Abstract
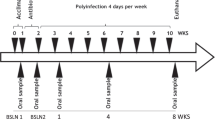
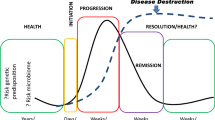
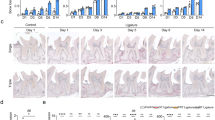
Periodontal disease (PD) is a common dental disease associated with the interaction between dysbiotic oral microbiota and host immunity. It is a prevalent disease, resulting in loss of gingival tissue, periodontal ligament, cementum and alveolar bone. PD is a major form of tooth loss in the adult population. Experimental animal models have enabled the study of PD pathogenesis and are used to test new therapeutic approaches for treating the disease. The ligature-induced periodontitis model has several advantages as compared with other models, including rapid disease induction, predictable bone loss and the capacity to study periodontal tissue and alveolar bone regeneration because the model is established within the periodontal apparatus. Although mice are the most convenient and versatile animal models used in research, ligature-induced periodontitis has been more frequently used in large animals. This is mostly due to the technical challenges involved in consistently placing ligatures around murine teeth. To reduce the technical challenge associated with the traditional ligature model, we previously developed a simplified method to easily install a bacterially retentive ligature between two molars for inducing periodontitis. In this protocol, we provide detailed instructions for placement of the ligature and demonstrate how the model can be used to evaluate gingival tissue inflammation and alveolar bone loss over a period of 18 d after ligature placement. This model can also be used on germ-free mice to investigate the role of human oral bacteria in periodontitis in vivo. In conclusion, this protocol enables the mechanistic study of the pathogenesis of periodontitis in vivo.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hajishengallis, G. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 35, 3–11 (2014).
Hajishengallis, G., Darveau, R. P. & Curtis, M. A. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 10, 717–725 (2012).
Richards, D. Review finds that severe periodontitis affects 11% of the world population. Evid. Based Dent. 15, 70–71 (2014).
Kassebaum, N. J. et al. Global burden of severe periodontitis in 1990-2010: a systematic review and meta-regression. J. Dent. Res. 93, 1045–1053 (2014).
Eke, P. I., Dye, B. A., Wei, L., Thornton-Evans, G. O. & Genco, R. J. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J. Dent. Res. 91, 914–920 (2012).
Tonetti, M. S., Jepsen, S., Jin, L. & Otomo-Corgel, J. Impact of the global burden of periodontal diseases on health, nutrition and wellbeing of mankind: a call for global action. J. Clin. Periodontol. 44, 456–462 (2017).
de Pablo, P., Chapple, I. L., Buckley, C. D. & Dietrich, T. Periodontitis in systemic rheumatic diseases. Nat. Rev. Rheumatol. 5, 218–224 (2009).
Pinho, M. M., Faria-Almeida, R., Azevedo, E., Manso, M. C. & Martins, L. Periodontitis and atherosclerosis: an observational study. J. Periodontal. Res. 48, 452–457 (2013).
Yoon, A. J., Cheng, B., Philipone, E., Turner, R. & Lamster, I. B. Inflammatory biomarkers in saliva: assessing the strength of association of diabetes mellitus and periodontal status with the oral inflammatory burden. J. Clin. Periodontol. 39, 434–440 (2012).
Colombo, A. P. et al. Clinical and microbiological features of refractory periodontitis subjects. J. Clin. Periodontol. 25, 169–180 (1998).
Levine, M., Lohinai, Z. & Teles, R. P. Low biofilm lysine content in refractory chronic periodontitis. J. Periodontol. 88, 181–189 (2017).
Oz, H. S. & Puleo, D. A. Animal models for periodontal disease. J. Biomed. Biotechnol. 2011, 754857, https://doi.org/10.1155/2011/754857 (2011).
Graves, D. T., Kang, J., Andriankaja, O., Wada, K. & Rossa, C. Jr. Animal models to study host-bacteria interactions involved in periodontitis. Front. Oral Biol. 15, 117–132 (2012).
Graves, D. T., Fine, D., Teng, Y. T., Van Dyke, T. E. & Hajishengallis, G. The use of rodent models to investigate host-bacteria interactions related to periodontal diseases. J. Clin. Periodontol. 35, 89–105 (2008).
Padial-Molina, M., Rodriguez, J. C., Volk, S. L. & Rios, H. F. Standardized in vivo model for studying novel regenerative approaches for multitissue bone-ligament interfaces. Nat. Protoc. 10, 1038–1049 (2015).
Abe, T. & Hajishengallis, G. Optimization of the ligature-induced periodontitis model in mice. J. Immunol. Methods 394, 49–54 (2013).
Weiner, G. S., DeMarco, T. J. & Bissada, N. F. Long term effect of systemic tetracycline administration on the severity of induced periodontitis in the rat. J. Periodontol. 50, 619–623 (1979).
Glowacki, A. J. et al. Prevention of inflammation-mediated bone loss in murine and canine periodontal disease via recruitment of regulatory lymphocytes. Proc. Natl. Acad. Sci. USA 110, 18525–18530 (2013).
