Abstract
Here we present a protocol for analyses of axon regeneration and density in unsectioned adult mouse spinal cord. This includes methods for injury and tracing of dorsal column sensory and corticospinal axons; clearing and staining of unsectioned spinal cord; visualization of axon degeneration and regeneration in cleared and uncleared specimens using two-photon microscopy; and either manual or semi-automatic analysis of axon density and regeneration in 3D space using Imaris and ImageJ software. This protocol can be used to elucidate the molecular and cellular mechanisms underlying nervous system degeneration and regeneration and to establish the therapeutic efficacy of candidate neuroregenerative treatments. Because tissue sectioning is not required, this protocol enables unambiguous evaluation of regeneration and greatly accelerates the speed at which analyses can be conducted. Surgical procedures take <30 min per mouse, with a wait period of 2 weeks between axonal injury and tracing and 2–8 weeks between tracing and tissue processing. Clearing and immunolabeling take ~1–2 weeks, depending on the size of the sample. Imaging and analysis can be performed in 1 d. All these procedures can be accomplished by a competent graduate student or experienced technician.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Hilton, B. J. & Bradke, F. Can injured adult CNS axons regenerate by recapitulating development? Development 144, 3417–3429 (2017).
Ramer, L. M., Ramer, M. S. & Bradbury, E. J. Restoring function after spinal cord injury: towards clinical translation of experimental strategies. Lancet Neurol. 13, 1241–1256 (2014).
Schwab, M. E. & Strittmatter, S. M. Nogo limits neural plasticity and recovery from injury. Curr. Opin. Neurobiol. 27, 53–60 (2014).
Galtrey, C. M. & Fawcett, J. W. The role of chondroitin sulfate proteoglycans in regeneration and plasticity in the central nervous system. Brain Res. Rev. 54, 1–18 (2007).
Blanquie, O. & Bradke, F. Cytoskeleton dynamics in axon regeneration. Curr. Opin. Neurobiol. 51, 60–69 (2018).
He, Z. & Jin, Y. Intrinsic control of axon regeneration. Neuron 90, 437–451 (2016).
Tuszynski, M. H. & Steward, O. Concepts and methods for the study of axonal regeneration in the CNS. Neuron 74, 777–791 (2012).
Curcio, M. & Bradke, F. Axon regeneration in the central nervous system: facing the challenges from the inside. Annu. Rev. Cell Dev. Biol. 34, 495–521 (2018).
Steward, O., Zheng, B. & Tessier‐Lavigne, M. False resurrections: distinguishing regenerated from spared axons in the injured central nervous system. J. Comp. Neurol. 459, 1–8 (2003).
Kim, S.-Y., Chung, K. & Deisseroth, K. Light microscopy mapping of connections in the intact brain. Trends Cogn. Sci. 17, 596–599 (2013).
Vigouroux, R. J., Belle, M. & Chédotal, A. Neuroscience in the third dimension: shedding new light on the brain with tissue clearing. Mol. Brain 10, 33 (2017).
Ertürk, A. et al. Three-dimensional imaging of the unsectioned adult spinal cord to assess axon regeneration and glial responses after injury. Nat. Med. 18, 166–171 (2012).
Ertürk, A. et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc. 7, 1983–1995 (2012).
Chung, K. et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013).
Yang, B. et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958 (2014).
Susaki, E. A. et al. Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat. Protoc. 10, 1709–1727 (2015).
Ruschel, J. et al. Systemic administration of epothilone B promotes axon regeneration after spinal cord injury. Science 348, 347–352 (2015).
Tedeschi, A. et al. The calcium channel subunit Alpha2delta2 suppresses axon regeneration in the adult CNS. Neuron 92, 419–434 (2016).
Ylera, B. et al. Chronically CNS-injured adult sensory neurons gain regenerative competence upon a lesion of their peripheral axon. Curr. Biol. 19, 930–936 (2009).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Omura, T. et al. Robust axonal regeneration occurs in the injured CAST/Ei mouse CNS. Neuron 86, 1215–1227 (2015).
