Abstract
Scale insects (Sternorrhyncha: Coccoidea) are one of the most invasive and agriculturally damaging insect groups. Their management and the development of new control methods are currently jeopardized by the scarcity of identification data, in particular in regions where no large survey coupling morphological and DNA analyses have been performed. In this study, we sampled 116 populations of armored scales (Hemiptera: Diaspididae) and 112 populations of soft scales (Hemiptera: Coccidae) in Chile, over a latitudinal gradient ranging from 18°S to 41°S, on fruit crops, ornamental plants and trees. We sequenced the COI and 28S genes in each population. In total, 19 Diaspididae species and 11 Coccidae species were identified morphologically. From the 63 COI haplotypes and the 54 28S haplotypes uncovered, and using several DNA data analysis methods (Automatic Barcode Gap Discovery, K2P distance, NJ trees), up to 36 genetic clusters were detected. Morphological and DNA data were congruent, except for three species (Aspidiotus nerii, Hemiberlesia rapax and Coccus hesperidum) in which DNA data revealed highly differentiated lineages. More than 50% of the haplotypes obtained had no high-scoring matches with any of the sequences in the GenBank database. This study provides 63 COI and 54 28S barcode sequences for the identification of Coccoidea from Chile.
Similar content being viewed by others
Introduction
Scale insects (Hemiptera: Coccoidea) are key pests on crops and ornamental plants worldwide. The three most important families in terms of economic damage and number of genera are Diaspididae (419 genera), Pseudococcidae (272 genera) and Coccidae (170 genera)1. Scale insects can cause economic damage directly, through sap sucking, the injection of toxins and the transmission of viruses that weaken the plant, lowering fruit quality and yield. Mealybugs (Pseudococcidae) and soft scales (Coccidae) may also cause indirect damage, by excreting honeydew onto the plant surface, thereby favoring the development of sooty mold fungi. Scale insects are also invasive pests2 for which quarantine is required in several markets, leading to the rejection of products during international trade. As a result, active inspections for these insects are carried out by national phytosanitary services, and their presence is monitored by most farm advisors. One of the principal problems in scale insect management concerns the difficulty of their identification. Morphological identification is possible but difficult: it requires high level of expertise and the examination of adult females, which are not always found in the field or in the commercial products. Moreover, even when adult females are examined, it is sometimes difficult to distinguish between very similar species3, 4. These difficulties have resulted in an overall lack of knowledge for fine-scale taxonomic identification5, 6 and, probably, widespread errors in management, with the implementation of control methods that are inappropriate for the species actually present in the target area7. In this context, the use of DNA barcoding has gradually increased over the last decade, as a means of disentangling complexes of cryptic species and surveying the biodiversity occurring in various regions of the world8, 9. Studies have been performed on the Diaspididae10,11,12, Pseudococcidae12,13,14,15,16 and Coccidae3, 17. Phylogenetic studies have also provided useful information for the barcoding of scale insects18, 19. However, most of these studies have focused on the insects of North America, Asia and Europe. Morphological studies have been performed on scale insects from South America20,21,22, but the marked lack of genetic information for scale insects from this continent is problematic for the efficient management of these pest species in this region. Previous DNA barcoding studies on scale insects have focused on the Pseudococcidae in Chile14, 23 and Brazil24. The Diaspididae and Coccidae families remain poorly characterized and underexplored.
We present here a survey of the Diaspididae and Coccidae over a latitudinal gradient in Chile, with the aim of providing a comprehensive genetic barcoding database. We collected scale insects from various fruit crops, ornamental plants and trees and shrubs growing next to crops and sequenced their COI and 28S genes. Our objectives were: (i) to survey the biodiversity of scales, (ii) to associate barcode sequences to each of the Chilean species identified on the basis of morphological characters; (iii) to explore the diversity of DNA sequences as a means of unraveling possible complexes of cryptic species. Chile is a particularly interesting location for studies of diversity via DNA barcoding. Its unique geography resembles that of an island and its environmental features create highly diverse agroecosystems and structured insect populations23. These conditions favor the establishment of new invading pests and the colonization of crops by native species, as observed for mealybugs infesting vineyards14, 25. Survey data are also of direct interest to stakeholders in Chile, such as the Chilean National Agricultural and Livestock Service (Servicio Agrícola y Ganadero), which carries out phytosanitary checks on exported products and prevents the introduction of new pests that might seriously damage Chilean agriculture.
Results
Coccidae
Eight Coccidae species were identified on the basis of morphology: Ceroplastes sinensis Del Guercio, Coccus hesperidum Linnaeus, Parasaissetia nigra (Nietner), Parthenolecanium corni (Bouché), Protopulvinaria pyriformis Cockerell, Pulvinariella mesembryanthemi (Vallot), Saissetia coffeae (Walker), Saissetia oleae (Olivier). Three other members of the Coccidae could not be identified to species level: two from the genus Ceroplastes (referred to as Ceroplastes sp. I and Ceroplastes sp. II in this article), and another that could not be identified beyond family level (referred to as unidentified Coccidae). For these three species, only immature stages were collected, precluding morphological identification to species level.
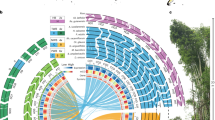
In total, 293 individuals from the Coccidae were processed for molecular analyses; 194 (66%) COI sequences and 220 (75%) 28S sequences were obtained. At least one gene was successfully sequenced in 253 individuals and both genes were sequenced in 161 individuals. For COI, we identified 23 different haplotypes and sequence length was 649 bp. All COI sequences had a bias toward low GC content (A = 40.6%, T = 37.6%, C = 14.8% and G = 7.0%), with a mean GC content of about 21.8% (range: 19.4–24.8%). For 28 S, 20 different haplotypes were identified and sequence length ranged from 788 bp in P. pyriformis to 818 bp in P. corni. No bias was observed for 28S sequences. Neighbor-joining trees were plotted for COI and 28S (see Figs 1 and 2, respectively, also including the Diaspididae found). The initial ABGD partition delineated 12 groups within the Coccidae while the recursive partition delineated 13 groups. The difference in the number of clusters with the morphological identification is due to the splitting of Coccus hesperidum into two and three groups, respectively.
