Abstract
A macroscopic description of a protein structure allows an understanding of the protein conformations in a more simplistic manner. Here, a new macroscopic approach that utilizes the joints of the protein secondary structures as a basic descriptor for the protein structure is proposed and applied to study the arrangement of secondary structures in helical membrane proteins. Two types of dihedral angle, Ω and λ, were defined based on the joint points of the transmembrane (TM) helices and loops, and employed to analyze 103 non-homologous membrane proteins with 3 to 14 TM helices. The Ω-λ plot, which is a distribution plot of the dihedral angles of the joint points, identified the allowed and disallowed regions of helical arrangement. Analyses of consecutive dihedral angle patterns indicated that there are preferred patterns in the helical alignment and extension of TM proteins, and helical extension pattern in TM proteins is varied as the size of TM proteins increases. Finally, we could identify some symmetric protein pairs in TM proteins under the joint-based coordinate and 3-dimensional coordinates. The joint-based approach is expected to help better understand and model the overall conformational features of complicated large-scale proteins, such as membrane proteins.
Similar content being viewed by others
Introduction
Protein structures are strongly related to their physical properties, such as folding, stability, and function. They also include information on how proteins have evolved and connected with each other. A study of the structural and conformational features of proteins is one of the most significant issues in protein science. Traditionally, many studies have examined protein structures with an all atom-based description1, 2. The Ramachandran’s plot1 with the backbone dihedral angle ϕ (N-Cα) and ψ (Cα-C) is a representative way of microscopic descriptions of the protein structure. The Ramachandran plot shows the allowed and disallowed values of the dihedral angles of amino-acid residues of polypeptide backbone chain. The plot provides an understanding of the local and global features of protein structures. For example, the Ramachandran plot is built in the PROCHECK3 and WHAT_CHECK4 to verify the stereochemical quality of the protein structure. Recently, it has been used widely to validate the protein structure generated from homology modeling5 within the frame of molecular dynamics simulations.
The protein geometry has also been studied at the coarse-grained level: Cα atom-based coordinates6,7,8,9, residue-based coordinates10, 11 and secondary structure-based coordinates12,13,14. The coarse-grained models for the protein structure enable an understanding of the protein conformations in more simplistic manner, which may have an advantage in studying large-scale proteins, such as multi-protein complex or membrane proteins with several transmembrane (TM) helices. This paper proposes a new macroscopic description method for the protein structures. In general, protein structures can be described using the secondary structures such as α helices, β sheets and loops as the basic units at the macroscopic level (Fig. 1a). Our new strategy is to use the joints of secondary structures as the basic constituents for a description of the protein structure and to study the protein conformational features by examining the 3-dimensional arrangement of the joints with their dihedral angles (Fig. 1b).
Schematic of the joint-based approach for macroscopic description of the protein structure. (a) Secondary structure-based description of the protein structure. The arrow means the sequence of amino acids of the protein with increasing residue numbers. (b) Newly devised joint-based description of the protein structure using the joints of secondary structures and their dihedral angles.
Here, the macroscopic description method is applied to study the conformational features of the membrane proteins. Membrane proteins have a range of cellular functions, such as signal transduction (protein kinase)15, 16, ion channeling (potassium channel)17,18,19, energy metabolism (voltage-dependent anion channel)20, and drug recognition (multidrug resistance protein)21, 22. Most membrane proteins are composed of transmembrane (TM) helices. Various cellular functions of the membrane proteins are quite relevant to their diverse conformations inside the lipid membrane. The complexity of the membrane protein can be understood within the frame of the statistical distribution of the conformations of the TM helices of the membrane proteins23. In this study, the conformation of the TM helices was investigated by the new description method using the structural joints at the macroscopic level. The non-homologous structures of the membrane proteins from Protein Data Bank were selected and analyzed using the joint-based method. Some common and interesting features of membrane proteins reflecting the conformational heterogeneity and specificity are suggested based on an analysis of the conformations of non-homologous membrane proteins with the dihedral angles of the joints.
Results
Macroscopic description of membrane protein structure using joint-based approach
Most membrane proteins with TM helices display a repetition of the TM helix and loop, as shown in Fig. 2a. To present the structure based on the joint approach, a set of joints associating the helices and loops were selected. In particular, the C-alpha carbon of the beginning and ending residues of each TM helix were considered as structural joining points, and employed as structural elements of the protein structure. The spatial arrangement of the joint points was determined by the dihedral angles between the two joint points. For example, 6 joints (P1, P2, P3, P4, P5 and P6) can be assigned for a protein composed of three helices (H1, H2 and H3) and two loops (L1 and L2) (Fig. 2a). The first dihedral angle involving four joints (P1, P2, P3 and P4) can be ascertained by measuring the angle between two planes made by (P1, P2, P3) and (P2, P3, P4). Similarly, the second dihedral angle can be found by applying the structural points (P2, P3, P4, P5), and the (P3, P4, P5 and P6) joints are used to determine the third, and so on. The dihedral angles are classified into two types: Ω and λ types. The first and third dihedral angles determined by the four joints in the Helix-Loop-Helix correspond to type Ω; they are denoted as Ω1 and Ω2, respectively. In a similar way, the dihedral angles determined by the four joints in a Loop-Helix-Loop, such as the second dihedral angle, correspond to the type λ, denoted as λ1. The conformation of TM helices of the membrane proteins can be represented by a set of two types of dihedral angles (Ω1, λ1, Ω2, λ2, Ω3 …) composed of a set of joints (P1, P2, P3, P4 …) at the macroscopic level. For the dihedral angles, the clockwise angle (from 0 to 180 degrees) was assigned as a positive value and counter-clockwise angle as a negative value (Fig. 2b). The algorithm to define the structural joints and the dihedral angles between the joint points is shown in detail (See Methods).
