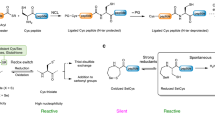
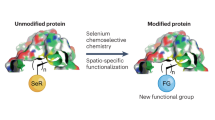
Abstract
Chemoselective peptide ligation methods have provided synthetic access to numerous proteins, including those bearing native post-translational modifications and unnatural labels. This protocol outlines the chemical synthesis of proteins using a recently discovered reaction (diselenide–selenoester ligation (DSL)) in a rapid, additive-free manner. After ligation, the products can be chemoselectively deselenized to produce native peptide and protein products. We describe methods for the synthesis of suitably functionalized peptide diselenide and peptide selenoester fragments via Fmoc-solid-phase peptide synthesis (SPPS) protocols, fusion of these fragments by DSL, and the chemoselective deselenization of the ligation products to generate native synthetic proteins. We demonstrate the method’s utility through the total chemical synthesis of the post-translationally modified collagenous domain of the hormone adiponectin via DSL–deselenization at selenocystine (the oxidized form of selenocysteine) and the rapid preparation of two tick-derived thrombin-inhibiting proteins by DSL–deselenization at β-selenoaspartate and γ-selenoglutamate. This method should find widespread use for the rapid synthesis of proteins, including cases in which other peptide ligation methods cannot be used (or cannot be used efficiently), e.g., at sterically hindered or deactivated acyl donors. The method’s speed and efficiency may render it useful in the generation of synthetic protein libraries. Each protein discussed can be synthesized within 15 working days from resin loading and can be readily produced by practitioners with master’s-level experience in organic chemistry. Each synthesis using these protocols was performed independently by two labs (one academic and one industrial), which attained comparable yields of the protein products.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The authors declare that all data supporting the findings of this study are available within the article and its supplementary information files
References
Dawson, P., Muir, T., Clark-Lewis, I. & Kent, S. Synthesis of proteins by native chemical ligation. Science 266, 776–779 (1994).
Zheng, J.-S., Tang, S., Qi, Y.-K., Wang, Z.-P. & Liu, L. Chemical synthesis of proteins using peptide hydrazides as thioester surrogates. Nat. Protoc. 8, 2483–2495 (2013).
Boll, E. et al. One-pot chemical synthesis of small ubiquitin-like modifier protein–peptide conjugates using bis(2-sulfanylethyl)amido peptide latent thioester surrogates. Nat. Protoc. 10, 269–292 (2015).
Harmand, T. J., Murar, C. E. & Bode, J. W. Protein chemical synthesis by α-ketoacid–hydroxylamine ligation. Nat. Protoc. 11, 1130–1147 (2016).
Tang, S. et al. Chemical synthesis of membrane proteins by the removable backbone modification method. Nat. Protoc. 12, 2554–2569 (2017).
Maity, S. K., Jbara, M., Mann, G., Kamnesky, G. & Brik, A. Total chemical synthesis of histones and their analogs, assisted by native chemical ligation and palladium complexes. Nat. Protoc. 12, 2293–2322 (2017).
Mitchell, N. J. et al. Rapid additive-free selenocystine–selenoester peptide ligation. J. Am. Chem. Soc. 137, 14011–14014 (2015).
Hackeng, T. M., Griffin, J. H. & Dawson, P. E. Protein synthesis by native chemical ligation: Expanded scope by using straightforward methodology. Proc. Natl. Acad. Sci. USA 96, 10068–10073 (1999).
Kulkarni, S. S., Sayers, J., Premdjee, B. & Payne, R. J. Rapid and efficient protein synthesis through expansion of the native chemical ligation concept. Nat. Rev. Chem 2, 0122 (2018).
Mitchell, N. J., Kulkarni, S. S., Malins, L. R., Wang, S. & Payne, R. J. One-pot ligation–oxidative deselenization at selenocysteine and selenocystine. Chem. Eur. J 23, 946–952 (2017).
Malins, L. R., Mitchell, N. J., McGowan, S. & Payne, R. J. Oxidative deselenization of selenocysteine: applications for programmed ligation at serine. Angew. Chem. Int. Ed. 54, 12716–12721 (2015).
