Abstract
The highly asymmetric outer membrane of Gram-negative bacteria functions in the defense against cytotoxic substances, such as antibiotics. The Mla pathway maintains outer membrane lipid asymmetry by transporting phospholipids between the inner and outer membranes. It comprises six Mla proteins, MlaFEDBCA, including the ABC transporter MlaFEDB, which functions via an unknown mechanism. Here we determine cryo-EM structures of Escherichia coli MlaFEDB in an apo state and bound to phospholipid, ADP or AMP-PNP to a resolution of 3.3–4.1 Å and establish a proteoliposome-based transport system that includes MlaFEDB, MlaC and MlaA–OmpF to monitor the transport direction of phospholipids. In vitro transport assays and in vivo membrane permeability assays combined with mutagenesis identify functional residues that not only recognize and transport phospholipids but also regulate the activity and structural stability of the MlaFEDB complex. Our results provide mechanistic insights into the Mla pathway, which could aid antimicrobial drug development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Electron microscopy density maps and atomic models have been deposited in the EMDB and PDB, respectively, with accession codes EMD-11547 and PDB 6ZY2 (apo MlaFEDB complex); EMD-11548 and PDB 6ZY3 (PE-bound MlaFEDB complex); EMD-11555 and PDB 6ZY9 (AMP-PNP-bound MlaFEDB complex); and EMD-11549 and PDB 6ZY4 (ADP-bound MlaFEDB complex). Source data are provided with this paper.
References
Raetz, C. R. H. & Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 (2002).
Whitfield, C. & Trent, M. S. Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128 (2014).
Rowlett, V. W. et al. Impact of membrane phospholipid alterations in Escherichia coli on cellular function and bacterial stress adaptation. J. Bacteriol. 199, e00849-16 (2017).
Sperandeo, P., Dehò, G. & Polissi, A. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim. Biophys. Acta 1791, 594–602 (2009).
Sperandeo, P., Martorana, A. M. & Polissi, A. Lipopolysaccharide biogenesis and transport at the outer membrane of Gram-negative bacteria. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1451–1460 (2017).
Bishop, R. E. Emerging roles for anionic non-bilayer phospholipids in fortifying the outer membrane permeability barrier. J. Bacteriol. 196, JB-02043 (2014).
Sperandeo, P., Martorana, A. M. & Polissi, A. In Bacterial Cell Walls and Membranes Vol. 92 (Ed. Kuhn, A.) 9–37 (Springer, 2019).
Dong, H., Tang, X., Zhang, Z. & Dong, C. Structural insight into lipopolysaccharide transport from the Gram-negative bacterial inner membrane to the outer membrane. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1461–1467 (2017).
Raetz, C. R. H., Reynolds, C. M., Trent, M. S. & Bishop, R. E. Lipid A modification systems in Gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329 (2007).
Bryant, C. E., Spring, D. R., Gangloff, M. & Gay, N. J. The molecular basis of the host response to lipopolysaccharide. Nat. Rev. Microbiol. 8, 8–14 (2010).
Dalebroux, Z. D. et al. Delivery of cardiolipins to the Salmonella outer membrane is necessary for survival within host tissues and virulence. Cell Host Microbe 17, 441–451 (2015).
Carpenter, C. D. et al. The Vps/VacJ ABC transporter is required for intercellular spread of Shigella flexneri. Infect. Immun. 82, 660–669 (2014).
Knowles, T. J., Scott-Tucker, A., Overduin, M. & Henderson, I. R. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat. Rev. Microbiol. 7, 206 (2009).
Sperandeo, P. et al. New insights into the Lpt machinery for lipopolysaccharide transport to the cell surface: LptA–LptC interaction and LptA stability as sensors of a properly assembled transenvelope complex. J. Bacteriol. 193, 1042–1053 (2011).
Narita, S. & Tokuda, H. Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides. FEBS Lett. 583, 2160–2164 (2009).
Okuda, S., Sherman, D. J., Silhavy, T. J., Ruiz, N. & Kahne, D. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat. Rev. Microbiol. 12, 337–345 (2016).
Gu, Y. et al. Structural basis of outer membrane protein insertion by the BAM complex. Nature 531, 64–69 (2016).