Joint-based description of membrane proteins with three helices and two loops. (a) Assignment of the Ω type and λ type dihedral angles. H1 to H3 are helices, L1 to L2 are loops, and P1 to P6 are joint points. Ω-type dihedral angles, such as Ω1, are defined by the four joint points in the Helix-Loop-Helix, such as P1, P2, P3, and P4. The λ-type dihedral angles, such as λ1, are defined by the four joint points in the Loop-Helix-Loop, such as P2, P3, P4 and P5. (b) Assignment of the positive and negative signs for dihedral angles. The positive (+) sign and negative (−) signs represent the clockwise and counter-clockwise angles, respectively, in the projections for the dihedral angles. The figures present the projections for Ω1 and λ1.
Dataset of target membrane proteins
A total of 103 non-homologous membrane proteins with 3 to 14 transmembrane (TM) helices were used as a dataset (Table 1). The dataset was obtained from the protein data bank (PDB) by characterizing all the resolved polytopic membrane spanning structures. The Methods section presents a detailed procedure to obtain the dataset. Briefly, (i) 2600 membrane proteins with X-ray crystal structures were collected from PDB, (ii) 959 proteins with only α helices were selected from them, and (iii) finally 103 non-homologous monomeric chains with 3 TM to 14 TM helices were selected from the 959 proteins. To validate how much the selected non-homologous protein dataset is complete, a structural homology detection study was performed using the 103 non-homologous proteins against whole 959 helical TM proteins, similarly to the previous study24. The selected 103 proteins could cover around 90 to 97% of the 959 protein structures depending on the RMSD threshold range (3 to 5 Å) for structural homology. This suggests that the selected dataset represents the whole dataset quite completely. The target dataset of the 103 protein structures were analyzed using the joint-based approach. Table S1 in Supplementary Information presents the Ω and λ type dihedral angles for the 103 non-homologous membrane proteins.
In this study, we measured the dihedral angles of joint-points of TM helical proteins and tried to analyze the macroscopic arrangement or extension of TM helices by simplifying them as straight lines between joints. Thus, it should be noted that the measured dihedral angles cannot reflect the exact microscopic structural features of transmembrane segments because the transmembrane helices include kinks and bends25, 26. However, it is known that the bending angles of most TM helices are known to be comparatively low (less than 20 degrees) due to the limited membrane space27, 28, which indicates that the joint-based dihedral angle data would provide us a macroscopic viewpoint of angles between TM helices.
Distribution of Ω and λ angles and their relevance to arrangement of helices
The dihedral angles between the joints are strongly related to the arrangements of the TM helices in membrane proteins at the macroscopic level. If the TM helices are simplified as straight lines of the joint points, as shown in the Fig. 2a, Ω type dihedral angles represent the arrangement of the TM helix region between the ith TM helix (Hi) and its adjacent i + 1th TM helix (Hi+1). The type λ dihedral angles also provides additional information of the relative arrangement between the ith TM helix (Hi) and i + 2th TM helix (Hi+2), considering that the i + 1th loop (Li+1) is attached to the i + 2th TM helix (Hi+2). Figure 3 shows specific examples of the relationship between the dihedral angles and helical arrangements. When the dihedral angle Ωi is close to 0°, helix Hi and the adjacent helix Hi+1 are in an anti-parallel arrangement (Fig. 3a). On the other hand, when the dihedral angle Ωi is close to ±180°, helix Hi and the adjacent helix Hi+1 are in parallel (Fig. 3b). When the dihedral angle λi is close to 0°, helix Hi+2 and helix Hi are in the same side with respect to helix Hi+1 (Fig. 3c). On the other hand, when the dihedral angle λi is close to ±180°, helix Hi+2 and helix Hi are in the opposite side with respect to helix Hi+1 (Fig. 3d).
Arrangements of the helices and loops depending on the Ω and λ dihedral angles. The central figure presents the front view of three consecutive helices in the membrane proteins. H(n) represents the helices and L(n) represents loops. (a) Front view of the arrangement of two adjacent helices, Hi and Hi+1 when Ωi = 0°, (b) Front view of the arrangement of two adjacent helices, Hi and Hi+1 when Ωi = ± 180°, (c) Top view of the arrangement of two adjacent loops, Li and Li+1, and three adjacent helices, Hi, Hi+1 and Hi+1, when λi = 0°, and (d) Top view of the arrangement of two adjacent loops, Li and Li+1, and three adjacent helices, Hi, Hi+1 and Hi+1, when λi = ±180°. These figures show the possible scenario depending on the dihedral angles rather than a real arrangement observed in TM proteins.
The distribution of dihedral angles for the type Ω and λ of the 103 non-homologous protein structures were analyzed using the (Ω, λ) plot (Fig. 4a). Such analysis was expected to play the role of the Ramachandran-plot, which may be used to determine the allowed and disallowed conformations of the TM helices for the 103 non-homologous membrane proteins. The Ω type dihedral angles were restricted to a very narrow region in the range of −50° to + 50°. On the other hand, the λ type dihedral angles were distributed in the entire region between −180° to + 180°. For quantitative analysis, the histograms for the respective dihedral angles were plotted (Fig. 4b and Supplementary Information Figure S2). In the case of the Ω type dihedral angles, more than 90% of the angles were in the range, −40° to + 40° (Fig. 4b). In particular, the two dominant dihedral angle distribution region were observed around −30° to −10° and +10° to +30°, showing a symmetrical bimodal distribution. For the λ type dihedral angles, however, no clear dominant angle distribution region like Ω type angles was observed (Supplementary Information Figure S2).
Distribution of the Ω type and λ type dihedral angles in the helical membrane proteins. (a) Ω-λ distribution plot for the helical membrane proteins. All Ω and λ type dihedral angles in the 103 non-homologous of membrane proteins are plotted together in the 2-D scatter plot (Ω type: x-axis, λ type: y-axis). (b) Histogram showing the overall distribution of the Ω type dihedral angles. Error bars were the standard deviations estimated by bootstrap method44, resampling the data 500 times with replacement and repeating the analysis.