Mitchell, N. J. et al. Accelerated protein synthesis via one-pot ligation-deselenization chemistry. Chem 2, 703–715 (2017).
Wang, X., Sanchez, J., Stone, M. J. & Payne, R. J. Sulfation of the human cytomegalovirus protein UL22A enhances binding to the chemokine RANTES. Angew. Chem. Int. Ed. 56, 8490–8494 (2017).
Malins, L. R. & Payne, R. J. Synthesis and utility of β-selenol-phenylalanine for native chemical ligation–deselenization chemistry. Org. Lett. 14, 3142–3145 (2012).
Sayers, J. et al. Construction of challenging proline–proline junctions via diselenide–selenoester ligation chemistry. J. Am. Chem. Soc. 140, 13327–13334 (2018).
Conibear, A. C., Watson, E. E., Payne, R. J. & Becker, C. F. W. Native chemical ligation in protein synthesis and semi-synthesis. Chem. Soc.Rev. 47, 9046–9068 (2018).
Gieselman, M. D., Xie, L. & van der Donk, W. A. Synthesis of a selenocysteine-containing peptide by native chemical ligation. Org. Lett. 3, 1331–1334 (2001).
Turer, A. T. & Scherer, P. E. Adiponectin: mechanistic insights and clinical implications. Diabetologia 55, 2319–2326 (2012).
Hanna, C. C., Kulkarni, S. S., Watson, E. E., Premdjee, B. & Payne, R. J. Solid-phase synthesis of peptide selenoesters via a side-chain anchoring strategy. Chem. Commun. 53, 5424–5427 (2017).
SteMarie, E. J., Ruggles, E. L. & Hondal, R. J. Removal of the 5-nitro-2-pyridine-sulfenyl protecting group from selenocysteine and cysteine by ascorbolysis. J. Pept. Sci. 22, 571–576 (2016).
Bruker. Instructions for Use: Bruker Guide to MALDI Sample Preparation. https://www.bruker.com/fileadmin/user_upload/8-PDFDocs/Separations_MassSpectrometry/InstructionForUse/8702557_IFU_Bruker_Guide_MALDI_Sample_Preparation_Revision_E.pdf (2015).
Acknowledgements
We acknowledge funding from the Australian Research Council Linkage Scheme (LP150100308) to K.W.C.-F. and R.J.P.
Author information
Authors and Affiliations
Contributions
S.S.K. and E.E.W. performed the experiments, compound characterization and data analysis at the University of Sydney, Sydney Australia. B.P. replicated the selenoesterification and ligation experiments, compound characterization and data analysis at Novo Nordisk A/S, Denmark. S.S.K., E.E.W., B.P., K.W.C.-F. and R.J.P. contributed to experimental design and writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Nature Protocols thanks Michio Iwaoka and other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Mitchell, N. J. et al. J. Am. Chem. Soc. 137, 14011–14014 (2015): https://doi.org/10.1021/jacs.5b07237
Mitchell, N. J. et al. Chem 2, 703–715 (2017): https://doi.org/10.1016/j.chempr.2017.04.003
Sayers, J. et al. J. Am. Chem. Soc. 140, 13327–13334 (2018): https://doi.org/10.1021/jacs.8b07877
Integrated supplementary information
Supplementary Fig. 1. Characterization of adiponectin (19–40) (C36Acm) selenoester—Payne Lab, University of Sydney.
a) Analytical reverse-phase HPLC trace and b) ESI mass envelope of reverse-phase HPLC purified Adiponectin selenoester (19-40). Rt 4.67 min (0 to 50% B over 5 min, λ = 214 nm); Calculated Mass [2M+3H]3+: 1646.8, [M+2H]2+: 1235.3; Mass Found (ESI+): 1646.1 [2M+3H]3+, 1234.9[M+2H]2+.