Han, L. et al. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat. Struct. Mol. Biol. 23, 192–196 (2016).
Noinaj, N. et al. Structural insight into the biogenesis of β-barrel membrane proteins. Nature 501, 385–390 (2013).
Bakelar, J., Buchanan, S. K. & Noinaj, N. The structure of the β-barrel assembly machinery complex. Science 351, 180–186 (2016).
Roman-Hernandez, G., Peterson, J. H. & Bernstein, H. D. Reconstitution of bacterial autotransporter assembly using purified components. Elife 3, e04234 (2014).
Silhavy, T. J., Kahne, D. & Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, a000414 (2010).
Villa, R. et al. The Escherichia coli Lpt transenvelope protein complex for lipopolysaccharide export is assembled via conserved structurally homologous domains. J. Bacteriol. 195, 1100–1108 (2013).
Botos, I. et al. Structural and functional characterization of the LPS transporter LptDE from Gram-negative pathogens. Structure 24, 965–976 (2016).
Dong, H., Zhang, Z., Tang, X., Paterson, N. G. & Dong, C. Structural and functional insights into the lipopolysaccharide ABC transporter LptB2FG. Nat. Commun. 8, 222 (2017).
Bishop, R. E. Ratcheting up lipopolysaccharide transport. Nature 567, 471–472 (2019).
Tran, A. X., Dong, C. & Whitfield, C. Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J. Biol. Chem. 285, 33529–33539 (2010).
Tang, X. et al. Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism. Nat. Commun. 10, 4175 (2019).
Dong, H. et al. Structural basis for outer membrane lipopolysaccharide insertion. Nature 511, 52–56 (2014).
Luo, Q. et al. Structural basis for lipopolysaccharide extraction by ABC transporter LptB2FG. Nat. Struct. Mol. Biol. 24, 469–474 (2017).
Qiao, S., Luo, Q., Zhao, Y., Zhang, X. C. & Huang, Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511, 108–111 (2014).
Li, Y., Orlando, B. J. & Liao, M. Structural basis of lipopolysaccharide extraction by the LptB2FGC complex. Nature 567, 486–490 (2019).
Owens, T. W. et al. Structural basis of unidirectional export of lipopolysaccharide to the cell surface. Nature 567, 550–553 (2019).
Powers, M. J. & Trent, M. S. Intermembrane transport: glycerophospholipid homeostasis of the Gram-negative cell envelope. Proc. Natl Acad. Sci. USA 116, 17147–17155 (2019).
Malinverni, J. C. & Silhavy, T. J. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc. Natl Acad. Sci. USA 106, 8009–8014 (2009).
Sutterlin, H. A. et al. Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc. Natl Acad. Sci. USA 113, E1565–E1574 (2016).
Baarda, B. I., Zielke, R. A., Le Van, A., Jerse, A. E. & Sikora, A. E. Neisseria gonorrhoeae MlaA influences gonococcal virulence and membrane vesicle production. PLoS Pathog. 15, e1007385 (2019).
Bernier, S. P., Son, S. & Surette, M. G. The Mla pathway plays an essential role in the intrinsic resistance of Burkholderia cepacia complex species to antimicrobials and host innate components. J. Bacteriol. 200, e00156-18 (2018).
Munguia, J. et al. The Mla pathway is critical for Pseudomonas aeruginosa resistance to outer membrane permeabilization and host innate immune clearance. J. Mol. Med. 95, 1127–1136 (2017).
Powers, M. J. & Trent, M. S. Phospholipid retention in the absence of asymmetry strengthens the outer membrane permeability barrier to last-resort antibiotics. Proc. Natl Acad. Sci. USA 115, E8518–E8527 (2018).
Thong, S. et al. Defining key roles for auxiliary proteins in an ABC transporter that maintains bacterial outer membrane lipid asymmetry. Elife 5, e19042 (2016).
Bishop, R. E. Phospholipid middle management. Nat. Microbiol. 4, 1608–1609 (2019).
Wong, L. H., Gatta, A. T. & Levine, T. P. Lipid transfer proteins: the lipid commute via shuttles, bridges and tubes. Nat. Rev. Mol. Cell Biol. 20, 85–101 (2019).