According to Fig. 4, the Ω type dihedral angles showed the preference in the narrow range of −40° to + 40° as a dominant accessible region, suggesting that two neighboring TM helices (Hi and Hi+1) tend to arrange in an anti-parallel manner, as shown in the Fig. 3a. In particular, the two major preferred regions around −30° to −10° and +10° to +30° with a relatively low frequency around 0° suggest that the most preferable arrangement of two neighboring helices is a slightly slanted anti-parallel arrangement. In the analysis of Ω angles (Fig. 4), an exceptional value (−173°) was observed, and identified as Ω7 of 3QNQA. The Ω value indicates that two consecutive helices (H7 and H8) of the protein are almost arranged in parallel as shown in Fig. 3b. This is the case that joint-based dihedral angle cannot reflect the exact structural features of transmembrane segments, which was mentioned above. It was confirmed that the H7 and H8 of the protein were a kinked helix and a short helix with long loop as a hugely bending TM segment, respectively, which resulted in such Ω value although the two consecutive TM regions are not parallel. On the other hand, the λ-type dihedral angles were distributed entirely in the all possible ranges of −180° to +180°. This suggests that helix Hi+2 can be arranged randomly between the same side (Fig. 3c) and opposite side (Fig. 3d) to helix Hi.
The dihedral angle analysis for the Ω-type suggested that the two adjacent TM helices prefer an anti-parallel orientation. Structurally, the helices that cross the hydrophobic lipid bilayer membranes prefer an anti-parallel arrangement. Thermodynamically, the anti-parallel arrangement of the two consecutive TM helices inside the lipid bilayer has stability by decreasing the internal energy due to a packing interaction. These are well-known features of helices in TM proteins29,30,31, which suggests that the joint-based approach is effective to explain the conformational features of TM proteins.
Local pattern of consecutive Ω or λ angles and their relevance to extension of helices
As shown above, measurements of the dihedral angle Ωi provides additional information about the arrangement of the neighboring TM helices (Hi and Hi+1). The relative arrangement of the two TM helices Hi and Hi+2 can be determined by measuring the dihedral angle λi. This suggests that measurements of the consecutive dihedral angles can allow a prediction of how the TM helices in the membrane proteins are arranged sequentially or extended. For example, the information of Ωi and Ωi+1 can determine the arrangement of Hi, Hi+1, and Hi+2, and the information of λi and λi+1 may allow a prediction of the relative positions of Hi+2 and Hi+3 to Hi and Hi+1. The helical extensions in the membrane proteins were examined through a joint-based approach using the local patterns of the consecutive dihedral angle clusters, such as Ωi-Ωi+1, and λi-λi+1.
All dihedral angles were categorized into two groups: positive (clockwise) and negative (counter-clockwise) signs in a simple manner. The dihedral angles of Ω and λ for the 103 non-homologous proteins (Supplementary Information Table S1) can be represented as two signs, i.e., positive and negative (Supplementary Information Table S2). Combinations of the two signs can generate four patterns, i.e., (+, +), (−, −), (+, −), and (−, +), for the two consecutive dihedral angle clusters, such as Ωi-Ωi+1 and λi-λi+1. In a similar manner, eight patterns can be generated for the three consecutive dihedral angle clusters, such as Ωi-Ωi+1-Ωi+2 and λi-λi+1-λi+2. All the frequencies of the patterns in the 103 non-homologous proteins were analyzed. For Ωi-Ωi+1 cluster, the frequency of the (+, +) pattern was the most dominant pattern (Fig. 5a), whereas frequency of the pattern (−, −) was the most dominant pattern for λi-λi+1 cluster (Fig. 5b). For the three consecutive dihedral angle clusters, i.e., Ωi-Ωi+1-Ωi+2 cluster and λi-λi+1-λi+2 cluster, (+, +, +) and (−, −, −) were the most dominant patterns among the 8 possible patterns (Fig. 5c and d), respectively. As mentioned previously, the λ-type dihedral angles were distributed in the entire range from −180° to 180° (Fig. 4a), which motivated us to divide the dihedral angle space further into four quadrants, i.e., −180° to −90° (denoted as −B), −90° to 0° (denoted as −A), 0° to 90° (denoted as A), and 90° to 180° (denoted as B), and the pattern of the λi-λi+1 cluster was examined more in detail. Among the 16 possible patterns for the λi-λi+1 cluster, the most dominant distribution was observed in the range, −90° to 0° and −90° to 0° (Fig. 6).
Frequencies of the patterns for consecutive Ω or λ type dihedral angles when Ω or λ type angles are categorized as (+) and (−). The bar diagrams show the observed numbers of (a) four different patterns of two consecutive Ω type angles, Ωi–Ωi+1 (b) four different patterns of two consecutive λ type angles, λi-λi+1 (c) eight different patterns of three consecutive Ω type angles, Ωi-Ωi+1-Ωi+2, and (d) eight different patterns of three consecutive λ type angles, λi-λi+1-λi+2. For (a) to (d), (i) Ω or λ type dihedral angles were split into two regions, i.e., (+) = 0° to 180°, (−) = 0° to −180°, (ii) four and eight patterns were generated from the combinations of two consecutive angles, and three consecutive angles, respectively, and (iii) finally, the number of patterns in the 103 non-homologous proteins were measured. Error bars were the standard deviations estimated by bootstrap method, resampling the data 500 times with replacement and repeating the analysis44.
Frequencies of the patterns for consecutive λ type dihedral angles when λ type dihedral angle was split into four regions. The bar diagram shows the observed numbers of the 16 different patterns of two consecutive λ type angles, λi–λi+1. Here, all λ type angles were split into four regions, i.e. A = 0° to 90°, B = 90° to 180°, −A = −90° to 0°, and −B = −90° to −180°; 16 patterns were generated from the combinations for two consecutive λ type angles, λi-λi+1, and finally the numbers of the patterns in the 103 non-homologous proteins were measured. Error bars were the standard deviations estimated by bootstrap method, resampling the data 500 times with replacement and repeating the analysis44.