Tedeschi, A., Omura, T. & Costigan, M. CNS repair and axon regeneration: using genetic variation to determine mechanisms. Exp. Neurol. 287, 409–422 (2017).
Geoffroy, C. G., Hilton, B. J., Tetzlaff, W. & Zheng, B. Evidence for an age-dependent decline in axon regeneration in the adult mammalian central nervous system. Cell Rep. 15, 238–246 (2016).
Geoffroy, C. G., Meves, J. M. & Zheng, B. The age factor in axonal repair after spinal cord injury: a focus on neuron-intrinsic mechanisms. Neurosci. Lett. 652, 41–49 (2017).
Willis, W. D. Jr & Coggeshall, R. E. Sensory Mechanisms of the Spinal Cord 3rd edn, Vol. 1: Primary Afferent Neurons and the Spinal Dorsal Horn (Springer Science & Business Media, New York, 2004).
Niu, J. et al. Modality-based organization of ascending somatosensory axons in the direct dorsal column pathway. J. Neurosci. 33, 17691–17709 (2013).
Faweett, J. & Keynes, R. J. Peripheral nerve regeneration. Annu. Rev. Neurosci. 13, 43–60 (1990).
Richardson, P. & Issa, V. Peripheral injury enhances central regeneration of primary sensory neurones. Nature 309, 791–793 (1984).
Neumann, S. & Woolf, C. J. Regeneration of dorsal column fibers into and beyond the lesion site following adult spinal cord injury. Neuron 23, 83–91 (1999).
Lu, P., Yang, H., Jones, L. L., Filbin, M. T. & Tuszynski, M. H. Combinatorial therapy with neurotrophins and cAMP promotes axonal regeneration beyond sites of spinal cord injury. J. Neurosci. 24, 6402–6409 (2004).
Scott, A. L. & Ramer, M. S. Schwann cell p75NTR prevents spontaneous sensory reinnervation of the adult spinal cord. Brain 133, 421–432 (2009).
Rigaud, M. et al. Species and strain differences in rodent sciatic nerve anatomy: implications for studies of neuropathic pain. Pain 136, 188–201 (2008).
Lemon, R. N. Descending pathways in motor control. Annu. Rev. Neurosci. 31, 195–218 (2008).
Nudo, R. & Masterton, R. Descending pathways to the spinal cord, III: sites of origin of the corticospinal tract. J. Comp. Neurol. 296, 559–583 (1990).
Lemon, R. N. & Griffiths, J. Comparing the function of the corticospinal system in different species: organizational differences for motor specialization? Muscle Nerve 32, 261–279 (2005).
Armand, J. The origin, course and terminations of corticospinal fibers in various mammals. Prog. Brain Res. 57, 329–360 (1982).
Fink, K. L., Strittmatter, S. M. & Cafferty, W. B. Comprehensive corticospinal labeling with mu-crystallin transgene reveals axon regeneration after spinal cord trauma in ngr1 −/− mice. J. Neurosci. 35, 15403–15418 (2015).
Steward, O., Zheng, B., Ho, C., Anderson, K. & Tessier‐Lavigne, M. The dorsolateral corticospinal tract in mice: an alternative route for corticospinal input to caudal segments following dorsal column lesions. J. Comp. Neurol. 472, 463–477 (2004).
Hilton, B. J. et al. Re-establishment of cortical motor output maps and spontaneous functional recovery via spared dorsolaterally projecting corticospinal neurons after dorsal column spinal cord injury in adult mice. J. Neurosci. 36, 4080–4092 (2016).
Wang, X. et al. Deconstruction of corticospinal circuits for goal-directed motor skills. Cell 171, 440–455.e14 (2017).
Porter, R. & Lemon, R. Corticospinal Function and Voluntary Movement (Oxford University Press, Oxford, UK, 1993).
Zukor, K. et al. Short hairpin RNA against PTEN enhances regenerative growth of corticospinal tract axons after spinal cord injury. J. Neurosci. 33, 15350–15361 (2013).
Anderson, M. A. et al. Astrocyte scar formation aids central nervous system axon regeneration. Nature 532, 195–200 (2016).