Supplementary Fig. 2. Characterization of adiponectin 19–40-SePh (C36Acm)—Dep. Research Chemistry, Novo Nordisk.
a) Analytical reverse-phase HPLC trace (C18 BEH) and b) ESI mass envelope of reverse-phase HPLC purified Adiponectin selenoester (19-40). Rt 2.93 min (5 to 30% B over 0.5 min followed by 30 to 50% B over 3.5 min, λ = 214 nm); Calculated Mass [M+2H]2+: 1235.1, [M+3H]3+: 823.7; Mass Found (ESI+): 1234.9 [M+2H]2+, 823.6 [M+3H]3+.
Supplementary Fig. 3. Characterization of adiponectin C-terminal diselenide dimer (41–107)—Payne Lab, University of Sydney.
a) Analytical reverse-phase HPLC trace and B) ESI mass envelope of reverse-phase HPLC purified Adiponectin diselenide dimer (41–107); Rt 3.87 min (0 to 50% B over 5 min, λ = 214 nm). Calculated Mass [2M+7H]7+: 1875.5 [2M+8H]8+: 1641.2, [2M+9H]9+: 1459.0, [2M+10H]10+: 1313.2, [2M+11H]11+: 1193.9, [2M+12H]12+: 1094.4, [2M+13H]13+: 1010.4, [2M+14H]14+: 938.3, [2M+15H]15+: 875.8, [2M+16H]16+: 821.1, [2M+17H]17+: 772.9, [2M+18H]18+: 730.0, [2M+19H]19+: 691.6, [2M+20H]20+: 657.0 Mass Found (ESI+): 1874.9 [2M+7H]7+, 1640.6 [2M+8H]8+, 1458.2 [2M+9H]9+, 1312.8 [2M+10H]10+, 1193.6 [2M+11H]11+, 1094.3 [2M+12H]12+, 1010.2 [2M+13H]13+, 938.0 [2M+14H]14+, 875.5 [2M+15H]15+, 820.9 [2M+16H]16+, 772.6 [2M+17H]17+, 729.9 [2M+18H]18+, 691.4 [2M+19H]19+, 656.8 [2M+20H]20+.
Supplementary Fig. 4. DSL preparation of adiponectin (19–107) (C36Acm)—Payne Lab, University of Sydney.
Analytical reverse-phase HPLC trace of the crude Adiponectin (19–107) (C36Acm) a) post ligation and b) post deselenization.
Supplementary Fig. 5. Characterization of adiponectin (19–107) (C36Acm)—Payne Lab, University of Sydney.
a) Analytical reverse-phase HPLC trace and b) ESI mass envelope of reverse-phase HPLC purified Adiponectin (19–107) (C36Acm); Rt 4.22 min (0 to 50% B over 5 min, λ = 214 nm). Calculated Mass [M+5H]5+: 1759.1, [M+6H]6+: 1466.1, [M+7H]7+: 1256.8, [M+8H]8+: 1099.8, [M+9H]9+: 977.7, [M+10H]10+: 880.1, [M+11H]11+: 800.1; Mass Found (ESI+): 1758.7 [M+5H]5+, 1466.1 [M+6H]6+, 1256.8 [M+7H]7+, 1099.9 [M+8H]8+, 977.8 [M+9H]9+, 880.2 [M+10H]10+, 800.2 [M+11H]11+.
Supplementary Fig. 6. Acm deprotection of adiponectin (19–107) (C36Acm)—Payne Lab, University of Sydney.
Analytical reverse-phase HPLC trace of the crude Adiponectin (19–107) post-Acm deprotection.
Supplementary Fig. 7. Characterization of purified adiponectin (19–107)—Payne Lab, University of Sydney.
a) Analytical reverse-phase HPLC trace and b) ESI mass envelope of reverse-phase HPLC purified Adiponectin (19–107); Rt 4.48 min (0 to 50% B over 5 min, λ = 214 nm). Calculated Mass [M+5H]5+: 1745.3, [M+6H]6+: 1454.6, [M+7H]7+: 1246.9, [M+8H]8+: 1091.2, [M+9H]9+: 970.1, [M+10H]10+: 873.2, [M+11H]11+: 793.9, [M+12H]12+: 727.8; Mass Found (ESI+): 1745.1 [M+5H]5+, 1454.2 [M+6H]6+, 1246.8 [M+7H]7+, 1091.1 [M+8H]8+, 970.0 [M+9H]9+, 873.1[M+10H]10+, 793.8 [M+11H]11+, 727.8 [M+12H]12+.