Figure 7a shows a schematic diagram of the membrane proteins with several TM helices observed in the front view. Figure 7b shows the schematic arrangement of three consecutive helices, i.e., Hi, Hi+1, and Hi+2, depending on the pattern of the Ωi-Ωi+1 cluster, observed in the side view. As shown in the schematic diagram, four different (+, +), (+, −), (−, +) and (−, −) patterns determine four different types of configurations between Hi and Hi+2 in parallel. The predominance of the (+, +) and (+, +, +) patterns in Ωi-Ωi+1 and Ωi-Ωi+1-Ωi+2 cluster analyses indicate that the membrane proteins favor the alternative packing of TM helices: zig-zag pattern. Figure 7c presents a schematic diagram of the membrane proteins with several TM helices observed in the top view and shows how the helices were extended depending on the pattern of the λi-λi+1 cluster. The (−, −) and (+, +) patterns suggest that the TM helices are extended in one direction with a zig-zag pattern. The (+, −) and (−, +) patterns, however, show that the TM helices are extended such that they are packed in a relatively compact space. According to the Fig. 5(b), (−, −) is the most preferred pattern, (−, +) and (+, −) patterns are next dominant patterns with similar frequency, and (+, +) is the least preferred pattern. The frequencies of the four patterns indicate that TM helices in the membrane proteins are extended by using zig-zag type extension and packing type extension almost equivalently. The large difference of frequency between (−, −) and (+, +) suggest that there is a significant directional bias in the zig-zag type extension, whereas there is no directional preference in the packing-type extension. The dominant pattern of (−90° to 0°, −90° to 0°) in the 16 possible patterns of the λi-λi+1 cluster (Fig. 6) suggests that there is also some angle preference between the TM helices in the extension of helices.
Relation of the dihedral angle patterns and helical arrangements or extensions. (a) Front view of the linearly ordered helical membrane proteins with the TM helices and loops. (b) Side view of the arrangement of three consecutive helices Hi-Hi+1-Hi+2 depending on the four different patterns of two consecutive Ω type angles, Ωi-Ωi+1. (c) Top view configuration of four consecutive helices, Hi-Hi+1-Hi+2-Hi+3, for the four different patterns of two consecutive λ type angles, λi-λi+1. Figure 7(a) and (b) were generated as follows. (1) The dihedral angles were computed by the original definition in the Fig. 2, (2) arrangement of helices was predicted based on the dihedral angles, and then (3) the predicted arrangement was drawn based on side view and top position. Zig-zag conformation denotes the helices are arranged or extended in the alternative direction as depicted in ++ and −− configurations.
As shown in Fig. 7, both (+, +) and (−, −) patterns are equivalent in the point that they present the zig-zag pattern in the helical alignment (Fig. 7b) and helical extension (Fig. 7c). The only difference is that the (+, +) pattern is for a helical alignment and (−, −) is for a helical extension. The biased zig-zag patterns in the helical alignment and extension may be relevant to the stereochemistry of the residue-residue interaction between the TM helices and the interaction between the loops and the environment.
Effect of TM helical position and number on the arrangement and extension of helices
As the number of TM helices increases, the arrangements of the TM helices in membrane proteins might be changed due to a change in the interaction energy term between the TM helices. To check this point, two kinds of analysis were carried out. First, the distributions of the dihedral angles for Ω and λ types were analyzed according to their relative position in the TM helices (Supplementary Information Figure S3(a) and (b)). The histograms for the distribution are shown (Supplementary Information Figure S1(a) and (b)). Ωn or λn (n = 1, 2, 3, 4 …) denotes the nth Ω or λ type dihedral angles in the 103 non-homologous proteins. The Ω type dihedral angles from Ω1 to Ω11 showed similar distributions and histograms in the range of approximately −50° to +50° (Supplementary Information Figure S3(a)). The λ type dihedral angles also exhibited a similar distribution and histogram patterns from λ1 to λ10, showing the distributions approximately in the entire ranges (Supplementary Information Figure S3(b)). These distribution patterns are similar to their overall distributions, as shown in Fig. 4. These results suggest that the relative arrangements of the two or three consecutive helices are not affected significantly by the relative position of the TM helices in the membrane. The distribution patterns of the terminal dihedral angles, such as Ω12, Ω13, λ11 and λ12 deviated substantially from the other ones, but an interpretation of such results may not be effective due to the insufficient sampling.
In addition, the frequencies of the four patterns for the Ωi-Ωi+1 or λi-λi+1 clusters were analyzed according to three different groups: proteins with 3–6 TM helices, proteins with 7–10 TM helices, and proteins with 11–14 TM helices. As shown in Fig. 8a, for the Ωi-Ωi+1 cluster, the (+, +) pattern shows up more frequently in the proteins with 3–6 TM helices. The pattern is roughly maintained as the TM number increases from 3–6 TM helices to 7–10 TM and 11–14 TM helices. These results indicate that the membrane proteins favor the alternative packing of TM helices in the helical arrangement regardless of their sizes. On the other hand, for λi-λi+1 cluster, (−, −) and (+, +) patterns show up less frequently than (+, −) and (−, +) patterns in the proteins with the 3–6 TM helices (Fig. 8b). As the number of TM helices increase, however, the (−, −) pattern becomes more dominant than other patterns. These results suggest that the TM helices prefer to be packed in their extension for small TM proteins, but zig-zag type extension plays an important role in the helical extension as the number of TM helices of the membrane proteins increases. Presumably, the zig-zag type extension of large TM proteins has the advantage of the efficient extension of the helices of large TM proteins in relatively narrow space inside the lipid bilayer.