Ekiert, D. C. et al. Architectures of lipid transport systems for the bacterial outer membrane. Cell 169, 273–285 (2017).
Chong, Z., Woo, W. & Chng, S.-S. Osmoporin OmpC forms a complex with MlaA to maintain outer membrane lipid asymmetry in Escherichia coli. Mol. Microbiol. 98, 1133–1146 (2015).
Abellón-Ruiz, J. et al. Structural basis for maintenance of bacterial outer membrane lipid asymmetry. Nat. Microbiol. 2, 1616–1623 (2017).
Kamischke, C. et al. The Acinetobacter baumannii Mla system and glycerophospholipid transport to the outer membrane. Elife 8, e40171 (2019).
Hughes, G. W. et al. Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system. Nat. Microbiol. 4, 1692–1705 (2019).
Ercan, B., Low, W.-Y., Liu, X. & Chng, S.-S. Characterization of interactions and phospholipid transfer between substrate binding proteins of the OmpC–Mla system. Biochemistry 58, 114–119 (2018).
Qian, H. et al. Structure of the human lipid exporter ABCA1. Cell 169, 1228–1239 (2017).
Locher, K. P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 (2016).
Lee, J.-Y. et al. Crystal structure of the human sterol transporter ABCG5/ABCG8. Nature 533, 561–564 (2016).
Yakushi, T., Masuda, K., Narita, S., Matsuyama, S. & Tokuda, H. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2, 212–218 (2000).
Grimm, J. et al. The inner membrane protein YhdP modulates the rate of anterograde phospholipid flow in Escherichia coli. Proc. Natl Acad. Sci. USA 117, 26907–26914 (2020).
Connerth, M. et al. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science 338, 815–818 (2012).
Liu, H. & Naismith, J. H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91 (2008).
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H. W. Semi-automated selection of cryo-EM particles in RELION-1.3. J. Struct. Biol. 189, 114–122 (2015).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Acknowledgements
We thank Y. Q. Wei and B. R. Dong for supporting the project and Y. Zhang for advice on cryo-sample preparation. This work was supported by grants from the National Key Research and Development Program of China (2017YFA0504803 and 2018YFA0507700 to X. Zhang and 2017YFC0840100 and 2017YFC00840101 to H.D.) and the Fundamental Research Funds for the Central Universities (2018XZZX001-13) to X. Zhang; the National Natural Science Foundation of China (31900039 to X.T. and 81971974 to H.D.); the National Clinical Research Centre for Geriatrics, West China Hospital (Z2018B01) to H.D.; Laboratory and Equipment Management, Zhejiang University (SJS201814) to S.C.; and a Wellcome Trust investigator award (WT106121MA) to C.D.
Author information
Authors and Affiliations
Contributions
H.D. and X.T. conceived and designed the experiments. X.T. and H.D. generated the constructs for protein expression. X.T., W.Q., Q.L. and K.Z. expressed and purified the proteins. Y.C., W.Q., Q.L., T.W., K.Z., Z.Z., C.Z., X.W., X. Zhu and J.C. performed mutagenesis and the ATPase and transport assays. X.T., H.D., W.Q., Q.L. and K.Z. prepared the samples. S.C., Z.J. and X. Zhang undertook data collection, processed electron microscopy data and performed structure determination. H.D., X.T. and C.D. carried out model building and refinement. H.D. and X.T. wrote the manuscript, and X. Zhang, S.C. and C.D. revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Anke Sparmann was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
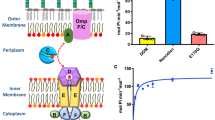
Extended Data Fig. 1 Determination of ATPase and PL transport activities of MlaFEDB.
