Abstract
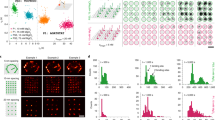
Atomic force microscopy (AFM)-based nanomechanical imaging provides a high-resolution approach for imaging biomolecules with nanometer resolution. Nevertheless, the lack of appropriate nanomechanical labels poses a limit to biological applications. Here, we describe how to generate a set of shape-resolved nanomechanical labels by exploiting self-assembled DNA origami technology. By designing ‘mediator’ strands that can hybridize with both the origami shapes and the target DNA, these origami shape IDs can be used to site-specifically label genomic DNA with high efficiency and high throughput. When DNA origami shape IDs are used to label target sequences containing two single-nucleotide polymorphisms (SNPs), this approach is capable of differentiating adjacent labeling sites separated by only 30 nucleobases (~10 nm) under AFM imaging. This resolution is a threefold improvement of that which can be obtained with imaging-based genotyping using super-resolution imaging. We further demonstrate how to use origami shape IDs for high-resolution genotyping of SNPs in disease-associated genes in patients. The entire protocol takes ~2 d to complete.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Rothemund, P. W. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297 (2006).
Shih, W. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414 (2010).
Andersen, E. S. et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 459, 73 (2009).
Ke, Y., Ong, L. L., Shih, W. M. & Yin, P. Three-dimensional structures self-assembled from DNA bricks. Science 338, 1177–1183 (2012).
Ke, Y., Lindsay, S., Chang, Y., Liu, Y. & Yan, H. Self-assembled water-soluble nucleic acid probe tiles for label-free RNA hybridization assays. Science 319, 180–183 (2008).
Zhang, H. et al. DNA origami-based shape IDs for single-molecule nanomechanical genotyping. Nat. Commun. 8, 14738 (2017).
Collins, F. S., Green, E. D., Guttmacher, A. E. & Guyer, M. S. A vision for the future of genomics research. Nature 422, 835–847 (2003).
Xiao, M. et al. Direct determination of haplotypes from single DNA molecules. Nat. Methods 6, 199 (2009).
Lam, E. T. et al. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat. Biotechnol. 30, 771–776 (2012).
Hell, S. W. Far-field optical nanoscopy. Science 316, 1153–1158 (2007).
Baday, M. et al. Multicolor super-resolution DNA imaging for genetic analysis. Nano Lett. 12, 3861 (2012).
Müller, D. J. & Dufrêne, Y. F. Atomic force microscopy as a multifunctional molecular toolbox in nanobiotechnology. Nat. Nanotechnol. 3, 261 (2008).
Hansma, H. G., Sinsheimer, R. L., Li, M. Q. & Hansma, P. K. Atomic force microscopy of single- and double-stranded DNA. Nucleic Acids Res. 20, 3585–3590 (1992).
Gross, L., Mohn, F., Moll, N., Liljeroth, P. & Meyer, G. The chemical structure of a molecule resolved by atomic force microscopy. Science 325, 1110 (2009).
Lü, J. H. et al. Positioning isolation and biochemical analysis of single DNA molecules based on nanomanipulation and single-molecule PCR. J. Am. Chem. Soc. 126, 11136 (2004).
Kufer, S. K., Puchner, E. M., Gumpp, H., Liedl, T. & Gaub, H. E. Single-molecule cut-and-paste surface assembly. Science 319, 594–596 (2008).
Zhang, Y. et al. Transfer of two-dimensional oligonucleotide patterns onto stereocontrolled plasmonic nanostructures through DNA-origami-based nanoimprinting lithography. Angew. Chem. 55, 8036 (2016).
Liu, H., Wang, J., Song, S., Fan, C. & Gothelf, K. V. A DNA-based system for selecting and displaying the combined result of two input variables. Nat. Commun. 6, 10089 (2015).
Schüller, V. J. et al. Cellular immunostimulation by CpG-sequence-coated DNA origami structures. ACS Nano 5, 9696 (2011).
Mikkilä, J. et al. Virus-encapsulated DNA origami nanostructures for cellular delivery. Nano Lett. 14, 2196 (2014).
Perrault, S. D. & Shih, W. M. Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability. ACS Nano 8, 5132–5140 (2014).
Langecker et al. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 338, 932–936 (2012).
Derr, N. D. et al. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science 338, 662 (2012).
Kuzyk, A. et al. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 483, 311–314 (2012).
Thacker, V. V. et al. DNA origami based assembly of gold nanoparticle dimers for surface-enhanced Raman scattering. Nat. Commun. 5, 3448 (2014).
Zhang, Z. et al. Asymmetric DNA origami for spatially addressable and index-free solution-phase DNA chips. Adv. Mater. 22, 2672–2675 (2010).
Lin, M. et al. Electrochemical detection of nucleic acids, proteins, small molecules and cells using a DNA-nanostructure-based universal biosensing platform. Nat. Protoc. 11, 1244 (2016).