Kajikawa, T. et al. Milk fat globule epidermal growth factor 8 inhibits periodontitis in non-human primates and its gingival crevicular fluid levels can differentiate periodontal health from disease in humans. J. Clin. Periodontol. 44, 472–483 (2017).
Martin, R., Bermudez-Humaran, L. G. & Langella, P. Gnotobiotic rodents: an in vivo model for the study of microbe-microbe interactions. Front. Microbiol. 7, 409 (2016).
Hajishengallis, G. et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10, 497–506 (2011).
Zenobia, C. et al. Commensal bacteria-dependent select expression of CXCL2 contributes to periodontal tissue homeostasis. Cell Microbiol. 15, 1419–1426 (2013).
Grover, M. & Kashyap, P. C. Germ-free mice as a model to study effect of gut microbiota on host physiology. Neurogastroenterol. Motil. 26, 745–748 (2014).
Rovin, S., Costich, E. R. & Gordon, H. A. The influence of bacteria and irritation in the initiation of periodontal disease in germfree and conventional rats. J. Periodontal. Res. 1, 193–204 (1966).
Xiao, E. et al. Diabetes enhances IL-17 expression and alters the oral microbiome to increase its pathogenicity. Cell Host Microbe 22, 120–128.e124 (2017).
Haara, O. et al. Ectodysplasin regulates activator-inhibitor balance in murine tooth development through Fgf20 signaling. Development 139, 3189–3199 (2012).
Martins, M. D. et al. Epigenetic modifications of histones in periodontal disease. J. Dent. Res. 95, 215–222 (2016).
Jiao, Y. et al. Induction of bone loss by pathobiont-mediated Nod1 signaling in the oral cavity. Cell Host Microbe 13, 595–601 (2013).
Hu, Y. et al. IL-21/anti-Tim1/CD40 ligand promotes B10 activity in vitro and alleviates bone loss in experimental periodontitis in vivo. Biochim. Biophys. Acta 1863, 2149–2157 (2017).
Chun, J., Kim, K. Y., Lee, J. H. & Choi, Y. The analysis of oral microbial communities of wild-type and toll-like receptor 2-deficient mice using a 454 GS FLX Titanium pyrosequencer. BMC Microbiol. 10, 101 (2010).
Abe, T. et al. Local complement-targeted intervention in periodontitis: proof-of-concept using a C5a receptor (CD88) antagonist. J. Immunol. 189, 5442–5448 (2012).
Baker, P. J., Dixon, M. & Roopenian, D. C. Genetic control of susceptibility to Porphyromonas gingivalis-induced alveolar bone loss in mice. Infect. Immun. 68, 5864–5868 (2000).
de Molon, R. S. et al. Long-term evaluation of oral gavage with periodontopathogens or ligature induction of experimental periodontal disease in mice. Clin. Oral Investig. 20, 1203–1216 (2016).
Genco, C. A., Cutler, C. W., Kapczynski, D., Maloney, K. & Arnold, R. R. A novel mouse model to study the virulence of and host response to Porphyromonas (Bacteroides) gingivalis. Infect. Immun. 59, 1255–1263 (1991).
Pouliot, M., Clish, C. B., Petasis, N. A., Van Dyke, T. E. & Serhan, C. N. Lipoxin A(4) analogues inhibit leukocyte recruitment to Porphyromonas gingivalis: a role for cyclooxygenase-2 and lipoxins in periodontal disease. Biochemistry 39, 4761–4768 (2000).
Zubery, Y. et al. Bone resorption caused by three periodontal pathogens in vivo in mice is mediated in part by prostaglandin. Infect. Immun. 66, 4158–4162 (1998).
Rath, H. et al. Biofilm formation by the oral pioneer colonizer Streptococcus gordonii: an experimental and numerical study. FEMS Microbiol. Ecol. 93, https://doi.org/10.1093/femsec/fix010 (2017).
Periasamy, S. & Kolenbrander, P. E. Central role of the early colonizer Veillonella sp. in establishing multispecies biofilm communities with initial, middle, and late colonizers of enamel. J. Bacteriol. 192, 2965–2972 (2010).
Guo, L., He, X. & Shi, W. Intercellular communications in multispecies oral microbial communities. Front. Microbiol. 5, 328, https://doi.org/10.3389/fmicb.2014.00328 (2014).
Heller, D., Silva-Boghossian, C. M., do Souto, R. M. & Colombo, A. P. Subgingival microbial profiles of generalized aggressive and chronic periodontal diseases. Arch. Oral Biol. 57, 973–980 (2012).
Zhou, P., Liu, J., Merritt, J. & Qi, F. A YadA-like autotransporter, Hag1 in Veillonella atypica is a multivalent hemagglutinin involved in adherence to oral Streptococci, Porphyromonas gingivalis, and human oral buccal cells. Mol. Oral Microbiol. 30, 269–279 (2015).