a, Coomassie brilliant blue staining of the purified MlaFEDB on SDS-PAGE. b, The relative ATPase activities of purified MlaFEDB in liposomes or detergent. c, d, The relative ATPase activities of MlaFEDB in the presence of ADP (c) or AMP-PNP (d). e, f, SDS-PAGE analysis of MlaA-OmpF (e) or MlaFEDB (f) constitution ratio and orientation ratio in proteoliposomes of E. coli polar lipids or POPC with or without proteinase K treatment. g, FRET scan of PL transport assay using MlaA-OmpF wildtype or mutant MlaA(∆Asn41-Phe42)-OmpF in retrograde direction. h, Thin-layer chromatogram (TLC) showing transported PLs from IM or OM proteoliposomes to apo-MlaC. i, SDS-PAGE analysis of proteins involved in the transported system of Fig. g. j, FRET scan of PL transport assay containing IM proteoliposomes and apo-MlaC only in the presence or absence of ATP and MgCl2. k, l, FRET scan of PL transport assay using OM proteoliposomes containing lipid A in different ratios to PLs for anterograde (k) and retrograde direction (l). m, Loading control of lipids A and MlaA protein by western blot. Data in a, e-m are representative results from n = 3 independent experiments. Data in b-d, g, j-l represents mean ± s.d. (n = 3 independent experiments). Uncropped images for all gels are available online. Source data for panels b-d and g, j-l are available in Supplementary Data Set 1.
Extended Data Fig. 2 Flowchart for cryo-EM single-particle data processing of PE bound MlaFEDB.
a, A micrograph of the single particles after drift correction and dose-weighting, 2D classifications, 3D classification, selections and 3D refinement. b, Angular distribution of the cryo-EM particles included in the final 3D reconstruction. c, Cryo-EM map coloured by local resolutions. d, Gold-standard FSC curves of the final EM maps. e, Values are plotted for the model versus the final map (FSC average, black), for the model that was refined into the first half-map and FSC calculated either for the same map (model vs first half-map, red) or for the second half-map (model vs second half map, blue). f, Cryo-EM density with the atomic model for TM1-TM5 and elbow helix of MlaE. The PE density from C1 (yellow) and C2 (blue) full maps are compared, which shows symmetry due to additive signal of two possible binding gestures of PE in either of the MlaE subunit.
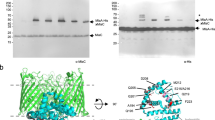
Extended Data Fig. 3 Interactions of MlaE to the peripasmic MlaD and the cytoplasmic MlaF.
a, Cartoon representation of PE-bound MlaFEDB complex. The colour scheme is the same as the Fig. 2. Each MlaE is surrounded by three TM segments from three adjacent MlaD subunits. b, MlaE interacts with the periplasmic domain of MlaD through the periplasmic loop 1 and 2. The interacting residues are labelled. c, The coupling helix residues of one MlaE unit interact with residues located at the groove of one MlaF unit. d, Two MlaD TM segments interact with the elbow helix of MlaE. e, The third MlaD TM segment interacts with TM1 and TM3 of MlaE.
Extended Data Fig. 4 Comparison of PE, ADP, AMP-PNP bound MlaFEDB structures to the apo structure of MlaFEDB.
The apo MlaFEDB is shown in green. The PE, ADP and AMP-PNP are shown in sphere. a, PE bound MlaFEDB complex is superimposed to the apo MlaFEDB complex. PE-bound MlaFEDB complex is coloured in blue. b, ADP-bound MlaFEDB structure is superimposed to the apo MlaFEDB. ADP-bound MlaFEDB is coloured in yellow. c, AMP-PNP bound MlaFEDB structure is superimposed to the apo MlaFEDB. AMP-PNP bound MlaFEDB is in orange. d, Cartoon representation of MlaFE structure of the complex.
Extended Data Fig. 5 PL binding residues in the cavity of MlaE.
a, Cartoon representation of apo MlaFEDB. b, 90° rotation along the y-axis relative to the left panel. c, Cytoplasmic view of apo MlaFEDB. d, Leaky expression of pTRC99a_MlaFEDCB in E. coli MlaE null strain, with or without MlaE mutations V77D, Y81E, E98R. e, PL transport assay by TLC showing transported PL to apo-MlaC using wildtype MlaFEDB and mutant MlaFE(Y81E)DB or MlaFE(E98R)DB constituted IM proteoliposomes in the absence of ATP. f, SDS-PAGE analysis of proteins involved in the PL transport assay of Fig e. Data in d-f are representative results from n = 3 independent experiments. Uncropped images for panels d-f are available as online.