Phylogenetic relationships between 63 haplotypes of Coccidae and Diaspididae based on COI sequences analyzed by neighbor-joining, with the K2P distance model, and Pseudococcus viburni (Hemiptera: Pseudococcidae) (GenBank accession: KJ530624) as the outgroup. Bootstrap values below 70% are not shown.
Phylogenetic relationships between 54 haplotypes of Coccidae and Diaspididae based on 28S sequences analyzed by neighbor-joining, with the K2P distance model, and Pseudococcus viburni (Hemiptera: Pseudococcidae) (GenBank accession: KU499443) as the outgroup. Bootstrap values below 70% are not shown.
Figure 3A shows the distribution of intraspecific (black) and interspecific (gray) pairwise distances according to the K2P nucleotide substitution model. We analyzed intra- and interspecific K2P distances with 11 species for COI and 8 species for 28S. The overall difference in the number of clusters between the two genes is due to (i) the merging of Ceroplastes sinensis and Ceroplastes sp I into a single cluster for 28S, and (ii) the unavailability of 28S sequences for Ceroplastes sp. II and the unidentified Coccidae. A barcode gap was observed for intra- and interspecific distances, for both genes. The intraspecific distances were between 0% and 2.0% for COI and between 0% and 0.6% for 28S. The interspecific distances ranged from 12.5% to 30.1% for COI and from 0.9% to 16.7% for 28S. The minimum interspecific distance for 28S (0.9%) was found between Ceroplastes sp. I and C. sinensis.
Species identifications corresponded to the scientific names of the best GenBank hits, with a percentage similarity exceeding 99.6% for the COI haplotypes of Parasaissetia nigra, Parthenolecanium corni, Protopulvinaria pyriformis and one of six haplotypes of Coccus hesperidum (Supplementary Data 2). Two haplotypes and their best GenBank hits were mismatched, with a percentage of similarity exceeding this threshold: two of the six COI haplotypes of Coccus hesperidum were matched with Unaspis yanonensis (Kuwana) (Hemiptera: Diaspididae) (GenBank accession: KP981079). For 28S, species identifications corresponded to the scientific names of the best GenBank hits, also with a percentage similarity exceeding 99.6% for the haplotypes of Parasaissetia nigra, Parthenolecanium corni, Protopulvinaria pyriformis and Saissetia coffeae. No mismatches with a percentage similarity exceeding this threshold were observed. The best GenBank hits for the nine 28S haplotypes of Saissetia oleae corresponded to Saissetia miranda (Cockerell & Parrott), with a percentage similarity of 98.7% to 99.4%. For the 28S haplotypes of Ceroplastes sp. I and C. sinensis, the best GenBank hit had a percentage similarity below 94%. The results of Blastn queries for each haplotype are provided in Supplementary Data 2.
The most widespread Coccidae were Coccus hesperidum, which was sampled from 20°S to 39°S (at less than 200 m above sea level), and Saissetia oleae, which was sampled from 19°S to 36°S (at elevations of up to 1,000 m above sea level) (Table 1). By contrast, Saissetia coffeae was found only in the northernmost regions of Chile (from 18°S to 29°S, at up to 830 m above sea level) and Pulvinariella mesembryanthemi was found only in the southern regions of Chile (from 39°S to 40°S). Parasaissetia nigra was found at only one site (30°S). The Coccidae with the largest numbers of host plants were S. oleae and C. hesperidum, which were collected on 26 host plants (from ten orders) and 12 host plants (from six orders), respectively (Supplementary Data 1 and 3). Ceroplastes sp. I was found on Vaccinium corymbosum (Ericaceae) and Berberis sp. (Berberidaceae), whereas Ceroplastes sp. II was collected from Schinus molle (Anacardiaceae), a tree endemic to South America. The complete list of host plants for each species is provided in Supplementary Data 1 and 3 (by host plant family and order).
Diaspididae
Seventeen Diaspididae species were identified morphologically: Aonidiella aurantii (Maskell), Aonidiella ensifera McKenzie, Aspidiotus nerii Bouché, Chrysomphalus dictyospermi (Morgan), Diaspidiotus ancylus Putnam, Diaspidiotus echinocacti (Bouché), Diaspidiotus perniciosus (Comstock), Diaspis chilensis Cockerell, Epidiaspis leperii Signoret, Furchadaspis zamiae (Morgan), Hemiberlesia lataniae (Signoret), Hemiberlesia rapax (Comstock), Hemiberlesia palmae (Cockerell), Lepidosaphes beckii (Newman), Lepidosaphes ulmi (Linnaeus), Melanaspis sitreana (Hempel), Pseudoparlatoria chilina (Lindinger). Two other members of the Diaspididae were not identified to species level. One belonged to genus Pseudoparlatoria and the other belonged to genus Aonidomytilus.
From 343 individuals of Diaspididae processed, 268 (78%) unambiguous sequences were obtained for COI and 275 (80%) unambiguous sequences were obtained for 28S. In total, at least one gene was successfully sequenced for 302 individuals and both genes were successfully sequenced for 241 individuals.