Frequencies of the patterns for two consecutive Ω or λ type dihedral angles depending on the proteins with a different number of TM helices. The 103 non-homologous proteins were categorized into three groups, i.e., proteins with 3 to 6 TM helices, 7 to 10 TM helices, and 11 to 14 TM helices. The bar diagrams show the observed numbers of (a) four different patterns of two consecutive Ω type angles, Ωi-Ωi+1, in each group, and (b) four different patterns of two consecutive λ type angles, λi-λi+1, in each group. Here, like (a) and (b) in the Fig. 5, (i) Ω or λ type angles were split into two regions, i.e. (+) = 0° to 180°, (−) = 0° to −180°, (ii) four patterns were generated from the combinations of two consecutive angles, and iii) finally, the number of the patterns in the three groups were measured. Error bars are generated by bootstrapping represent standard deviation, resampling the data 500 times with replacement and repeating the analysis44.
Identification of symmetric pairs in TM proteins
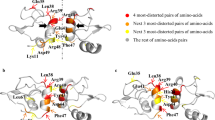
The analyses of consecutive Ω and λ angles shown in the Fig. 5 also indicate that there are many local symmetric configurations in the arrangement helices of membrane proteins. For example, (+, +, +) presents the symmetric configuration of (−, −, −). This observation motivated us to explore the existence of a symmetric configuration in the level of global TM protein structure. For this, we first assigned the proteins showing symmetric configurations based on λ angle signs from the whole TM protein dataset, and then selected protein pairs showing roughly symmetric configuration at a level of macroscopic 3-dimensional structure by visual inspection. λ angle sign was focused in the first step because λ angles showed more significant variations compared to Ω angles (Fig. 4) and therefore they may affect the 3-dimensional protein structure more. The proteins showing symmetric configurations based on λ angle signs were identified only in 3–6 TM proteins, and presented in Supplementary Information Table S3. In the dataset of 7 to 14 TM proteins, any protein pairs showing symmetrical property of λ angle signs were not detected, and further investigation for structural symmetry was not executed. Supplementary Information Table S4 shows the protein pairs exhibiting symmetry based on macroscopic 3-dimensional structure. Briefly, among the nine 3TM proteins in the whole dataset, two proteins (3ZE5A and 5AJIA) were identified to exhibit macroscopic 3-dimensional symmetry against three proteins (4O9PA, 3RKOA, and 1YQ3C). In the thirteen 4TM proteins, two proteins (4WD8A and 5DRIA) showed a symmetrical structure against one protein (1Q90A). In the nine 5TM proteins, 4A2NB and 3WVFA were identified as symmetric structural pairs. In 6TM proteins, some protein pairs showing symmetric configuration of λ angle signs were detected, but they were not structurally symmetric. Supplementary Information Figure S4(a) and (b) illustrates λ angle patterns and macroscopic helical arrangements of the three representatives symmetric protein pairs. These results indicate that there are protein pairs showing symmetric structural property in TM proteins, although the formation of symmetric pairs was not a general feature of TM proteins and observed only in small TM proteins. Further studies should be performed to understand the formation of such symmetrical pairs, but this study demonstrates that the joint-based approach can be efficiently used to find out some macroscopic structural patterns of TM proteins.
Discussion
Examining the structural and conformational features of proteins in nature efficiently is still a challenging task because of their structural complexity and diversity. A macroscopic description of the protein structure offers a more simplistic way to understand structurally heterogeneous proteins, which can be complementary to the microscopic description method. In this report, a new macroscopic description method, i.e. joint-based description method, was introduced. The primary feature of the approach is to use a joint of secondary structures as the basic element for a description of the protein structure, whereas most developed protein structure description methods utilize physical entities, such as atoms, amino acids, and secondary structures. We performed the analyses of TM structures using the new joint-based approach, and found out some interesting conformational features or patterns in TM proteins. For example, we identified the allowed and disallowed regions of helical arrangement, variation of helical extension pattern depending on TM protein sizes and the possibility of structurally symmetric pairs in TM proteins. This study revealed a possible way to examine the arrangements of physical entities by investigating those of the joints between physical entities at the macroscopic levels. This study focused on membrane proteins, but the joint-based description method is expected to be applied to examine the conformational features of other classes of proteins and find the new features of protein structures in nature.
We note that there is no large difference in the angle distributions according to the configuration of TM helices for both the Ω-type and λ-type (Supplementary Information Figure S3a and b). The TM helices positioning in the membrane proteins can be restricted significantly inside the lipid bilayer as the number of TM helices increases, which might result in the variability of the dihedral angle distributions of Ω and λ-type according to their configurations. Surprisingly, these results suggest that the local arrangement of two consecutive TM helices is not affected too much by the position of the TM helices. On the other hand, an analyses of the patterns for Ωi-Ωi+1 and λi-λi+1 clusters revealed a clear preference of the zig-zag pattern in the packing and extension of the TM helices for the membrane proteins with high TM numbers (Fig. 8a and b). Presumably, this suggests that a zig-zag pattern is an optimized form required for the efficient TM helix-packing geometry inside the lipid bilayer. Overall, these results indicate that the membrane protein structure formations in the lipid membrane environment are controlled more significantly by an extension of the TM helix structures rather than the local arrangements.
A symmetric pair in the molecular geometry has been popularly observed in natural small molecules. Representative examples are the existence of stereoisomerism of amino acids and monosaccharides. Our joint-based approach allowed us to catch that there are some geometrically symmetric pairs in TM proteins. This suggests that the symmetric properties such as stereoisomerism observed in small molecules can exist in the level of global protein structures. Of course, this study was very limited to TM proteins and therefore further analyses should be performed against more expanded protein dataset. Our joint-based approach for protein structure is expected to be efficiently used in such studies.
Protein conformational diversity is closely associated with its functions. From the macroscopic analyses of TM topology in terms of Ω and λ angles, we could observe some structural features which can be related to functions. For example, the unique dihedral space (more ++ dyad signatures for Ω and more −− dyad signatures for λ angle) can be related to channeling activity. It has been reported that 11–14 TMH proteins, where “zig-zag” conformation is the most common, are mostly transporters and this conformation is required to form a channel for ion transport32. In a similar way, 7TM GPCR protein families showed dihedral angle deviations that can be also related to functional features. For the 7TM GPCR, 3 rd to 5 th omega angles showed significant differences than other omega angles where the most functionally important structural changes occurs according to the previous studies33, 34. These indicate that the joint-based macroscopic approach for protein structures can be used in the study on the structure/function linkage.
