, B., Evans, J. D., Chen, Y. P., Gauthier, L. & Neumann, P. Dead or alive: Deformed wing virus and Varroa destructor reduce the life span of winter honeybees. Appl. Environ. Microbiol. 78, 981–987 (2012).
van Dooremalen, C. et al. Winter survival of individual honey bees and honey bee colonies depends on level of Varroa destructor infestation. PLoS One 7, e36285 (2012).
Ongus, J. R. et al. Complete sequence of a picorna-like virus of the genus Iflavirus replicating in the mite Varroa destructor. J. Gen. Virol. 85, 3747–3755 (2004).
Mordecai, G. J. et al. Superinfection exclusion and the long-term survival of honey bees in Varroa-infested colonies. ISME J. 10, 1–10 (2016).
McMahon, D. P. et al. Elevated virulence of an emerging viral genotype as a driver of honeybee loss. Proc. R. Soc. B Biol. Sci. doi:10.1098/rspb.2016.0811 (2016)
Benaets, K. et al. Covert deformed wing virus infections have long-term deleterious effects on honeybee foraging and survival. doi:10.1098/rspb.2016.2149 (2017).
Zioni, N., Soroker, V. & Chejanovsky, N. Replication of varroa destructor virus 1 (VDV-1) and a varroa destructor virus 1-deformed wing virus recombinant (VDV-1-DWV) in the head of the honey bee. Virology. 417, 106–112 (2011).
Radzevičiūtė, R. et al. Replication of honey bee-associated RNA viruses across multiple bee species in apple orchards of Georgia, Germany and Kyrgyzstan. J. Invertebr. Pathol. doi:10.1016/j.jip.2017.04.002 (2017).
Strauss, U. et al. Seasonal prevalence of pathogens and parasites in the savannah honeybee (Apis mellifera scutellata). J. Invertebr. Pathol. 114, 45–52 (2013).
Mordecai, G. J., Wilfert, L., Martin, S. J., Jones, I. M. & Schroeder, D. C. Diversity in a honey bee pathogen: first report of a third master variant of the Deformed Wing Virus quasispecies. ISME J. 10, 1–10 (2016).
van der Zee, R. et al. Results of international standardised beekeeper surveys of colony losses for winter 2012-2013: analysis of winter loss rates and mixed effects modelling of risk factors for winter loss. J. Apic. Res. 53, 19–34 (2014).
Salvy, M. et al. Modifications of the cuticular hydrocarbon profile of Apis mellifera worker bees in the presence of the ectoparasitic mite Varroa jacobsoni in brood cells. Parasitology. 122, 145–159 (2001).
Bailey, L. & Woods, R. D. Three previously undescribed viruses from the honey bee. J. Gen. Virol. 25, 175–186 (1974).
Martin, S. J. The role of varroa and viral pathogens in the collapse of honeybee colonies: A modelling approach. J. Appl. Ecol 38, 1082–1093 (2001).
Bailey, L. & Gibbs, a. J. Acute infection of bees with Paralysis Virus. J. Insect Pathol. 6, 395–407 (1964).
Schroeder, D. C. & Martin, S. J. Deformed wing virus: the main suspect in unexplained honeybee deaths. Virulence. 3, 589–598 (2012).
Wilfert, L. et al. Deformed Wing Virus is a recent global epidemic in honeybees driven by Varroa mites. Science. 351, 594–597 (2016).
Moore, J. et al. Recombinants between Deformed wing virus and Varroa destructor virus-1 may prevail in Varroa destructor-infested honeybee colonies. J. Gen. Virol. 92, 156–61 (2011).
Wang, H. et al. Sequence recombination and conservation of Varroa destructor virus-1 and Deformed wing virus in field collected honey bees (Apis mellifera). PLoS One 8, 1–9 (2013).
Ryabov, E. V. et al. A virulent strain of deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Pathog. 10, e1004230 (2014).
Dalmon, A. et al. Evidence for positive selection and recombination hotspots in Deformed wing virus (DWV). Sci. Rep. 7, 41045, doi:10.1038/srep41045 (2017).
Cornman, R. S. et al. Population-genomic variation within RNA viruses of the Western honey bee, Apis mellifera, inferred from deep sequencing. BMC Genomics. 14, 154 (2013).
Becher, M. A., Osborne, J. L., Thorbek, P., Kennedy, P. J. & Grimm, V. Towards a systems approach for understanding honeybee decline: A stocktaking and synthesis of existing models. J. Appl. Ecol. 50, 868–880 (2013).
Imdorf, A., Buehlmann, G., Gerig, L., Kilchenmann, V. & Wille, H. Überprüfung der Schätzmethode zur Ermittlung der Brutfläche und der Anzahl Arbeiterinnen in Freifliegenden Bienenvölkern. Apidologie. 18, 137–146 (1987).
Frey, E. & Rosenkranz, P. Autumn invasion rates of Varroa destructor (Mesostigmata: Varroidae) into honey bee (Hymenoptera: Apidae) colonies and the resulting increase in mite populations. J. Econ. Entomol. 107, 508–515 (2014).
de Smet, L. et al. BeeDoctor, a versatile MLPA-based diagnostic tool for screening bee viruses. PLoS One 7 (2012).
de Miranda, J. R. et al. Standard methods for virus research in Apis mellifera. J. Apic. Res. 52, 1–48 (2013).
R Core Team R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing (2014).
Barton, K. MuMIn: Multi-model inference. R package version 1.15.6. Available at: https://cran.r-project.org/web/packages/MuMIn/index.html (2016).
Burnham, K. & Anderson, D. Model selection and multimodel inference: a practical information-theoretic approach. (Springer, 2002).
Acknowledgements
We thank S. Erler for assistance during the sampling, J.R. de Miranda and L. De Smet for supplying qRT-PCR and MLPA positive controls respectively, in the form of plasmids, and the anonymous referees for their valuable comments, which helped strengthen the manuscript. This work was supported by funds from BBSRC, NERC, Defra, the Scottish Government and the Wellcome Trust as part of the UK Insect Pollinators Initiative (IPI grant BB/I000100/1), the Federal Ministry of Food, Agriculture and Consumer Protection (Germany): Fit Bee project (grant 511-06.01-28-1-71.007-10), and the EU: BeeDoc (grant 244956).
Author information
Authors and Affiliations
Contributions
M.E.N., D.P.M., V.D. and R.J.P. conceived the study. E.F. and P.R. conducted and designed the field experiment. M.E.N., D.P.M. and V.D. carried out the laboratory molecular work. M.E.N. and D.P.M. analysed the data and drafted the manuscript; all co- authors contributed to data interpretation and to the writing of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Natsopoulou, M.E., McMahon, D.P., Doublet, V. et al. The virulent, emerging genotype B of Deformed wing virus is closely linked to overwinter honeybee worker loss. Sci Rep 7, 5242 (2017). https://doi.org/10.1038/s41598-017-05596-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-05596-3
This article is cited by
-
Transmission of deformed wing virus between Varroa destructor foundresses, mite offspring and infested honey bees
Parasites & Vectors (2022)
-
A derived honey bee stock confers resistance to Varroa destructor and associated viral transmission
Scientific Reports (2022)
-
Viral communities in the parasite Varroa destructor and in colonies of their honey bee host (Apis mellifera) in New Zealand
Scientific Reports (2022)
-
Viral load, not food availability or temperature, predicts colony longevity in an invasive eusocial wasp with plastic life history
Scientific Reports (2021)
-
Genetic variations and relationships between deformed wing virus strains infesting honey bees based on structural proteins
Biologia (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.