Extended Data Fig. 6 Interactions of MlaB to MlaF using AMP-PNP bound structure of MlaFEDB.
a, Cartoon representation of structure of the AMP-PNP bound MlaFEDB complex. b, 90° rotation along the y-axis relative to the left panel. c, Structure of MlaB showing the STAS domain, consisting of three α-helices and four β-strands. d, The interactions between one MlaB and one MlaF subunit. e, 90° rotation along the y-axis relative to the left panel. f, SDS-PAGE analysis of proteins of wildtype and MlaF binding residue mutants MlaFEDB(W29E), MlaFEDB(Y88E) and MlaFEDB(T52A). g, ATPase activity of Mutant MlaFEDB(T52A) in both detergent and liposomes. h, FRET scan of PL transport using mutant MlaFEDB(T52A) or wildtype MlaFEDB IM proteoliposomes in the presence of ATP and MgCl2 for retrograde direction. Data f, h are representative results from n = 3 independent experiments. Data in g, h presents mean ± s.d. (n = 3 independent experiments). Uncropped images for panel f are available online. Source data for panels g and h are available in Supplementary Data Set 1.
Extended Data Fig. 7 Functional residues characterization of MlaD.
a, Coomassie brilliant blue staining of purified Mla proteins with MlaD truncations. Purified MlaFEB was incubated with TM truncated MlaD(ΔTM) or full length MlaD to form complex in vitro, which were purified using affinity columns and analyzed on SDS-PAGE along with wild-type MlaFEDB and MlaFEB. b, The relative ATPase activities of wild-type, MlaFEB and in vitro formed MlaFEDB were measured in detergent and liposomes. Soluble MlaD(ΔTM) was added into MlaFEB constructed systems and its effect to ATPase activity was also analyzed. c, FRET scan of PL transport assay using wildtype MlaFEDB or MlaFEB complex incubated with truncated MlaD(ΔTM) or full length MlaD for retrograde direction. Soluble MlaD(ΔTM) was added into MlaFEB constructed systems and its effect to transport activity was also analyzed. d, FRET scan of PL transport assay using wildtype MlaFEDB or MlaFEDB mutants MlaFED(ΔL143-G153)B for retrograde direction. e, Leaky expression of pTRC99a_MlaFEDCB containing MlaD mutation L143E, I147E, F150E or Y152E. f, SDS-PAGE analysis of purified MlaFEDB complexes with MlaD mutation I143E, I147, F150E or Y152E. The mutants MlaD I143E, I147E and F150E lost the SDS-resistance hexameric form. Data are representative results from n = 3 independent experiments. Data in b represents mean ± s.d. (n = 4 independent experiments). Data in c, d present mean of triplicates ± s.d. (n = 3). Uncropped images for panel a, e and f are available online. Source data for panels c and d are available in Supplementary Data Set 1.
Extended Data Fig. 8 Effect of nucleotide binding mutants of MlaF on ATPase activity and in vitro PL transport.
a, The size-exclusion chromatogram of purified MlaFEDB complexes with MlaF single catalytic mutants of F16A, R18A, K47A, E170A and H203A. The mutants were eluted almost at the same time as the wild-type MlaFEDB, indicating that the mutants have the intact structure as the wild-type. b, The relative ATPase activities of MlaFEDB wild-type and mutants in detergent and liposomes. c, SDS-PAGE analysis of purified wildtype and the mutants. d, FRET scan of PL transport using MlaFEDB mutant IM proteoliposomes containing single mutation on MlaF K47A, E170A or H203A, wildtype OM proteoliposomes and MlaC for retrograde direction. The catalytic and ATP binding residue mutants abolished retrograde transport of PLs. e, Amino acid sequence alignment of MlaF and LptB. MlaF and LptB have 24.54% amino acid identity. The C-terminal tail of MlaF is longer than that of LptB. f, Dimeric MlaF is superimposed into the dimeric LptB. MlaF has a long C-terminal tail that interacts with the other MlaF molecule, but LptB does not. Data in a, c and d are representative results from n = 3 independent experiments. Data in b represents mean ± s.d. (n = 3 independent experiments). An uncropped image for panel c is available online. Source data for panels b and d are available in Supplementary Data Set 1.