For COI, 40 different haplotypes were found. The length of the sequence alignment was 649 bp for 36 haplotypes. Four haplotypes of three species differed: both ends of the sequences of Furchadaspis zamiae were unreadable (resulting in a sequence length of 445 bp – HCOI11 and HCOI12). The sequences of Hemiberlesia rapax III presented an insertion of 24 repetitions of ‘TAA’ and had length of 656 bp. The sequences of Aspidiotus nerii III presented two insertions of 227 bp and 4 bp and had length of 880 bp. Both sequences were suspected to be COI-like pseudogene and they were not taken into account in the following analyses. All COI sequences had a bias toward low GC content (A = 39.8%, T = 41.6%, C = 11.9% and G = 6.7%), with a mean GC content of about 18.6% (range: 16.9–20.0%). A neighbor-joining tree was plotted for COI (Fig. 1, also including the Coccidae). Two monophyletic lineages of Aspidiotus nerii (I and II) and a complex of two cryptic species of Hemiberlesia rapax (I and II) were observed. For 28S, we identified 34 different haplotypes. The length of high-quality sequence ranged from 706 bp to 720 bp for species of the tribe Diaspidini and from 721 to 736 bp for species of the tribe Aspidiotini. A neighbor-joining tree was plotted for 28S (Fig. 2, also including the Coccidae). The initial ABGD partition delineated 19 groups within the Diaspididae due to the subdivision of H. rapax into two groups and the unsuccessful PCR for Pseudoparlatoria sp. The recursive partition delineated 23 groups due to the subdivision of H. lataniae and D. chilensis into two groups, and A. nerii into three groups.
We determined the distribution of the intraspecific (black) and interspecific (gray) pairwise distances according to the K2P nucleotide substitution model (Fig. 3B). For these analyses, we used 19 species for 28S and 20 species for COI. Both Hemiberlesia rapax and Aspidiotus nerii was subdivided into two groups. A barcode gap was observed for intra- and interspecific distances, for both genes. The intraspecific distance ranged from 0% to 1.7% for COI and from 0% to 0.4% for 28S. The interspecific distance ranged from 6.3% to 27.9% for COI and from 1.5% to 17.4% for 28S. The interspecific distance for COI interspecific distance ranged from 7.7% to 8.2% within the two linages of A. nerii, and from 6.3% to 6.5% within the cryptic species of H. rapax. The 28S interspecific distance ranged from 0 to 0.01% within the two linages of A. nerii, and was 0.3% within the cryptic species of H. rapax.
For 18 out of 40 COI haplotypes, the species identification corresponded to the scientific names of the best GenBank hit with a percentage of similarity over 90.8% (Supplementary Data 2). The lowest values corresponded to the haplotypes of Aspidiotus nerii I (~93%) and Hemiberlesia lataniae (~91%). Without taking into account these haplotypes, the percentage of similarity was over 98.5%. For 20 out of 34 28S haplotypes, the species identification corresponded to the scientific names of the best GenBank hit with a percentage of similarity over 99.3%. Three GenBank hits mismatched with a percentage of similarity higher than this threshold: Aonidomytilus sp. matched with Dactylaspis sp. (100%); Aonidiella aurantii with Aonidiella sp. (100%) and Hemiberlesia palmae with Abgrallapsis cyanophylli (99.4%). The result of Blastn request for each haplotype is provided in Supplementary Data 2.
Each lineage of A. nerii had specific COI haplotypes, but shared at least the haplotype H28S33 (n = 83). Individuals of A. nerii I (n = 33) displayed the haplotype HCOI28 or HCOI29. Individuals of A. nerii II (n = 49) displayed five haplotypes: HCOI30 to HCOI34. Individuals of A. nerii III (n = 15) had the COI-like pseudogene. Contrary to A. nerii, each cryptic species of H. rapax complex displayed specific haplotypes of COI and 28S. Individuals of H. rapax I (n = 13) displayed the haplotype HCOI21 and the haplotypes H28S20 or H28S21. No individuals of H. rapax II (n = 9) were sequenced for both genes and they displayed the haplotypes HCOI37 to HCOI39, or the haplotype H28S15. Individuals of H. rapax III (n = 7) with the COI-like pseudogene displayed the haplotypes H28S16 to H28S19.
The Diaspididae with the widest distribution were Aspidiotus nerii sampled from 19°S to 41°S and Hemiberlesia rapax sampled from 18°S to 36°S (Table 1). Lineage I and II of A. nerii had comparable distribution from 19° to 33°S (up to 510 m above sea level) and from 20° to 41°S (up to 1280 m above sea level) respectively, while the lineage III (COI-like pseudogene) of A. nerii ranged from 29° to 33°S (from 120 to 790 m above sea level). Each cryptic species of H. rapax displayed different geographical patterns: H. rapax I was sampled from 28°S to 36°S (up to 540 m above sea level); H. rapax II from 18° to 35°S (up to 830 m above sea level); H. rapax III (COI-like pseudogene) from 30° to 36°S (from 130 to 1180 m above sea level). The other species were found on the central zone of Chile (29°S to 34°S, Regions IV to VI) except the genus Pseudoparlatoria and Aonidomytilus only found in the southern regions (from 39° to 41°S) and H. palmae only found in the North of Chile (18°S).