Supplementary Fig. 8. MALDI-TOF of purified adiponectin (19–107)—Payne Lab, University of Sydney.
Calculated Mass for C372H588N114O125S2 [M+H]+: 8721.5; Mass Found: 8722.0 [M+H]+.
Supplementary Fig. 9. DSL preparation of adiponectin (19–107) (C36Acm)—Dep. Research Chemistry, Novo Nordisk.
Analytical reverse-phase HPLC (C18 BEH) of a) Crude DSL mixture during preparation of Adiponectin (19–107) (C36Acm) after 10 min b) Crude deselenization mixture of Adiponectin (19–107) (C36Acm) after 16 h (5% B over 0.5 min followed by 5 to 60% B over 3.5 min, λ = 214 nm).
Supplementary Fig. 10. Characterization of adiponectin (19–107) (C36Acm)—Dep. Research Chemistry, Novo Nordisk.
a) Analytical reverse-phase HPLC trace (C18 BEH) and b) ESI mass envelope of reverse-phase HPLC purified Adiponectin (19–107) (C36Acm). Rt 2.48 min (5 to 20% B over 0.5 min followed by 20 to 40% B over 3.5 min, λ = 214 nm); Calculated Mass [M+5H]5+: 1759.5, [M+6H]6+: 1466.4, [M+7H]7+: 1257.1, [M+8H]8+: 1100.1, [M+9H]9+: 977.9, [M+10H]10+: 880.2, [M+11H]11+: 800.3, [M+12H]12+: 733.7; Mass Found (ESI+): 1759.3 [M+5H]5+, 1466.2 [M+6H]6+, 1256.9 [M+7H]7+, 1099.9 [M+8H]8+, 977.8 [M+9H]9+, 880.1 [M+10H]10+, 800.2 [M+11H]11+, 733.6 [M+12H]12+.
Supplementary Fig. 11. Acm deprotection of adiponectin (19–107) (C36Acm)—Dep. Research Chemistry, Novo Nordisk.
a) Analytical reverse-phase HPLC trace (C18 CSH) and b) ESI mass envelope of reverse-phase HPLC purified Adiponectin (19–107). Rt 2.04 min (5% B over 0.5 min followed by 5 to 60% B over 3.5 min, λ = 214 nm); Calculated Mass [M+5H]5+: 1745.3, [M+6H]6+: 1454.6, [M+7H]7+: 1246.9, [M+8H]8+: 1091.2, [M+9H]9+: 970.0, [M+10H]10+: 873.1, [M+11H]11+: 793.9, [M+12H]12+: 727.8; Mass Found (ESI+): 1744.9 [M+5H]5+, 1454.4 [M+6H]6+, 1246.8 [M+7H]7+, 1091.1 [M+8H]8+, 970.0[M+9H]9+, 873.1 [M+10H]10+, 793.8 [M+11H]11+, 727.7 [M+12H]12+.
Supplementary Fig. 12. Characterization of haemathrin-1 N-terminal selenoester (1–23)—Payne Lab, University of Sydney.
a) Analytical reverse-phase HPLC trace and B) ESI mass envelope of reverse-phase HPLC purified Haemathrin-1 selenoester (1–28). Rt 3.91 min (0 to 50% B over 5 min, λ = 214 nm); Calculated Mass [2M+3H]3+: 1856.9, [M+2H]2+: 1393.0, [M+3H]3+: 929.0, [M+4H]4+: 697.0, [M+5H]5+: 557.8, [M+6H]6+: 465.0; Mass Found (ESI+): 1856.2 [2M+3H]3+, 1392.2 [M+2H]2+, 928.5 [M+3H]3+, 696.7 [M+4H]4+, 557.5 [M+5H]5+, 464.7 [M+6H]6+.