Chalmers, N. I., Palmer, R. J., Jr., Cisar, J. O. & Kolenbrander, P. E. Characterization of a Streptococcus sp.-Veillonella sp. community micromanipulated from dental plaque. J. Bacteriol. 190, 8145–8154 (2008).
Lima, B. P., Shi, W. & Lux, R. Identification and characterization of a novel Fusobacterium nucleatum adhesin involved in physical interaction and biofilm formation with Streptococcus gordonii. Microbiologyopen 6, https://doi.org/10.1002/mbo3.444 (2017).
Doi, H. et al. Potency of umbilical cord blood- and Wharton’s jelly-derived mesenchymal stem cells for scarless wound healing. Sci. Rep. 6, 18844, https://doi.org/10.1038/srep18844 (2016).
Salvi, G. E., Beck, J. D. & Offenbacher, S. PGE2, IL-1beta, and TNF-alpha responses in diabetics as modifiers of periodontal disease expression. Ann. Periodontol. 3, 40–50 (1998).
Korte, D. L. & Kinney, J. Personalized medicine: an update of salivary biomarkers for periodontal diseases. Periodontol. 2000 70, 26–37 (2016).
Cekici, A., Kantarci, A., Hasturk, H. & Van Dyke, T. E. Inflammatory and immune pathways in the pathogenesis of periodontal disease. Periodontol. 2000 64, 57–80 (2014).
Jun, H. K., Jung, Y. J. & Choi, B. K. Treponema denticola, Porphyromonas gingivalis, and Tannerella forsythia induce cell death and release of endogenous danger signals. Arch. Oral Biol. 73, 72–78 (2017).
Rock, K. L. & Kono, H. The inflammatory response to cell death. Annu. Rev. Pathol. 3, 99–126 (2008).
Violant, D., Galofre, M., Nart, J. & Teles, R. P. In vitro evaluation of a multispecies oral biofilm on different implant surfaces. Biomed. Mater. 9, 035007, https://doi.org/10.1088/1748-6041/9/3/035007 (2014).
Foster, J. S. & Kolenbrander, P. E. Development of a multispecies oral bacterial community in a saliva-conditioned flow cell. Appl. Environ. Microbiol. 70, 4340–4348 (2004).
Acknowledgements
This study was supported by grants T90DE021986 and F32DE026688 (Y.J.), an IBM Junior Faculty Development Award from the University of North Carolina at Chapel Hill and grants K01DE027087 (J.M.), 5R01DE023836 (S.O.) and 5R01DE025207 (M.H.S.). We thank W.V. Giannobile of the University of Michigan School of Dentistry for constructive suggestions and helpful discussion. We thank B. Huang and J. Ashley of the Histology Research Core Facility at UNC for histological embedding and sectioning. We thank the Microscope Core facility for the images. We thank the UNC Biomedical Research Imaging Center for microCT scanning. We thank the Germ Free & Gnotobiotic Mouse Facilities at the University of Michigan and the Gnotobiotic Rodent Core Facility at the University of North Carolina for assistance with the human bacterial infection experiment. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Y.J. and J.M. designed the experiments. Y.J., J.M. and M.S.G. performed the in vivo experiments. L.J. performed H&E and MPO staining and analysis. M.Z.M. performed TRAP staining and analysis. M.S.G. performed qRT-PCR and analysis using the RT2 Profiler Array. S.Z. and L.S. performed the microCT analysis. Y.J., J.M., T.M. and M.S.G. developed the murine dental bed and ligature holder. J.M. and Y.J. wrote and edited the manuscript together. S.O., N.I. and M.H.S. helped to develop the model and edited the manuscript. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
1. Jiao, Y. et al. Cell Host Microbe 13, 595–601 (2013): https://doi.org/10.1016/j.chom.2013.04.005
Integrated supplementary information
Supplementary Figure 1
Typical colony morphology for S. gordonii, F. nucleatum and V. parvula on a TSA plate.
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1
Supplementary Manual
Detailed top view of blueprints required for manufacture of the murine dental bed
Rights and permissions
About this article
Cite this article
Marchesan, J., Girnary, M.S., Jing, L. et al. An experimental murine model to study periodontitis. Nat Protoc 13, 2247–2267 (2018). https://doi.org/10.1038/s41596-018-0035-4
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-018-0035-4
This article is cited by
-
Association between the systemic immune inflammation index and periodontitis: a cross-sectional study
Journal of Translational Medicine (2024)
-
Association between healthy lifestyle combinations and periodontitis in NHANES
BMC Oral Health (2024)
-
Tim4 deficiency reduces CD301b+ macrophage and aggravates periodontitis bone loss
International Journal of Oral Science (2024)
-
A diet rich in omega-3 fatty acid improves periodontitis and tissue destruction by MMP2- and MMP9-linked inflammation in a murine model
Odontology (2024)
-
Intervention with extracellular matrix metalloproteinase inducer in osteoclasts attenuates periodontitis-induced bone resorption
Odontology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.