The Diaspididae found on the greatest number of host plants were Aspidiotus nerii and Hemiberlesia rapax (considering all three groups for both species), collected on 18 host plants (from nine orders) and 13 host plants (from seven orders), respectively (Supplementary Data 1 and 3). Within the three lineages of A. nerii, A. nerii I was found on seven host plants among which Aristotelia chilensis (Elaeocarpaceae) and Persea americana (Lauraceae) (9 out of 10 sites) were specific to this first lineage. A. nerii II was found on 13 host-plants among which six were specific (Citrus spp. (Rutaceae), Annona cherimola (Annonaceae), Macadamia sp. (Proteaceae), Nerium oleander (Apocynaceae), Quillaja saponaria (Quillajaceae), Peumus boldo (Monimiaceae)) to this second lineage. A. nerii III was found on six ornamental and South American trees among which Euonymus sp. (Celastraceae) and Schinus molle were specific to this third lineage. Within the cryptic species of H. rapax, H. rapax I was found on 6 host-plants among which Citrus sinensis and Nerium oleander were specific to this first cryptic species. H. rapax II found on 6 host-plants among which only Acacia retinodes (Fabaceae) was specific to this second cryptic species. H. rapax III was found on five host-plants each shared with at least one of the other cryptic species. The complete list of host plants for each species is provided in Supplementary Data 1 and 3 (by host plant family and order).
Discussion
This survey reported COI and 28S barcode sequences for 11 putative species of Coccidae and 19 of Diaspididae from Chile, among which eight Coccidae and 17 Diaspididae species could be assigned a name based on morphological examination. Except for Parasaissetia nigra which is a new mention for Chile, all these species were already reported in the country21, 26,27,28. Three species of Coccidae could not be morphologically identified because only immature stages were collected and the GenBank matches were not associated with high similarity percentages. Two species of Diaspididae were not morphologically identified at species levels and belonged to the genus Pseudoparlatoria and Aonidomytilus, and were found on native plant hosts. In Chile, only Aonidomytilus espinosai (Porter) is mentioned for this genus1. The K2P distance between the 28S sequences of A. espinosai (GenBank accession: DQ145290 and DQ145291) and the haplotype H28S07 was 7.1%. This value is superior to the maximum intraspecific distance observed (0.4%) but it is included within the range of the interspecific distance (from 1.5% to 17.4%). This species of Aonidomytilus is probably a new species for Chile. No reference sequence was available for Pseudoparlatoria. Four others endemic species of Diaspididae were collected: Aonidiella ensifera, Diaspis chilensis, Melanaspis sitreana, Pseudoparlatoria chilina 26. They were collected mainly on endemic plants, such as Quillaja saponaria or Nothofagus spp. (Nothofagaceae), and on Hedera helix (Araliaceae) and Olea europaea (Oleaceae) for A. ensifera. For Coccidae, sequences of the recently recorded Pulvinariella mesembryanthemi 21 were provided.
Most COI and 28S sequences are new barcodes for the morphologically identified species. More than 50% of haplotypes (39 for COI and 28 for 28S) match with sequences from the GenBank database with low percentage of similarity, probably due to the scarcity of sequence data for these groups, especially in South America. The previous molecular systematic studies included few individuals from Argentina, Colombia and Chile (n = 1) for Diaspididae18, 19, 29, 30 and from Colombia for Coccidae31. Futhermore, the COI region we sequenced (the Folmer region of COI, i.e. the “Barcode region”) does not overlap with the COI region sequenced in a set of previous studies, notably those using a fragment overlaping the 5′ end of COI and the 3′ end of COII. However, the low amount of GenBank matches at 28S can hardly be explained by the use of different 28S regions since the one sequenced in this study and in previous ones overlap over more than 650 bp. In addition, we noticed an inconsistency between the sequences deposited by Wei et al. (data deposited on GenBank without reference to a published article) as Unaspis yanonensis (Diaspididae) and the haplotypes HCOI16, HCOI18, HCOI19 and HCOI20 identified in our study as Coccus hesperidum (Coccidae). It is difficult to deal with inconsistencies between databases when all studies do not carry out morphological vouchering of specimens from which DNA has been extracted. To enable future revisions of the samples processed in this study, we deposited all our slide-mounted vouchers in the collections of ANSES (Montferrier sur Lez, France) and/or MNHN (Santiago, Chile).
These new sequences will be useful to future identification of these species. However, we noticed that the COI sequences gave more relevant information for the delineation of the Ceroplastes species as observed by Deng et al.17 on six Ceroplastes species from China. For COI, we obtained for each species one distinct haplotype while for 28S only two species were successfully sequenced. Moreover, the interspecific distance within COI haplotypes of the three species ranged from 12.5% to 17.5% which is superior to the minimum interspecific distance observed (12.5% - Fig. 3A) and congruent with the average intraspecific distance (12.2%) found by Deng et al.17. Overall, the combination of these COI and 28S markers has repeatedly proved useful for routine identification and DNA barcoding of Coccidae, Diaspididae and Pseudococcidae13, 15, 32, with 28S providing high rates of successful PCR and signal to easily distinguish most species and COI providing more polymorphism to distinguish closely related species and feed large DNA-barcodes databases relying on the COI Folmer region. However, for large-scale systematics studies, one may choose to switch to multiplex PCR using a set of mitochondrial and nuclear markers followed by next generation sequencing33.
Another result was the remarkably low G-C content observed in the COI gene for both Diaspididae (18.8%) and Coccidae (21.9%). This low G-C content of COI gene is common for the scale insects: 14.9% in Pseudococcidae12, 18.2% in Diaspididae12 and 20.4% in Ceroplastes17. This average content is lower than the value of 27.7–39.5% in other orders insects9.