The joint-based approach is expected to be used for predicting the conformations of the transmembrane helices, a problem that can arise in low-resolution electron microscopy. In addition, it can be used for validating low resolution models of TM proteins similar to the previous studies such as “CaBLAM” method35, 36. As a further study, we have a plan to perform the applications of our approach to structural prediction and validation studies such as k-fold and leave-one-out cross validation based on machine-learning algorithm. For these applications, the joint-based dihedral angle determination method should also be further standardized since it can be sensitive to some factors such as definition of helices and accuracy of protein models.
Another potential of the joint-based metric is that it can be applied to new coordinates for molecular dynamics (MD) simulation of TM proteins at large scale. Membrane proteins are dynamic entities with partial folding and unfolding23. The computational time for folding and unfolding of complex membrane proteins at atomistic level is thus immense. Coarse-grained models such as MARTINI model37, 38 have been applied to MD simulation for the folding of membrane proteins within lipid bilayer, but they still have many limitations in computational time. In our joint-representation, a TM helix is treated as one unit of “rigid-body” at more coarse-grained model. Thus, a force field based on joint-representation can reduce the computational time scale to simulate the folding/unfolding of membrane proteins with large number of TM helices in lipid bilayer using molecular dynamics simulation. One of our long-term purposes is to develop an effective metric for such large scale coarse-grained MD simulation based on the joint-based approach.
Methods
Collection of structural dataset
First, with the aid of PDB, a search was made for membrane proteins with X-ray crystal structures and approximately 2600 structures were found. Only α helix containing proteins were then collected and separated to approximately 959 hits. The dataset of 511 refined structures having sequence identity less than 90%, including with resolution (≤3.5 Å) was selected for the unique proteins containing both homologous and non-homologous protein chains. Nearly 160 proteins with the sequence identity less than 30% with ≤3.5 Å resolution structures were extracted using PICESES server39, 40 and grouped as non-homologous membrane proteins. Helical proteins were classified according to the TM numbers from 3TM to 14TM. 55 protein structures in the same superfamily were treated as remote homologous and were expelled from the list. When choosing a monomer, only one conformation was considered where more than one conformation is available for the same superfamily. 103 protein chains were finally identified as a training dataset. To validate the completeness of the selected dataset, we performed DALI search using the selected 103 non-homologous proteins and examined how many structural homologs of the 103 proteins were detected in the whole 959 proteins. The 103 structures detected 89.7%, 96.2%, and 97.5% of the 959 proteins when the threshold of RMSD for structural homology was set to 3.0 Å, 4.0 Å, and 5.0 Å, respectively.
Determination of structural joint points
To select the structural joints, the amino acid position was scrutinized visually for Cα XYZ coordinates from the corresponding PDB file. The written PYTHON program read each protein structure for the “HELIX” in the PDB files to detect their each helix residue and output their amino acid positions and Cα XYZ coordinates. In addition, specialized databases for the TM helices were also cross checked for their beginning and ending residue position numbers. For each individual protein, the specialized databases, such as OPM41, PDBTM42, and TMPad43, were referred to classify their SSE (Secondary Structure Element) topologies and were used to identify their helical and loop segments based on the coordinates obtained from PDB. To select the fixed joint points, we majorly relied on OPM helical segments annotation with the help of manual inspections to avoid ambiguities. Such specified residue coordinates for each secondary structure, i.e., helices, were treated as the structural joining points to represent protein macroscopically. Table 1 lists the PDB codes and the corresponding topology of the membrane proteins. The listed coordinates of the structural joints represent each SSE; their continuous adjacent joint points were chosen for each helix and loop. While establishing a connection of these joints residues, a new description of the overall protein structure was portrayed.
Dihedral angle calculation of Ω and λ types
The filtered PDB structures were parsed and Cα XYZ coordinates preselected from each joint were exploited for the dihedral measurements. The first dihedral angle involving four joints P1, P2, P3 and P4 can be ascertained by measuring the angle between the two planes made by P1, P2, P3 and P2, P3, P4. Similarly, the second dihedral angle can be found by applying the structural points (P2, P3, P4, and P5), and the (P3, P4, P5 and P6) joints are used to determine the third. Initially, for the set of four xyz coordinate points that define a dihedral angle, the algorithm calculates three vectors, namely, \(\overrightarrow{{V}_{1}}={P}_{1}-{P}_{2}\), \(\overrightarrow{{V}_{2}}={P}_{2}-{P}_{3}\) and \(\overrightarrow{{V}_{3}}={P}_{3}-{P}_{4}\), where \(\overrightarrow{{V}_{n}}={P}_{x}-{P}_{y}\) is the vector from point x to point y. \(\overrightarrow{{{\rm{V}}}_{1}}\) and \(\overrightarrow{{{\rm{V}}}_{2}}\) defines the 1st plane (Orthogonal frame, Mn), whereas \(\overrightarrow{{{\rm{V}}}_{2}}\) and \(\overrightarrow{{{\rm{V}}}_{3}}\) does the 2nd plane. The angle between these planes reflects the dihedral angle between the helices (or loops), which is designated as Ω (or λ). The normal unit vector to this plane was calculated by taking the cross product of these two vectors: \(\overrightarrow{{{\rm{N}}}_{321}}=\frac{\overrightarrow{{{\rm{V}}}_{1}}={{\rm{P}}}_{1}-{{\rm{P}}}_{2}\ast \overrightarrow{{{\rm{V}}}_{2}}={{\rm{P}}}_{2}-{{\rm{P}}}_{3}}{|\overrightarrow{{{\rm{V}}}_{1}}={{\rm{P}}}_{1}-{{\rm{P}}}_{2}\ast \overrightarrow{{{\rm{V}}}_{2}}={{\rm{P}}}_{2}-{{\rm{P}}}_{3}|}\). The normal unit vector to the plane defined by the second, third and fourth joint coordinates \(\overrightarrow{{N}_{321}}\) was calculated in an analogous manner. The angle between such planes reflects the dihedral angle between the helices, which is designated as Ω. The arctan2 of Ω is calculated using the following relation: dihedral_1 = np.arctan(Y 1, X 1) These are combined and the angle is calculated using the arctan2 function. Such measurements are converted from radians to degrees within the range of −180° to 0° to 180° using the following equation, In_degrees_1 = dihedral_1 ∗ 180°/π to facilitate the analysis. The resulting number of dihedral angles for each protein is directly proportional to the number of helices and loops present in them. A python script was developed in house and executed for the dihedral angle calculation using the Spyder python interface.