Supplementary Fig. 13. Characterization of haemathrin-1 N-terminal selenoester (1–23)—Dep. Research Chemistry, Novo Nordisk.
a) Analytical reverse-phase HPLC trace (C18 BEH) and b) ESI mass envelope of reverse-phase HPLC purified Haemathrin-1 selenoester (1–23). Rt 2.21 min (5 to 20% B over 0.5 min followed by 20 to 40% B over 3.5 min, λ = 214 nm); Calculated Mass [M+2H]2+: 1392.7, [M+3H]3+: 928.8, [M+4H]4+: 696.8, [M+5H]5+: 557.7; Mass Found (ESI+): 1392.5 [M+2H]2+, 928.6 [M+3H]3+, 696.7 [M+4H]4+, 557.6 [M+5H]5+.
Supplementary Fig. 14. Characterization of haemathrin-1 C-terminal diselenide dimer (24–59)—Payne Lab, University of Sydney.
a) Analytical reverse-phase HPLC trace and b) ESI mass envelope of reverse-phase HPLC purified Haemathrin-1 diselenide dimer (29–59); Rt 3.41 min (0 to 50% B over 5 min, λ = 215 nm). Calculated Mass [2M+5H]5+: 1658.3, [2M+6H]6+: 1382.1, [2M+7H]7+: 1184.8, [2M+8H]8+: 1036.8, [2M+9H]9+: 921.7, [2M+10H]10+: 829.6, [2M+11H]11+: 754.3, [2M+12H]12+: 691.5; Mass Found (ESI+); 1657.5 [2M+5H]5+, 1381.4 [2M+6H]6+, 1184.3 [2M+7H]7+, 1036.4 [2M+8H]8+, 921.3 [2M+9H]9+, 829.3 [2M+10H]10+, 754.0 [2M+11H]11+, 691.2 [2M+12H]12+.
Supplementary Fig. 15. Characterization of haemathrin-2 N-terminal selenoester (1–28)—Payne Lab, University of Sydney.
a) Analytical reverse-phase HPLC trace and B) ESI mass envelope of reverse-phase HPLC purified Haemathrin-2 selenoester (1–28). Rt 3.91 min (0 to 50% B over 5 min, λ = 214 nm); Calculated Mass [M+2H]2+: 1701.3, [M+3H]3+: 1134.5, [M+4H]4+: 851.2, [M+5H]5+: 681.1, [M+6H]6+: 567.8, [M+7H]7+: 486.8; Mass Found (ESI+): 1701.1 [M+2H]2+, 1134.4 [M+3H]3+, 851.0 [M+4H]4+, 681.1 [M+5H]5+, 567.7 [M+6H]6+, 486.8 [M+7H]7+.
Supplementary Fig. 16. Characterization of haemathrin-2 N-terminal selenoester (1–28)—Dep. Research Chemistry, Novo Nordisk.
a) Analytical reverse-phase HPLC trace (C18 BEH) and b) ESI mass envelope of reverse-phase HPLC purified Haemathrin-2 selenoester (1–28). Rt 2.72 min (5 to 15% B over 0.5 min followed by 15 to 35% B over 3.5 min, λ = 214 nm); Calculated Mass [M+2H]2+: 1701.3, [M+3H]3+: 1134.5, [M+4H]4+: 851.2, [M+5H]5+: 681.1; Mass Found (ESI+): 1701.1 [M+2H]2+, 1134.4 [M+3H]3+, 851.0 [M+4H]4+, 681.0 [M+5H]5+.