Extended Data Fig. 9 Functional interacting residues between MlaB and MlaF.
a, Residues from the C-terminal tail of MlaF show interactions with the opposite MlaF and MlaB. b, SDS-PAGE analysis of purified MlaFEDB wildtype or mutant containing single mutation of the signature motif residues mutants E144A, S146S, R151A of MlaF, and C-terminal residues Y256D and H262D of MlaF. c, The relative ATPase activity of MlaFEDB mutants in detergent and in liposomes. d, FRET scan of PL transport assay using wildtype MlaFEDB or mutant IM proteoliposomes, wildtype OM proteoliposome and MlaC for retrograde direction. e, Cellular sensitivity to chlorpromazine by the mutant MlaF(Y256D)EDB or MlaF(H262D)EDB, showing no cellular effect. f, Leaky expression of pTRC99a_MlaFEDCB carrying mutations on C-terminal residues. g, SDS-PAGE analysis of purified MlaFEDB and mutant with MlaF being truncated at the C-terminal tail MlaF(ΔI247-S269)EDB. h, Leaky expression of pTRC99a_MlaFEDCB carrying mutations on signature motif residues. Data in b, d-h are representative results from n = 3 independent experiments. Data in c represents mean ± s.d. (n = 3 independent experiments). Uncropped images are available online. Source data for panels c and d are available in Supplementary Data Set 1.
Extended Data Fig. 10 Structures of reported ABC transporters in apo and AMP-PNP bound states.
a, Human ABCA1 lipid exporter, apo ABCA1 (PDB code: 5XJY). The ABCA1 has limited structural similarity to MlaFEDB (Dali server search with a Z score of 10). b, E. coli vitamin B12 importer, apo BtuCDF (PDB code: 2QI9), BtuCDF in complex with AMP-PNP (PDB code: 4FI3). BtuCDF binding AMP-PNP causes conformational changes. c, E. coli lipopolysaccharide transporter, apo LptB2FGC (PDB code: 6S8N), LptB2FGC in complex with AMP-PNP (PDB code: 6S8G). LptB has some structural similarity to MlaF. LptB2FGC binding AMP-PNP causes significant conformational changes. d, human ABCG2 multidrug transporter, apo (PDB code: 6MIJ), ABCG2 E211Q mutant in complex with ATP (PDB code: 6HZM). ATP binding ABCG2 causes conformational changes. e, human mitochondrial ABC transporter ABCB10, apo (PDB code: 3ZDQ), in complex with AMP-PNP (PDB code: 4AYW).
Supplementary information
Supplementary Information
Supplementary Figs. 1–3.
Supplementary Dataset 1
Statistical source data.
Source data
Source Data Fig. 1
Unprocessed gels
Source Data Fig. 3
Unprocessed gel
Source Data Extended Data Fig. 1
Unprocessed gels and unprocessed western blots
Source Data Extended Data Fig. 5
Unprocessed gels
Source Data Extended Data Fig. 6
Unprocessed gels
Source Data Extended Data Fig. 7
Unprocessed gels
Source Data Extended Data Fig. 8
Unprocessed gels
Source Data Extended Data Fig. 9
Unprocessed gels
Rights and permissions
About this article
Cite this article
Tang, X., Chang, S., Qiao, W. et al. Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB. Nat Struct Mol Biol 28, 81–91 (2021). https://doi.org/10.1038/s41594-020-00532-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-020-00532-y
This article is cited by
-
Structure of an endogenous mycobacterial MCE lipid transporter
Nature (2023)
-
Molecular mechanism of phospholipid transport at the bacterial outer membrane interface
Nature Communications (2023)
-
The Mla system of diderm Firmicute Veillonella parvula reveals an ancestral transenvelope bridge for phospholipid trafficking
Nature Communications (2023)
-
A conserved membrane protein negatively regulates Mce1 complexes in mycobacteria
Nature Communications (2023)
-
Structural basis of BAM-mediated outer membrane β-barrel protein assembly
Nature (2023)