For the Diaspididae, the oleander scale A. nerii and the greedy scale H. rapax displayed high diversity of haplotypes and contrasted geographical and host plant distribution patterns. Three lineages of A. nerii were observed at COI: the lineage I was found on 90% of the avocado trees whereas Citrus trees only hosted the lineage II; the lineage III (COI-like pseudogene) showed the smallest distribution within the central regions (temperate climate) of Chile. A. nerii is a cosmopolitan and polyphagous pest that comprises multiple cryptic parthenogenetic and sexual taxa34, 35. The Chilean diversity of A. nerii could be the result of local adaptation or of multiple introductions of the different lineages from various source populations. As parthenogenetic lineages are known to occur in the A. nerii complex34, 35, locating parthenogenetic lineages in Chile could be an advantage for national biocontrol manufacturers as rearing parthenogenetic scales generally enable higher population growth rates and subsequently more cost-efficient production of parasitoids. Further investigations should be dedicated to look for this trait within the Chilean lineages due to low-quality comparison with GenBank sequences. Direct observation or testing the presence of the endosymbiont Cardinium 35 could be two ways to detect parthenogenesis within A. nerii lineages. The cryptic species complex of H. rapax also displayed different lineages with distinct host distributions. Citrus trees only hosted H. rapax I while H. rapax III shared its host plants with at least another cryptic species. In contrast to A. nerii, the diversity of H. rapax could be explained by the fact that South America is the native region of the genus Hemiberlesia 36.
For the Coccidae, the brown scale, Coccus hesperidum, presented high diversity for COI with six haplotypes and an COI intraspecific distance up to 2.0% which was the maximum value obtained for this distance (Fig. 3A). Moreover, the initial ABGD partition suggested two groups: the group A (n = 16) with the haplotype HCOI48 to HCOI50 and the group B (n = 5) with the haplotypes HCOI45 to HCOI47 (Fig. 1). The group A had the greater distribution (20°S to 39°S) and number of host-plants (9 species) than the group B sampled from 29°S to 33°S on three host-plants. Furthermore, the unique haplotype of 28S matched at 99.3% with a sequence from Coccus formicarii (GenBank accession: JX866687). The study of Lin et al.31 suggested the genus Coccus might not be monophyletic. New studies of this group including South American samples may provide insights into the species delineation in this genus. The black scale, Saissetia oleae, also presented high diversity with six haplotypes for COI and nine haplotypes of 28S. The 28S intraspecific distance reached 0.6% which was the maximum value for this distance (Fig. 3A). The black scale is a polyphagous species, widely distributed around the world and one of the most notorious Coccidae pests in Chile37, 38. We collected this species in most surveyed regions (from 19°S to 36°S) and on more than 26 host plants. The haplotype H28S36 and H28S37 (Fig. 2) were not found on the northern regions of Chile (up to 32°S). However, few individuals sequenced came from these northern regions and we cannot conclude the existence of geographical pattern. Moreover, there is no haplotype pattern of host plant structuration for Saissetia oleae.
Occurrence of cryptic species or association of some lineages with certain host plants could have repercussion on their pest status and their biological control using parasitoids. Indeed, some hymenopteran parasitoids are known to be highly host specific39, 40 and consequently pest management through biocontrol may have to be adjusted for each lineage separately. Even though this study did not have the objective of determining pest species, we could establish an overview of the scale insects collected by crop. The complete list of hosts for each species is provided in Supplementary Data 1. Further studies, focused on each crop, are necessary to determine the scale insects species and their diversity in order to improve the biological control of these pests identifying adequate parasitoids and/or predators.
This study brings new barcode sequences from cosmopolitan and endemic species of Coccidae and Diaspididae from Chile. These new sequences of COI and 28S of South America are now available for further worldwide phylogenetic studies. One species, Parasaissetia nigra is a new mention for Chile. The study unraveled high diversity in the genera Hemiberlesia and also likely cryptic lineages within Aspidiotus nerii and Coccus hesperidum.
Methods
Sample collection
In total, 116 populations of Diaspididae and 112 populations of Coccidae were sampled along a gradient running from Arica in northern Chile (18°S) to Frutillar in the south (41°S), between January 2015 and February 2016 (Fig. 4). The minimum distance between two sampled sites was 1 km. We collected up to 10 adult females from the Diaspididae or immature instars and young females from the Coccidae from crops, indigenous or endemic trees and shrubs and ornamental plants. The collected insects were preserved in 95% ethanol and stored at −20 °C for molecular analysis and morphological identification. Details about the samples collected including sampling locations, host plants, and dates, are available in Supplementary Data 1.
DNA extraction and amplification
When possible, we analyzed two individuals from each sample collected. DNA was extracted from 343 specimens of Diaspididae and 293 of Coccidae, with the DNeasy Tissue Kit (QIAGEN, Hilden, Germany). The extraction was performed without crushing the body of the insect, and it was therefore possible to recover the specimen for its subsequent morphological identification. The procedure used followed the manufacturer’s guidelines, but with two minor changes to improve DNA extraction: cell lysis was carried out over a period of six to eight hours, and we used two elution steps (2 × 50 μl of AE buffer)32.
We amplified two different loci, chosen for analysis on the basis of their suitability for DNA barcoding, population genetics and phylogenetic studies: the HCO-LCO region of the mitochondrial locus encoding cytochrome oxidase subunit I (COI) and the D2 region of the nuclear 28S gene32. For COI, we used the primers PCO-F1 (5′ CCTTCAACTAATCATAAAAATATYAG 3′) and Lep-R1 (5′ TAAACTTCTGGATGTCCAAAAAATCA 3′); and for 28S, we used the primers C-28SLong-F (5′ GAGAGTTMAASAGTACGTGAAAC 3′) and C-28SLong-R (5′ TCGGARGGAACCAGCTACTA 3′). PCR was performed with a 23 μl reaction mixture and 2 μl of diluted DNA (1–20 ng). The reaction mixture contained 12.5 μl of 1X QIAGEN Multiplex PCR buffer and each of the primers required at a concentration of 0.2 μM. PCR was carried out as follows: initial denaturation at 95 °C for 15 minutes, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing for 90 s at a temperature of 48 °C (COI) or 58 °C (28S), depending on the primers, elongation at 72 °C for 60 s, and then a final extension phase at 72 °C for 10 minutes. The final products were separated by electrophoresis with the QIAxcel Advanced System (QIAGEN, Hilden, Germany) for quality control. The, PCR products were then sent to Genewiz (UK, Essex), for bidirectional sequencing by capillary electrophoresis on an ABI 3130XL automatic sequencer (Applied Biosystems, Foster City, CA, USA). The consensus sequences were provided by Genewiz. Chromatograms were visualized with SeqTrace 0.9.041 to check nucleotide variations. The sequences were deposited in GenBank (COI accession numbers: KY084933 to KY085394 and 28S accession numbers: KY085395 to KY085889 – Supplementary Data 1).