Zhang, Z. et al. A DNA-origami chip platform for label-free snp genotyping using toehold-mediated strand displacement. Small 6, 1854–1858 (2010).
Subramanian, H. K., Chakraborty, B., Sha, R. & Seeman, N. C. The label-free unambiguous detection and symbolic display of single nucleotide polymorphisms on DNA origami. Nano Lett. 11, 910–913 (2011).
Nangreave, J., Yan, H. & Liu, Y. Studies of thermal stability of multivalent DNA hybridization in a nanostructured system. Biophys. J. 97, 563–571 (2009).
Chen, P. et al. Gold nanoparticles for high-throughput genotyping of long-range haplotypes. Nat. Nanotechnol. 6, 639–644 (2011).
Li, H. et al. Nanoparticle PCR: nanogold-assisted PCR with enhanced specificity. Angew. Chem. 44, 5100 (2005).
Woolley, A. T., Guillemette, C., Li, C. C., Housman, D. E. & Lieber, C. M. Direct haplotyping of kilobase-size DNA using carbon nanotube probes. Nat. Biotechnol. 18, 760 (2000).
Acknowledgements
This work was supported by the Ministry of Science and Technology of China (2017YFA0205302, 2016YFA0201200 and 2016YFA0400900), the NSFC (61771253 and 21390414), the Program for Changjiang Scholars and Innovative Research Team in University (IRT 15R37), the Natural Science Foundation of Jiangsu Province (BK20151504) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, YX03001).
Author information
Authors and Affiliations
Contributions
C.F. and Lianhui W. supervised the projects; J.C., H.Z., Y.X., H.L., Lianhui W. and C.F. designed and conducted the experiments; J.C., H.Z., Y.X., H.L., Lihua W. and C.F. analyzed the data; and J.C., Y.X., Q.L. and C.F. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
DNA origami-based shape IDs for single-molecule nanomechanical genotyping: https://doi.org/10.1038/ncomms14738
A DNA-origami chip platform for label-free SNP genotyping using toehold-mediated strand displacement: https://doi.org/10.1002/smll.201000908
Gold nanoparticles for high-throughput genotyping of long-range haplotypes: https://doi.org/10.1038/nnano.2011.141
Asymmetric DNA origami for spatially addressable and index-free solution-phase DNA chips: https://doi.org/10.1002/adma.201000151
Integrated supplementary information
Supplementary Fig. 1 Step-by-step schematic illustration of the procedure.
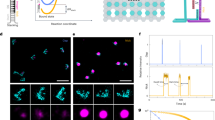
(a) M13mp18 single-strand DNA was annealed with staples in a Peltier thermal cycler (a-1). Then the PCR mixture was purified by agarose gel electrophoresis (a-2) and subsequently extracted using Freeze ’N Squeeze DNA gel extraction spin columns (a-3). (b) phiX 174 DNA was first annealed with mediator primer and extends to dsDNA (b-1). Then the PCR product was purified by agarose gel electrophoresis (b-2) and subsequently extracted using Freeze ’N Squeeze DNA gel extraction spin columns (b-3). The purified target dsDNA hybridized with DNA origami shape IDs (b-4). (c) The process of amplification (c-1), digestion (c-2), annealing and extension (c-3) were used to deal with linear (patient sample) DNA. Then the target DNA was purified by agarose gel electrophoresis (c-4), extracted using Freeze ’N Squeeze DNA gel extraction spin columns (c-5) and hybridized with DNA origami shape IDs (c-6). Finally, all products including DNA origami shape IDs and the mixture products of hybridization were imaged with AFM
Supplementary Fig. 2 Design of “mediator” DNA strands.
The M-strand consists of three blocks, M1, M2 and M3. The M1 block is 20-base long, which is used to hybridize with the target DNA. M1 block is set at the 3′ end of the M-strand and the first base at 3′ end is the complementary base to the target SNP. M2 block is a spacer with five “T”s. M3 block is also 20-base long, which can be recognized by M3’ on the DNA origami shape IDs. Adapted from ref. 6, Springer Nature
Supplementary Fig. 3 Site-specific single labeling of phiX 174 by cross-shaped IDs.
(a) Cross-shaped ID with one M3’strand at the corner point. (b) Cross-shaped ID with one M3’strand at the edge middle point. (c) Cross-shaped ID with one M3’strand at the inner point of the origami. Scale bar: 200 nm. Adapted from ref. 6, Springer Nature
Supplementary Fig. 4 Site-specific single labeling of phiX 174 by triangular-shaped IDs.
(a) Triangular-shaped ID with one M3’strand at the corner point. (b) Triangular-shaped ID with one M3’strand at the edge middle point. (c) Triangular-shaped ID with one M3’strand at the inner point of the origami. Scale bar: 200 nm. Adapted from ref. 6, Springer Nature
Supplementary Fig. 5 Site-specific single labeling of phiX 174 by rectangular-shaped IDs.
