Abstract
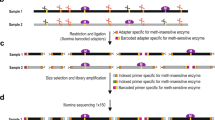
Whole-genome bisulfite sequencing (WGBS) has been widely used to quantify cytosine DNA methylation frequency in an expanding array of cell and tissue types. Because of the denaturing conditions used, this method ultimately leads to the measurement of methylation frequencies at single cytosines. Hence, the methylation frequency of CpG dyads (two complementary CG dinucleotides) can be only indirectly inferred by overlaying the methylation frequency of two cytosines measured independently. Furthermore, hemi-methylated CpGs (hemiCpGs) have not been previously analyzed in WGBS studies. We recently developed in silico strand annealing (iSA), a bioinformatics method applicable to WGBS data, to resolve the methylation status of CpG dyads into unmethylated, hemi-methylated, and methylated. HemiCpGs account for 4–20% of the DNA methylome in different cell types, and some can be inherited across cell divisions, suggesting a role as a stable epigenetic mark. Therefore, it is important to resolve hemiCpGs from fully methylated CpGs in WGBS studies. This protocol describes step-by-step commands to accomplish this task, including dividing alignments by strand, pairing alignments between strands, and extracting single-fragment methylation calls. The versatility of iSA enables its application downstream of other WGBS-related methods such as nasBS-seq (nascent DNA bisulfite sequencing), ChIP-BS-seq (ChIP followed by bisulfite sequencing), TAB-seq, oxBS-seq, and fCAB-seq. iSA is also tunable for analyzing the methylation status of cytosines in any sequence context. We exemplify this flexibility by uncovering the single-fragment non-CpG methylome. This protocol provides enough details for users with little experience in bioinformatic analysis and takes 2–7 h.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
WGBS dataset (GSM1386021) is available at SRA.
References
Xu, C. & Corces, V. G. Nascent DNA methylome mapping reveals inheritance of hemimethylation at CTCF/cohesin sites. Science 359, 1166–1170 (2018).
Law, J. A. & Jacobsen, S. E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220 (2010).
Statham, A. L. et al. Bisulfite sequencing of chromatin immunoprecipitated DNA (BisChIP-seq) directly informs methylation status of histone-modified DNA. Genome Res. 22, 1120–1127 (2012).
Brinkman, A. B. et al. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 22, 1128–1138 (2012).
Meissner, A. et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 33, 5868–5877 (2005).
Kivioja, T. et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 9, 72–74 (2011).
Yu, M. et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380 (2012).
Booth, M. J. et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336, 934–937 (2012).
Song, C. X. et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell 153, 678–691 (2013).
Laird, C. D. et al. Hairpin-bisulfite PCR: assessing epigenetic methylation patterns on complementary strands of individual DNA molecules. Proc. Natl. Acad. Sci. USA 101, 204–209 (2004).
Arand, J. et al. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genet. 8, e1002750 (2012).
Zhao, L. et al. The dynamics of DNA methylation fidelity during mouse embryonic stem cell self-renewal and differentiation. Genome Res. 24, 1296–1307 (2014).
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Ramirez, F., Dundar, F., Diehl, S., Gruning, B. A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Holmes, E. E. et al. Performance evaluation of kits for bisulfite-conversion of DNA from tissues, cell lines, FFPE tissues, aspirates, lavages, effusions, plasma, serum, and urine. PLoS ONE 9, e93933 (2014).
Miura, F. et al. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 40, e136 (2012).
Wang, L. et al. Programming and inheritance of parental DNA methylomes in mammals. Cell 157, 979–991 (2014).
Acknowledgements
This work was supported by U.S. Public Health Service Award 5P01 GM085354. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
C.X. and V.G.C. conceived the project; C.X. designed and streamlined the protocol; C.X. and V.G.C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related link
Key reference using this protocol
Xu, C. & Corces, V. G. Science 359, 1166–1170 (2018): https://doi.org/10.1126/science.aan5480
Code availability
An interactive shell script is available at https://github.com/chxu02/iSA.
Supplementary information
Rights and permissions
About this article
Cite this article
Xu, C., Corces, V.G. Resolution of the DNA methylation state of single CpG dyads using in silico strand annealing and WGBS data. Nat Protoc 14, 202–216 (2019). https://doi.org/10.1038/s41596-018-0090-x
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-018-0090-x
This article is cited by
-
Epigenetic regulation in metabolic diseases: mechanisms and advances in clinical study
Signal Transduction and Targeted Therapy (2023)
-
Statistical and bioinformatic analysis of hemimethylation patterns in non-small cell lung cancer
BMC Cancer (2021)
-
Simultaneously measuring the methylation of parent and daughter strands of replicated DNA at the single-molecule level by Hammer-seq
Nature Protocols (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.