Supplementary Fig. 17. Characterization of haemathrin-2 C-terminal diselenide dimer (29–59)—Payne Lab, University of Sydney.
a) Analytical reverse-phase HPLC trace and b) ESI mass envelope of reverse-phase HPLC purified Haemathrin-2 diselenide dimer (29–59); Rt 3.61 min (0 to 50% B over 5 min, λ = 215 nm). Calculated Mass [M+2H]2+: 1773.3, [2M+5H]5+: 1418.4, [M+3H]3+: 1182.5, [2M+7H]7+: 1013.5, [M+4H]4+: 887.2, [2M+9H]9+: 788.5, [M+5H]5+: 709.9, [2M+11H]11+: 645.3; Mass Found (ESI+); 1774.3 [M+2H]2+, 1419.4 [2M+5H]5+, 1182.9 [M+3H]3+, 1014.0 [2M+7H]7+, 887.3 [M+4H]4+, 788.7 [2M+9H]9+, 709.9 [M+5H]5+, 645.6 [2M+11H]11+.
Supplementary Fig. 18. Characterization of haemathrin-2 C-terminal diselenide dimer (29–59)—Dep. Research Chemistry, Novo Nordisk.
a) Analytical reverse-phase HPLC trace (C18 BEH) and b) ESI mass envelope of reverse-phase HPLC purified Haemathrin-2 diselenide (29–59). Rt 2.48 min (5 to 15% B over 0.5 min followed by 15 to 35% B over 3.5 min, λ = 214 nm); Calculated Mass [2M+4H]4+: 1772.8, [2M+5H]5+: 1418.4, [2M+6H]6+: 1182.2, [2M+7H]7+: 1013.4, [2M+8H]8+: 886.9, [2M+9H]9+: 788.5, [2M+10H]10+: 709.7; Mass Found (ESI+): 1772.6 [2M+4H]4+, 1418.3 [2M+5H]5+, 1182.1 [2M+6H]6+, 1013.4 [2M+7H]7+, 887.0 [2M+8H]8+, 788.4 [2M+9H]9+, 709.8 [2M+10H]10+.
Supplementary Fig. 19. DSL preparation of haemathrin-1 (1–59)—Payne Lab, University of Sydney.
Analytical reverse-phase HPLC trace of the crude Haemathrin-1 A) post ligation and B) post deselenization.
Supplementary Fig. 20. Characterization of haemathrin-1 (1–59)—Payne Lab, University of Sydney.
a) Analytical reverse-phase HPLC trace and b) ESI mass envelope of reverse-phase HPLC purified Haemathrin-1; Rt 3.41 min (0 to 50% B over 5 min, λ = 214 nm). Calculated Mass [M+4H]4+: 1673.5, [M+5H]5+: 1339.0, [M+6H]6+: 1116.0, [M+7H]7+: 956.7, [M+8H]8+: 837.3, [M+9H]9+: 744.3, [M+10H]10+: 670.0, [M+11H]11+: 609.2; Mass Found (ESI+): 1673.1 [M+4H]4+, 1338.8 [M+5H]5+, 1115.8 [M+6H]6+, 965.5 [M+7H]7+, 837.1 [M+8H]8+, 744.2 [M+9H]9+, 669.9 [M+10H]10+, 609.1 [M+11H]11+.
Supplementary Fig. 21. MALDI-TOF of haemathrin-1 (1–59)—Payne Lab, University of Sydney.
Calculated Mass for C279H445N91O101 [M+H]+: 6690.1; Mass Found: 6690.1 [M+H]+.
Supplementary Fig. 22. DSL preparation of haemathrin-1 (1–59)—Dep. Research Chemistry, Novo Nordisk.
Analytical reverse-phase HPLC (C18 BEH) of a) Crude DSL mixture during preparation of Haemathrin-1 after 10 min b) Crude deselenization mixture of Haemathrin-1 after 10 min (5% B over 0.5 min followed by 5 to 60% B over 3.5 min, λ = 214 nm).
Supplementary Fig. 23. Characterization of haemathrin-1 (1–59)—Dep. Research Chemistry, Novo Nordisk.
a) Analytical reverse-phase HPLC trace (C18 BEH) and B) ESI mass envelope of reverse-phase HPLC purified Haemathrin-1. Rt 2.77 min (5 to 10% B over 0.5 min followed by 10 to 30% B over 3.5 min, λ = 214 nm); Calculated Mass [M+4H]4+: 1673.3, [M+5H]5+: 1338.9, [M+6H]6+: 1115.9, [M+7H]7+: 956.9, [M+8H]8+: 837.2, [M+9H]9+: 744.3, [M+10H]10+: 669.9, [M+11H]11+: 609.1; Mass Found (ESI+): 1673.2 [M+4H]4+, 1338.7 [M+5H]5+, 1115.8 [M+6H]6+, 956.5 [M+7H]7+, 837.1 [M+8H]8+, 744.2 [M+9H]9+, 669.9 [M+10H]10+, 609.1 [M+11H]11+.