Wang, Z., Maunze, B., Wang, Y., Tsoulfas, P. & Blackmore, M. G. Global connectivity and function of descending spinal input revealed by 3D microscopy and retrograde transduction. J. Neurosci. 38, 10566–10581 (2018).
Zhang, F. et al. Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nat. Protoc. 5, 439–456 (2010).
Roth, B. L. DREADDs for neuroscientists. Neuron 89, 683–694 (2016).
Zingg, B. et al. AAV-mediated anterograde transsynaptic tagging: mapping corticocollicular input-defined neural pathways for defense behaviors. Neuron 93, 33–47 (2017).
Belle, M. et al. A simple method for 3D analysis of immunolabeled axonal tracts in a transparent nervous system. Cell Rep. 9, 1191–1201 (2014).
Renier, N. et al. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910 (2014).
Al-Ali, H., Beckerman, S. R., Bixby, J. L. & Lemmon, V. P. In vitro models of axon regeneration. Exp. Neurol. 287, 423–434 (2017).
Hammarlund, M. & Jin, Y. Axon regeneration in C. elegans. Curr. Opin. Neurobiol. 27, 199–207 (2014).
Brace, E. & DiAntonio, A. Models of axon regeneration in Drosophila. Exp. Neurol. 287, 310–317 (2017).
Laskowski, C. J. & Bradke, F. In vivo imaging: a dynamic imaging approach to study spinal cord regeneration. Exp. Neurol. 242, 11–17 (2013).
Gomis-Rüth, S., Stiess, M., Wierenga, C. J., Meyn, L. & Bradke, F. Single-cell axotomy of cultured hippocampal neurons integrated in neuronal circuits. Nat. Protoc. 9, 1028–1037 (2014).
Stiess, M. et al. Axon extension occurs independently of centrosomal microtubule nucleation. Science 327, 704–707 (2010).
Gomis-Rüth, S., Wierenga, C. J. & Bradke, F. Plasticity of polarization: changing dendrites into axons in neurons integrated in neuronal circuits. Curr. Biol. 18, 992–1000 (2008).
Pernet, V. & Schwab, M. E. Lost in the jungle: new hurdles for optic nerve axon regeneration. Trends Neurosci. 37, 381–387 (2014).
Gyllensten, L. & Malmfors, T. Myelinization of the optic nerve and its dependence on visual function—a quantitative investigation in mice. Development 11, 255–266 (1963).
Bray, E. R. et al. 3D visualization of individual regenerating retinal ganglion cell axons reveals surprisingly complex growth paths. eNeuro 4, ENEURO.0093-17.2017 (2017).
Luo, X. et al. Three-dimensional evaluation of retinal ganglion cell axon regeneration and pathfinding in whole mouse tissue after injury. Exp. Neurol. 247, 653–662 (2013).
Parrinello, S. et al. EphB signaling directs peripheral nerve regeneration through Sox2-dependent Schwann cell sorting. Cell 143, 145–155 (2010).
Silasi, G. & Murphy, T. H. Stroke and the connectome: how connectivity guides therapeutic intervention. Neuron 83, 1354–1368 (2014).
Raff, M. C., Whitmore, A. V. & Finn, J. T. Axonal self-destruction and neurodegeneration. Science 296, 868–871 (2002).
Dominici, C. et al. Floor-plate-derived netrin-1 is dispensable for commissural axon guidance. Nature 545, 350–354 (2017).
Kim, J.-E., Li, S., GrandPré, T., Qiu, D. & Strittmatter, S. M. Axon regeneration in young adult mice lacking Nogo-A/B. Neuron 38, 187–199 (2003).
Jayaprakash, N. et al. Optogenetic interrogation of functional synapse formation by corticospinal tract axons in the injured spinal cord. J. Neurosci. 36, 5877–5890 (2016).
Hellal, F. et al. Microtubule stabilization reduces scarring and causes axon regeneration after spinal cord injury. Science 331, 928–931 (2011).
Liu, K. et al. PTEN deletion enhances the regenerative ability of adult corticospinal neurons. Nat. Neurosci. 13, 1075–1081 (2010).
Steward, O. et al. Regenerative growth of corticospinal tract axons via the ventral column after spinal cord injury in mice. J. Neurosci. 28, 6836–6847 (2008).