Phylogenetic analysis
We considered sequences differing by one or more nucleotides to correspond to different haplotypes. Haplotype alignment was performed with MEGA version 742 with the CLUSTALW method43. Blastn queries against NCBI GenBank, neighbor-joining tree generation and bootstrapping were performed on the haplotypes of each gene with the R44: libraries “ape”45, “ade4”46, and “phangorn”47. The intraspecific and interspecific pairwise distances were calculated with the R package ‘adhoc’48. The Kimura two-parameter nucleotide substitution model (K2P)49 was used for all analyses. Species delineation was performed on the concatened 28S and COI sequences with the online version of Automatic Barcode Gap Discovery (ABGD)50, with a prior maximal distance P = 0.001 and a Jukes-Cantor MinSlope distance of 1.0.
Morphological identification
In the laboratory, we mounted one to ten specimens per COI and 28S haplotype on slides, as follows: specimens were macerated in 10% KOH, washed in distilled water, stained in a mixture of fuschin acid, distilled water, glycerol and lactic acid, dehydrated in acetic acid and lavender oil and mounted in Canada balsam32. All the slide-mounted specimens were deposited in the ANSES collection at Montferrier-sur-Lez and most were also deposited in the entomological collection of the Chilean National Museum of Natural History (MNHN) of Santiago, Chile (Supplementary Data 1). Several taxonomic and ecological articles were used for the identification of species and establishment of their distribution37, 51,52,53,54,55.
References
García Morales, M. et al. ScaleNet: A literature-based model of scale insect biology and systematics. Database. http://scalenet.info, doi:10.1093/database/bav118 (2016).
Miller, D. R., Miller, G. L., Hodges, G. S. & Davidson, J. A. Introduced scale insects (Hemiptera: Coccoidea) of the United States and their impact on U.S. agriculture. Proc. Entomol. Soc. Washingt. 107, 123–158 (2005).
Wang, X.-B. et al. DNA barcoding of common soft scales (Hemiptera: Coccoidea: Coccidae) in China. Bull. Entomol. Res. 105, 1–10 (2015).
Gwiazdowski, R. A., Vea, I. M., Andersen, J. C. & Normark, B. B. Discovery of cryptic species among North American pine-feeding Chionaspis scale insects (Hemiptera: Diaspididae). Biol. J. Linn. Soc. 104, 47–62, doi:10.1111/bij.2011.104.issue-1 (2011).
Gullan, P. J. & Cook, L. G. Phylogeny and higher classification of the scale insects (Hemiptera: Sternorrhyncha: Coccoidea). Zootaxa 425, 413–425 (2007).
Gullan, P. J., Downie, D. A. & Steffan, S. A. A new pest species of the mealybug genus Ferrisia fullaway (Hemiptera: Pseudococcidae) from the United States. Ann. Entomol. Soc. Am. 96, 723–737 (2003).
Varela, L., Smith, R., Battany, M. & Bentley, W. Grape, obscure or vine—which mealybug is it; why should you care. Pr. Winer. Vineyard 27, 37–46 (2006).
Sun, S. et al. DNA barcoding reveal patterns of species diversity among northwestern Pacific molluscs. Sci. Rep. 6, 33367, doi:10.1038/srep33367 (2016).
Hebert, P. D. N., Cywinska, A., Ball, S. L. & deWaard, J. R. Biological identifications through DNA barcodes. Proc. R. Soc. London B Biol. Sci. 270, 313–321, doi:10.1098/rspb.2002.2218 (2003).
Campbell, A. M., Lawrence, A. J., Hudspath, C. B. & Gruwell, M. E. Molecular identification of diaspididae and elucidation of non-native species using the genes 28s and 16s. Insects 5, 528–538, doi:10.3390/insects5030528 (2014).
Sethusa, M. T. et al. DNA barcode efficacy for the identification of economically important scale insects (Hemiptera: Coccoidea) in South Africa. African J. Entomol. 22, 1–10 (2014).
Park, D.-S., Suh, S.-J., Hebert, P. D. N., Oh, H.-W. & Hong, K.-J. DNA barcodes for two scale insect families, mealybugs (Hemiptera: Pseudococcidae) and armored scales (Hemiptera: Diaspididae). Bull. Entomol. Res. 101, 429–34, doi:10.1017/S0007485310000714 (2011).
Abd-Rabou, S. et al. Identification of mealybug pest species (Hemiptera: Pseudococcidae) in Egypt and France, using a DNA barcoding approach. Bull. Entomol. Res. 102, 515–523, doi:10.1017/S0007485312000041 (2012).
Correa, M. C. G., Germain, J.-F., Malausa, T. & Zaviezo, T. Molecular and morphological characterization of mealybugs (Hemiptera: Pseudococcidae) from Chilean vineyards. Bull. Entomol. Res. 102, 524–530, doi:10.1017/S0007485312000053 (2012).