Supplementary Fig. 24. DSL preparation of haemathrin-2 (1–59)—Payne Lab, University of Sydney.
Analytical reverse-phase HPLC trace of crude Haemathrin-2 post a) post ligation and b) post deselenization.
Supplementary Fig. 25. Characterization of haemathrin-2 (1–59)—Payne Lab, University of Sydney.
a) Analytical reverse-phase HPLC trace and b) ESI mass envelope of reverse-phase HPLC purified Haemathrin-2; Rt 3.63 min (0 to 50% B over 5 min, λ = 214 nm). Calculated Mass [M+4H]4+: 1678.3, [M+5H]5+: 1342.8, [M+6H]6+: 1119.2, [M+7H]7+: 959.5, [M+8H]8+: 839.7, [M+9H]9+: 746.5, [M+10H]10+: 671.9, [M+11H]11+: 610.9; Mass Found (ESI+): 1677.9 [M+4H]4+, 1342.5 [M+5H]5+, 1118.9 [M+6H]6+, 959.3 [M+7H]7+, 839.5 [M+8H]8+, 746.3 [M+9H]9+, 671.7 [M+10H]10+, 610.8 [M+11H]11+. * Indicates artefact masses due to dimerization.
Supplementary Fig. 26. MALDI-TOF of haemathrin-2 (1–59)—Payne Lab, University of Sydney.
Calculated Mass for C280H446N88O104 [M+H]+: 6709.1; Mass Found: 6709.6 [M+H]+.
Supplementary Fig. 27. DSL preparation of haemathrin-2 (1–59)—Dep. Research Chemistry, Novo Nordisk.
Analytical reverse-phase HPLC (C18 BEH) of a) Crude DSL mixture during preparation of Haemathrin-2 after 10 min b) Crude deselenization mixture of Haemathrin-2 after 10 min (5% B over 0.5 min followed by 5 to 60% B over 3.5 min, λ = 214 nm).
Supplementary Fig. 28. Characterization of haemathrin-2 (1–59)—Dep. Research Chemistry, Novo Nordisk.
a) Analytical reverse-phase HPLC trace (C18 CSH) and b) ESI mass envelope of reverse-phase HPLC purified Haemathrin-2 Rt 1.78 min (5% B over 0.5 min followed by 5 to 60% B over 3.5 min, λ = 214 nm); Calculated Mass [M+4H]4+: 1678.1, [M+5H]5+: 1342.7, [M+6H]6+: 1119.0, [M+7H]7+: 959.3, [M+8H]8+: 839.5, [M+9H]9+: 746.4, [M+10H]10+: 671.8; Mass Found (ESI+): 1677.9 [M+4H]4+, 1342.5 [M+5H]5+, 1119.0 [M+6H]6+, 959.3 [M+7H]7+, 839.5 [M+8H]8+, 746.3 [M+9H]9+, 671.8 [M+10H]10+.
Supplementary information
Supplementary Information
Supplementary Figures 1–28 and Supplementary Text
Rights and permissions
About this article
Cite this article
Kulkarni, S.S., Watson, E.E., Premdjee, B. et al. Diselenide–selenoester ligation for chemical protein synthesis. Nat Protoc 14, 2229–2257 (2019). https://doi.org/10.1038/s41596-019-0180-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0180-4
This article is cited by
-
Selenium chemistry for spatio-selective peptide and protein functionalization
Nature Reviews Chemistry (2024)
-
Chemical Synthesis of Proteins Containing 300 Amino Acids
Chemical Research in Chinese Universities (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.