Analyses of consecutive dihedral angle patterns
To perform the conformational search based on the signature patterns, the preferred orientations among various combinations of consecutive dihedral angles were counted statistically. For the 103 structures selected, each structure was presented by the Ωn-λn-Ωn+1-λn+1 dihedral angle sets, as summarized in Supplementary Information Table S1; n stands for Helix numbers in the protein structure. The calculated dihedral angles were converted to positive (+ve) and negative (−ve) signatures to represent the conformations, as given in Supplementary Information Table S2. A consecutive Ω-Ω pattern was selected for each fold as Ωn-Ωn+1. Grouped Ωn-Ωn+1 should be a consecutive, adjacent set, and no fixed order, whereas non-consecutive Ωn-Ωn+2 were not considered. For example, the Ω-Ω pattern angles were selected from Ω1-λ1-Ω2-λ2-Ω3-λ3 to Ωn-λn as any consecutive Ωn-Ωn. To make more defined distribution patterns, the consecutive Ωn-Ωn+1 and Ωn-Ωn+1-Ωn+2 were also tested.
References
Ramachandran, G. N., Ramakrishnan, C. & Sasisekharan, V. Stereochemistry of Polypeptide-Chain Configurations. Curr Sci India 59, 813–817 (1990).
Dunbrack, R. L. & Karplus, M. Backbone-Dependent Rotamer Library for Proteins - Application to Side-Chain Prediction. J Mol Biol 230, 543–574, doi:10.1006/jmbi.1993.1170 (1993).
Laskowski, R. A., Macarthur, M. W., Moss, D. S. & Thornton, J. M. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr 26, 283–291, doi:10.1107/S0021889892009944 (1993).
Hooft, R. W. W., Vriend, G., Sander, C. & Abola, E. E. Errors in protein structures. Nature 381, 272–272, doi:10.1038/381272a0 (1996).
Shahlaei, M. et al. Homology modeling of human CCR5 and analysis of its binding properties through molecular docking and molecular dynamics simulation. Bba-Biomembranes 1808, 802–817, doi:10.1016/j.bbamem.2010.12.004 (2011).
Flocco, M. M. & Mowbray, S. L. C-Alpha-Based Torsion Angles - a Simple Tool to Analyze Protein Conformational-Changes. Protein Sci 4, 2118–2122 (1995).
Dewitte, R. S. & Shakhnovich, E. I. Pseudodihedrals - Simplified Protein Backbone Representation with Knowledge-Based Energy. Protein Sci 3, 1570–1581 (1994).
Kleywegt, G. J. Validation of protein models from C-alpha coordinates alone. J Mol Biol 273, 371–376, doi:10.1006/jmbi.1997.1309 (1997).
Madan, B., Seo, S. Y. & Lee, S. G. Structural and sequence features of two residue turns in beta-hairpins. Proteins 82, 1721–1733, doi:10.1002/prot.24526 (2014).
Kolodny, R., Koehl, P., Guibas, L. & Levitt, M. Small libraries of protein fragments model native protein structures accurately. J Mol Biol 323, 297–307, doi:10.1016/S0022-2836(02)00942-7 (2002).
Kolinski, A. & Skolnick, J. Reduced models of proteins and their applications. Polymer 45, 511–524, doi:10.1016/j.polymer.2003.10.064 (2004).
Gong, H. P. & Rose, G. D. Does secondary structure determine tertiary structure in proteins? Proteins 61, 338–343, doi:10.1002/prot.20622 (2005).
Mizuguchi, K. & Go, N. Comparison of Spatial Arrangements of Secondary Structural Elements in Proteins. Protein Eng 8, 353–362, doi:10.1093/protein/8.4.353 (1995).
Koch, I., Lengauer, T. & Wanke, E. An algorithm for finding maximal common subtopologies in a set of protein structures. J Comput Biol 3, 289–306, doi:10.1089/cmb.1996.3.289 (1996).
Schenk, P. W. & Snaar-Jagalska, B. E. Signal perception and transduction: the role of protein kinases. Bba-Mol Cell Res 1449, 1–24, doi:10.1016/S0167-4889(98)00178-5 (1999).
Stock, A. M., Robinson, V. L. & Goudreau, P. N. Two-component signal transduction. Annu Rev Biochem 69, 183–215, doi:10.1146/annurev.biochem.69.1.183 (2000).
Doyle, D. A. et al. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science 280, 69–77, doi:10.1126/science.280.5360.69 (1998).
Lu, Z., Klem, A. M. & Ramu, Y. Ion conduction pore is conserved among potassium channels. Nature 413, 809–813, doi:10.1038/35101535 (2001).
Yellen, G. The voltage-gated potassium channels and their relatives. Nature 419, 35–42, doi:10.1038/nature00978 (2002).
Mccabe, E. R. B. Microcompartmentation of Energy-Metabolism at the Outer Mitochondrial-Membrane - Role in Diabetes-Mellitus and Other Diseases. J Bioenerg Biomembr 26, 317–325, doi:10.1007/Bf00763103 (1994).
Edgar, R. & Bibi, E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J Bacteriol 179, 2274–2280 (1997).