Beltrà, A., Soto, A. & Malausa, T. Molecular and morphological characterisation of Pseudococcidae surveyed on crops and ornamental plants in Spain. Bull. Entomol. Res. 102, 165–172, doi:10.1017/S0007485311000514 (2012).
Malausa, T. et al. Genetic structure and gene flow in French populations of two Ostrinia taxa: host races or sibling species? Mol. Ecol. 16, 4210–4222, doi:10.1111/mec.2007.16.issue-20 (2007).
Deng, J. et al. DNA barcoding of six Ceroplastes species (Hemiptera: Coccoidea: Coccidae) from China. Mol. Ecol. Resour. 12, 791–796, doi:10.1111/men.2012.12.issue-5 (2012).
Andersen, J. C. et al. A phylogenetic analysis of armored scale insects (Hemiptera: Diaspididae), based upon nuclear, mitochondrial, and endosymbiont gene sequences. Mol. Phylogenet. Evol. 57, 992–1003, doi:10.1016/j.ympev.2010.05.002 (2010).
Morse, G. E. & Normark, B. B. A molecular phylogenetic study of armoured scale insects (Hemiptera: Diaspididae). Syst. Entomol. 31, 338–349, doi:10.1111/sen.2006.31.issue-2 (2006).
Gavrilov-zimin, I. A. & Przhiboro, A. A. A new genus and new species of felt scales (Homoptera: Coccinea: Eriococcidae) from tierra del fuego (Chile). Graellsia 72, 1–4 (2016).
Kondo, T. & Gullan, P. J. The Coccidae (Hemiptera: Coccoidea) of Chile, with descriptions of three new species and transfer of Lecanium resinatum Kieffer & Herbst to the Kerriidae. Zootaxa 15, 1–15 (2010).
Willink Granara, M. & Claps, L. Cochinillas (Hemiptera: Coccoidea) Presentes en Plantas Ornamentales de la Argentina. Neotrop. Entomol. 32, 625–637 (2003).
Correa, M. C. G. et al. Mealybug species from Chilean agricultural landscapes and main factors influencing the genetic structure of Pseudococcus viburni. Sci. Rep. 5, 16483, doi:10.1038/srep16483 (2015).
Pacheco Da Silva, V. C. et al. Molecular and morphological identification of mealybug species (Hemiptera: Pseudococcidae) in Brazilian vineyards. PLoS One 9, 1–13, doi:10.1371/journal.pone.0103267 (2014).
Correa, M. et al. A new species of Pseudococcus (Hemiptera: Pseudococcidae) belonging to the ‘Pseudococcus maritimus’ complex from Chile: Molecular and morphological description. Zootaxa 46–54 (2011).
Claps, L. E., Wolff, V. R. S. & Gonzalez, R. H. Catálogo de las especies de Diaspididae (Herniptera: Coccoidea) nativas de Argentina, Brasil y Chile. Insecta mundi 13, 346 (1999).
Nakahara, S. Checklist of the armored scales (Homoptera: Diaspididae) of the conterminous United States. (1982).
Charlin, R. C. Distribución geografica, plantas hospederas y nuevas identificaciones de coccidos para Chile. Rev. Peru. Entomol 15, 215–218 (1972).
Ross, L., Hardy, N. B., Okusu, A. & Normark, B. B. Large population size predicts the distribution of asexuality in scale insects. Evolution. 67, 196–206, doi:10.1111/j.1558-5646.2012.01784.x (2013).
Rugman-Jones, P. F., Morse, J. G. & Stouthamer, R. Rapid molecular identification of armored scale insects (Hemiptera: Diaspididae) on Mexican ‘Hass’ avocado. J. Econ. Entomol. 102, 1948–1953, doi:10.1603/029.102.0527 (2009).
Lin, Y. P., Kondo, T., Gullan, P. & Cook, L. G. Delimiting genera of scale insects: Molecular and morphological evidence for synonymising Taiwansaissetia Tao, Wong and Chang with Coccus Linnaeus (Hemiptera: Coccoidea: Coccidae). Syst. Entomol. 38, 249–264, doi:10.1111/j.1365-3113.2012.00664.x (2013).
Malausa, T. et al. DNA markers to disentangle complexes of cryptic taxa in mealybugs (Hemiptera: Pseudococcidae). J. Appl. Entomol. 135, 142–155, doi:10.1111/jen.2011.135.issue-1-2 (2011).
Cruaud, P., Rasplus, J., Rodriguez, L. J. & Cruaud, A. High-throughput sequencing of multiple amplicons for barcoding and integrative taxonomy. Nat. Publ. Gr. 7, 1–12, doi:10.1038/srep41948 (2017).
Andersen, J. C., Normark, B. B., Morse, G. E. & Gruwell, M. E. Cryptic diversity in the Aspidiotus nerii complex in Australia. Ann. Entomol. Soc. Am. 103, 844–854, doi:10.1603/AN10060 (2010).
Provencher, L. M., Morse, G. E., Weeks, A. R. & Normak, B. B. Parthenogenesis in the Aspidiotus nerii complex (Hemiptera: Diaspididae): a single origin of a worldwide, polyphagous lineage associated with Cardinium bacteria. Ann. Entomol. Soc. Am. 98, 629–635 (2005).
Williams, D. J. Some South American scale insects (Homoptera: Coccoidea) on Nothofagus. J. Nat. Hist. 19, 249–258 (1985).
González, R. H. Insectos coccoideos plagas de cultivos frutales en Chile (Hemiptera: Coccoidea). (Editorial universitaria, 2016).