(a) Rectangular-shaped ID with one M3’strand at the corner point. (b) Rectangular-shaped ID with one M3’strand at the edge middle point. (c) Rectangular-shaped ID with one M3’strand at the inner point of the origami. Scale bar: 200 nm. Adapted from ref. 6, Springer Nature
Supplementary Fig. 6 Gel electrophoresis of the phiX174 DNA template’s asymmetric extension assisted by AuNPs.
The phiX174 DNA templates were extended by a perfectly matched primer (line T) and one-base mismatched primer (line F) assisted under different concentrations of AuNP. Line M, DL15,000 maker. Adapted from ref. 6, Springer Nature
Supplementary Fig. 7 Gel electrophoresis of regenerated dsDNA from ssDNA for the 4.6-kb AMD samples.
4.6-kb long-range PCR products of four different AMD samples (lanes 1–4) were digested with lambda exonuclease to produce the corresponding ssDNA (lanes 5–8, respectively). After primer extension, dsDNA samples were regenerated (lanes 9–12, respectively). Line M, DL15,000 maker. Adapted from ref. 6, Springer Nature
Supplementary Fig. 8 Gel electrophoresis of regenerated dsDNA from ssDNA for the 12-kb p53 gene samples.
12-kb long-range PCR product of one DNA samples (lanes 1) was digested with lambda exonuclease to produce the corresponding ssDNA (lanes 2). After primer extension, dsDNA samples were regenerated (lanes 3). Line M, DL15,000 maker. Adapted from ref. 6, Springer Nature
Supplementary Fig. 9 Direct haplotyping of 4.6-kb region samples on chromosome 3 and a 12-kb region of the p53 gene from the Han Chinese population by shape IDs.
(a) Upper, schematic showing haplotype and relative locations of two SNPs (rs17038640 (C/T) and rs4676487 (C/T)) on a 4.6-kb region. Lower, for each SNP locus, the triangular- and the STV-decorated triangular-shape IDs are used to label the alleles C and T, respectively. The shape IDs in the AFM images show that the genomic DNA of one sample is homozygous at both loci (C–C), and the genomic DNA of the other three samples is heterozygous at both loci (T–T and C–C). Scale bar, 100 nm. (b) Single-blinded test for haplotyping P53 gene. Upper, schematic showing a 12-kb region of the human p53 gene located on chromosome 17. Each origami shape ID corresponds to a specific allele. For example, for SNP 1, A and C allele correspond to cross-origami with or without STVs, respectively. Middle, AFM images for the haplotypes of these three SNPs. Haplotype 1 contains A–G–T and haplotype 2 contains C–C–C, which are also verified by capillary sequencing data. Lower, when the first SNP was A on one sequence, the other SNPs obtained from independent capillary sequencing were G and T, which was consistent with that from the nanomechanical imaging in haplotype 1. Scale bar, 200 nm. Adapted from ref. 6, Springer Nature
Supplementary Fig. 10 Schematic showing the gel electrophoresis of a 34-kb region of an AMD sample.
Upper: schematic showing a 34-kb region AMD sample, which is on chromosome 10 from the Han Chinese population. It consists of seven SNP alleles that are split into four overlapping fragments. Each fragment contains two or three SNP loci, which have at least one SNP at the ends as “joint locus”. The “joint locus” is heterozygously shared by its adjacent fragment and used for subsequent fragment connection. Lower: gel electrophoresis of regenerated dsDNA from ssDNA. Four fragments in 34 kb were generated by long-range PCR of one AMD sample (lanes 1, 4, 7 and 10) and were digested with lambda exonuclease to produce the corresponding ssDNA (lanes 2, 5, 8 and 11, respectively). After primer extension, dsDNA samples were regenerated (lanes 3, 6, 9 and 12, respectively). Line M, DL15,000 marker. Adapted from ref. 6, Springer Nature
Supplementary Fig. 11 Direct haplotyping of AMD-related 34-kb region samples by shape IDs.
The long 34-kb region was split into four overlapping fragments. The haplotypes of each fragment can be directly read by shape IDs. For example, in fragment 2, the two haplotypes of C–T–C and T–G–T correspond to SNPs 2, 3 and 4, respectively. Connection of the four fragments reconstructs continuous haplotypes for this 34-kb region. That is, haplotype 1 contains G–C–T–C–A–G–C, and haplotype 2 contains T–T–G–T–G–A–T. All the seven SNPs are heterozygous in the sample, and two haplotypes were read simultaneously using AFM, which were also supported by the capillary sequencing results. Adapted from ref. 6, Springer Nature
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11, Supplementary Tables 1–3
Rights and permissions
About this article
Cite this article
Chao, J., Zhang, H., Xing, Y. et al. Programming DNA origami assembly for shape-resolved nanomechanical imaging labels. Nat Protoc 13, 1569–1585 (2018). https://doi.org/10.1038/s41596-018-0004-y
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-018-0004-y
This article is cited by
-
Designer DNA nanostructures for viral inhibition
Nature Protocols (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.