Acknowledgements
We thank S. Stern, B. Schaffran, S. Cerqueira, G. Duncan, A. Husch, and T. Van Rossum for helpful feedback on the protocol. We also thank the DZNE Light Microscope Facility (LMF), the Image and Data Analysis Facility (IDAF), and the Animal Research Facility (ARF) for technical support. We particularly thank K. Keppler (DZNE LMF) and C. Moehl (DZNE IDAF) for their help. We are grateful to J. Liu and W. Tetzlaff (UBC) for helpful discussions of mouse models of spinal cord injury. B.J.H. is supported by a Wings for Life (WfL) Aguayo-Tator Mentoring Fellowship and a non-stipendiary European Molecular Biology Organization (EMBO) Long-Term Fellowship (ALTF 28-2017). A.T. is supported by the Craig H. Neilsen Foundation, the Marina Romoli Onlus Association and Discovery Themes on Chronic Brain Injury. F.B. is supported by the Deutsches Zentrum für Neurodegenerative Erkrankungen (DZNE), the International Foundation for Research in Paraplegia, WfL, the Deutsche Forschungsgemeinschaft, ERANET AXON REPAIR, and ERANET RATER SCI. F.B. is a member of the excellence cluster ImmunoSensation2 (EXC2151–390873048) and the SFB 1089 and is a recipient of the Roger de Spoelberch Prize.
Author information
Authors and Affiliations
Contributions
B.J.H., O.B., A.T., and F.B. conceived and designed the protocol. B.J.H., O.B., and A.T. performed the experiments. B.J.H. and O.B. wrote the manuscript. F.B. supervised the project and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Journal peer review information: Nature Protocols thanks Kai Liu and other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Tedeschi, A. et al. Neuron 92, 419−434 (2016): https://doi.org/10.1016/j.neuron.2016.09.026
Ertürk, A. et al. Nat. Med. 18, 166−171 (2012): https://doi.org/10.1038/nm.2600
Integrated supplementary information
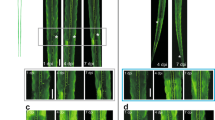
Supplementary Figure 1 Comparison of analysis in unsectioned vs. sectioned spinal cord.
(a) One hour after thoracic incomplete lateral hemisection spinal cord injury, AAV1-CMV-GFP was injected into the left sciatic nerve and a conditioning lesion (n = 4) or sham procedure (n = 3) was performed. Four weeks after injury, animals were perfused and unsectioned spinal cords were cleared and imaged. Unsectioned spinal cords were then cut into 60 µm sagittal sections and stained for GFAP. Images were taken of 2–3 consecutive sections containing the highest density of GFP+ sensory axons using a Leica 8 Confocal microscope and axon regeneration was analyzed on these sections. Images are representative of the extent of axon regeneration for each group and are of the same samples with and without sectioning. Scale bars, 200 µm. (b) Inset of a. Arrowheads point to regenerating axons rostral to the lesion site after a conditioning lesion. Scale bar, 100 µm. (c) Quantification of regenerating axons and comparison of sectioned vs. unsectioned analysis. Sham n = 3 and PNL n = 4.
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1
Supplementary Video 1
Volume rendering of conditioned sensory axons regenerating in unsectioned and uncleared spinal cord.
Supplementary Video 2
Rotation and ortho-slicing of the sample.
Supplementary Video 3
Volume rendering of corticospinal axons in a cleared sample with a spinal cord injury.
Supplementary Data 1
Raw data in Imaris file format of GFP+ sensory axons regenerating past a spinal cord injury site.
Rights and permissions
About this article
Cite this article
Hilton, B.J., Blanquie, O., Tedeschi, A. et al. High-resolution 3D imaging and analysis of axon regeneration in unsectioned spinal cord with or without tissue clearing. Nat Protoc 14, 1235–1260 (2019). https://doi.org/10.1038/s41596-019-0140-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0140-z
This article is cited by
-
Longitudinal interrogation of sympathetic neural circuits and hemodynamics in preclinical models
Nature Protocols (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.