Ripa, R. & Larral, P. Manejo de plagas en paltos y cítricos. 23 (INIA, 2008).
Deng, J. et al. Cophylogenetic relationships between Anicetus parasitoids (Hymenoptera: Encyrtidae) and their scale insect hosts (Hemiptera: Coccidae). BMC Evol. Biol. 13, 275, doi:10.1186/1471-2148-13-275 (2013).
Chesters, D. et al. The integrative taxonomic approach reveals host specific species in an encyrtid parasitoid species complex. PLoS One 7, 1–11, doi:10.1371/journal.pone.0037655 (2012).
Stucky, B. J. Seqtrace: A graphical tool for rapidly processing DNA sequencing chromatograms. J. Biomol. Tech. 23, 90–93, doi:10.7171/jbt.12-2303-004 (2012).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for biggest datasets. Mol. Biol. Evol. 33, 1870–1874, doi:10.1093/molbev/msw054 (2016).
Thompson, J., Higgins, D. & Gibson, T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalities and weight matrix choice. Nucleic Acids Res. 22, 4673–4680, doi:10.1093/nar/22.22.4673 (1994).
R Core Team. R: A language and environment for statistical computing. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Version 3.2.5 (2015).
Paradis, E., Claude, J. & Strimmer, K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, (289–290 (2004).
Dray, S. & Dufour, A. B. The ade4 package: implementing the duality diagram for ecologists. J. Stat. Softw. 22, 1–20, doi:10.18637/jss.v022.i04 (2007).
Schliep, K. P. Phangorn: phylogenetic analysis in R. Bioinformatics 27, 592–593, doi:10.1093/bioinformatics/btq706 (2011).
Sonet, G. et al. Adhoc: an R package to calculate ad hoc distance thresholds for DNA barcoding identification. Zookeys 336, 329–335, doi:10.3897/zookeys.365.6034 (2013).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120, doi:10.1007/BF01731581 (1980).
Puillandre, N., Lambert, A., Brouillet, S. & Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 21, 1864–1877, doi:10.1111/j.1365-294X.2011.05239.x (2012).
Williams, D. J. & Watson, G. W. The Scale Insects of the Tropical South Pacific Region. Pt. 3: The Soft Scales (Coccidae) and Other Families. (1990).
Williams, D. J. & Watson, G. W. The Scale Insects of the Tropical South Pacific Region. Pt. 1. The Armoured Scales (Diaspididae). (1988).
Lepage, H. S. & Giannotti, O. Notas Coccidológicas. Arq. Inst. Biol. (Sao. Paulo). 14, 338–340 (1943).
Porter, D. E. Un Cóccido nuevo para la Rep. Argentina. Rev. Chil. Hist. Nat. 32, 269 (1928).
Cockerell, T. D. A. A new species of Coccidae of the genus Diaspis. Actes la Société Sci. du Chili 5, 6–7 (1895).
Minka, T. P.and Deckmyn, A. maps: Draw Geographical Maps. R package version 3.1.1. http://CRAN.R-project.org/package=maps (2016).
Acknowledgements
The authors would like to thank all the growers that allowed us to access into their orchards to collect samples. We are grateful to Paula Molina and Angela Romo for their technical help in the laboratory. We are also grateful to the persons who helped us collect samples through Chile: Hugo Herrera, Francisco Bugueño, Sebastian Catalan and Isaac Vera from Xilema S.P.A. (Quillota); Claudio Salas from Instituto de Investigaciones Agropecuarias (Intihuasi - La Serena); Marta Godoy from Cabilfrut (Ovalle); Leonardo Fuentes from Pontificia Universidad Católica de Valparaíso (Quillota); Alda Romero from Pontificia Universidad Católica de Chile (Santiago); Jean-Claude Malausa from Institut National de la Recherche Agronomique (Sophia-Antipolis, France). Miguel Gómez helped in plant identification (Pontificia Universidad Católica de Chile, Santiago). This project was supported by the EU FP7 Marie Curie IAPP Project “Colbics” #324475 and by the “Fundación para la Innovación Agraria” (FIA) #PYT-2015-0230.
Author information
Authors and Affiliations
Contributions
All authors reviewed the manuscript. A.P., C.M., M.T., Z.T. participated to the writing of the article. M.T., G.G., K.P., Z.T. participated to the formulation of the goals and aim of the experiment. A.P., A.J., G.G., K.P. participated to the field collection. CD realized all the molecular laboratory experiments. GJF realized all the morphological identification. A.P., M.T. analyzed the results. M.T., C.M. participated to the acquisition of the financial support for the project.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Accession Codes: From KY084933 to KY085394 for COI sequences and from KY085395 to KY085889 for 28S sequences.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Amouroux, P., Crochard, D., Germain, JF. et al. Genetic diversity of armored scales (Hemiptera: Diaspididae) and soft scales (Hemiptera: Coccidae) in Chile. Sci Rep 7, 2014 (2017). https://doi.org/10.1038/s41598-017-01997-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-01997-6
This article is cited by
-
Melanaspis corticosa: a new insect pest of olive trees in Europe
Phytoparasitica (2023)
-
Variation in defensive traits against herbivores of native and invasive populations of Carpobrotus edulis
Biological Invasions (2023)
-
An invasive population of Roseau Cane Scale in the Mississippi River Delta, USA originated from northeastern China
Biological Invasions (2022)
-
First report of the rhodesgrass mealybug Antonina graminis (Maskell, 1897) (Hemiptera: Coccomorpha: Pseudococcidae) in Chile, with key to genera of Pseudococcidae
International Journal of Tropical Insect Science (2021)
-
Genetic variability on worldwide populations of the scale insect Pulvinariella mesembryanthemi
Biological Invasions (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.