Borst, P., Evers, R., Kool, M. & Wijnholds, J. A family of drug transporters: The multidrug resistance-associated proteins. J Natl Cancer I 92, 1295–1302, doi:10.1093/jnci/92.16.1295 (2000).
von Heijne, G. Membrane-protein topology. Nat Rev Mol Cell Bio 7, 909–918, doi:10.1038/nrm2063 (2006).
Zhang, Y., Hubner, I. A., Arakaki, A. K., Shakhnovich, E. & Skolnick, J. On the origin and highly likely completeness of single-domain protein structures. Proceedings of the National Academy of Sciences of the United States of America 103, 2605–2610, doi:10.1073/pnas.0509379103 (2006).
Efremov, R. G., Vereshaga, Y. A., Volynsky, P. E., Nolde, D. E. & Arseniev, A. S. Association of transmembrane helices: what determines assembling of a dimer? Journal of computer-aided molecular design 20, 27–45 (2006).
Bocharov, E. V., Volynsky, P. E., Pavlov, K. V., Efremov, R. G. & Arseniev, A. S. Structure elucidation of dimeric transmembrane domains of bitopic proteins. Cell adhesion & migration 4, 284–298 (2010).
Wilman, H. R., Shi, J. & Deane, C. M. Helix kinks are equally prevalent in soluble and membrane proteins. Proteins: Structure, Function, and Bioinformatics 82, 1960–1970 (2014).
Riek, R. P., Rigoutsos, I., Novotny, J. & Graham, R. M. Non-α-helical elements modulate polytopic membrane protein architecture. J Mol Biol 306, 349–362 (2001).
Chou, K. C., Carlacci, L., Maggiora, G. M., Parodi, L. A. & Schulz, M. W. An Energy-Based Approach to Packing the 7-Helix Bundle of Bacteriorhodopsin. Protein Sci 1, 810–827 (1992).
Gilson, M. K. & Honig, B. Destabilization of an Alpha-Helix-Bundle Protein by Helix Dipoles. Proceedings of the National Academy of Sciences of the United States of America 86, 1524–1528, doi:10.1073/pnas.86.5.1524 (1989).
Chou, K. C., Maggiora, G. M., Nemethy, G. & Scheraga, H. A. Energetics of the Structure of the 4-Alpha-Helix Bundle in Proteins. Proceedings of the National Academy of Sciences of the United States of America 85, 4295–4299, doi:10.1073/pnas.85.12.4295 (1988).
Dai, J. & Zhou, H.-X. General rules for the arrangements and gating motions of pore-lining helices in homomeric ion channels. Nature communications 5, doi:10.1038/ncomms5641 (2014).
Niv, M. Y., Skrabanek, L., Filizola, M. & Weinstein, H. Modeling activated states of GPCRs: the rhodopsin template. Journal of computer-aided molecular design 20, 437–448 (2006).
Rosenbaum, D. M., Rasmussen, S. G. & Kobilka, B. K. The structure and function of G-protein-coupled receptors. Nature 459, 356–363 (2009).
Richardson, J. S., Prisant, M. G. & Richardson, D. C. Crystallographic model validation: from diagnosis to healing. Current opinion in structural biology 23, 707–714 (2013).
Richardson, J. S. & Richardson, D. C. Doing molecular biophysics: Finding, naming, and picturing signal within complexity. Annual review of biophysics 42, 1, doi:10.1146/annurev-biophys-083012-130353 (2013).
Marrink, S. J., Risselada, H. J., Yefimov, S., Tieleman, D. P. & de Vries, A. H. The MARTINI force field: Coarse grained model for biomolecular simulations. J Phys Chem B 111, 7812–7824, doi:10.1021/jp071097f (2007).
de Jong, D. H. et al. Improved Parameters for the Martini Coarse-Grained Protein Force Field. J Chem Theory Comput 9, 687–697, doi:10.1021/ct300646g (2013).
Wang, G. L. & Dunbrack, R. L. PISCES: a protein sequence culling server. Bioinformatics 19, 1589–1591, doi:10.1093/bioinformatics/btg224 (2003).
Wang, G. L. & Dunbrack, R. L. PISCES: recent improvements to a PDB sequence culling server. Nucleic Acids Res 33, W94–W98, doi:10.1093/nar/gki402 (2005).
Lomize, M. A., Lomize, A. L., Pogozheva, I. D. & Mosberg, H. I. OPM: Orientations of proteins in membranes database. Bioinformatics 22, 623–625, doi:10.1093/bioinformatics/btk023 (2006).
Tusnady, G. E., Dosztanyi, Z. & Simon, I. PDB_TM: selection and membrane localization of transmembrane proteins in the protein data bank. Nucleic Acids Res 33, D275–D278, doi:10.1093/nar/gki002 (2005).
Lo, A., Cheng, C. W., Chiu, Y. Y., Sung, T. Y. & Hsu, W. L. TMPad: an integrated structural database for helix-packing folds in transmembrane proteins. Nucleic Acids Res 39, D347–D355, doi:10.1093/nar/gkq1255 (2011).
Markus, M. T. & Groenen, P. J. F. An introduction to the bootstrap. Psychometrika 63, 97–101 (1998).
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2015R1D1A1A01056766 & 2015R1D1A1A01061125).
Author information
Authors and Affiliations
Contributions
J.T. and S.G.L. designed research; J.T. performed research; J.T., S.W. and S.G.L. analyzed data; J.T., S.W. and S.G.L. wrote the paper. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Thangappan, J., Wu, S. & Lee, SG. Joint-based description of protein structure: its application to the geometric characterization of membrane proteins. Sci Rep 7, 1056 (2017). https://doi.org/10.1038/s41598-017-01011-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-01011-z
This article is cited by
-
Comparative Analysis of TM and Cytoplasmic β-barrel Conformations Using Joint Descriptor
Scientific Reports (2018)
-
Measuring the Conformational Distance of GPCR-related Proteins Using a Joint-based Descriptor
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.